Submitted:
15 April 2024
Posted:
16 April 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Results
Discussion
Acknowledgments
Materials and Methods
Strains and Growth Conditions
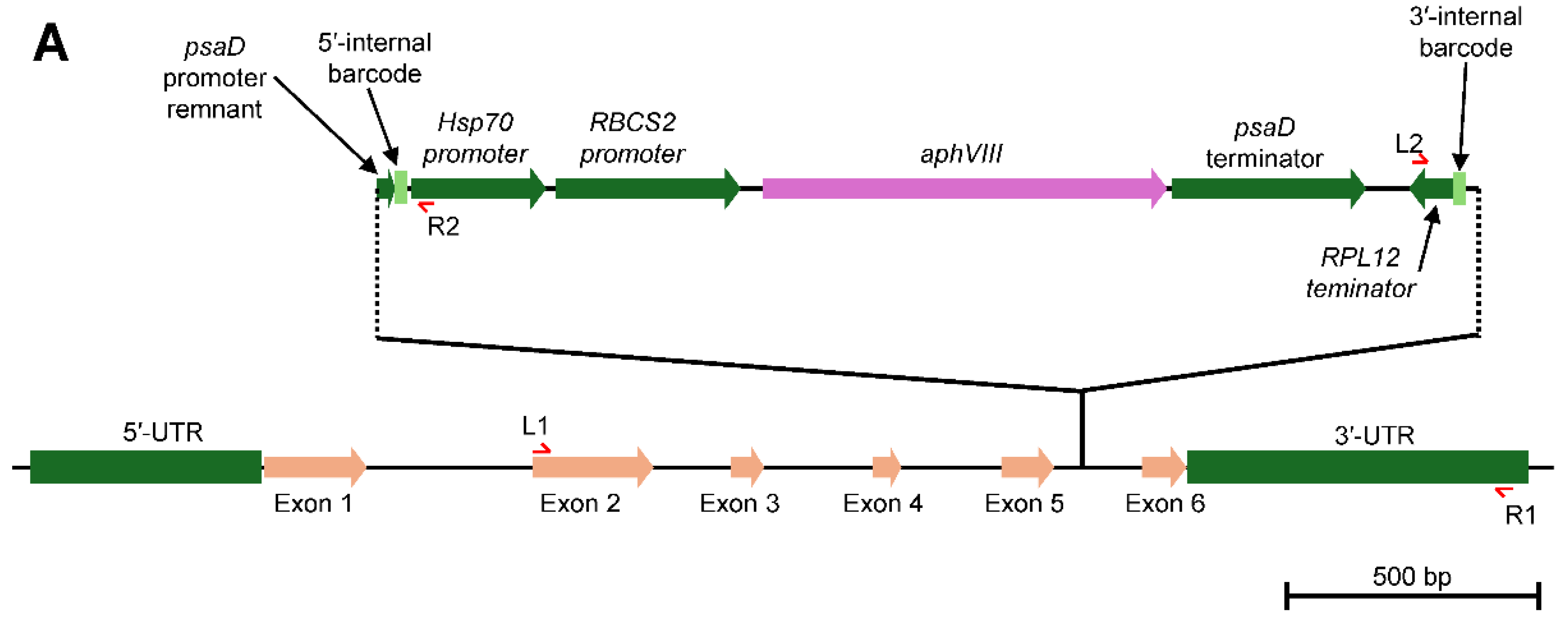
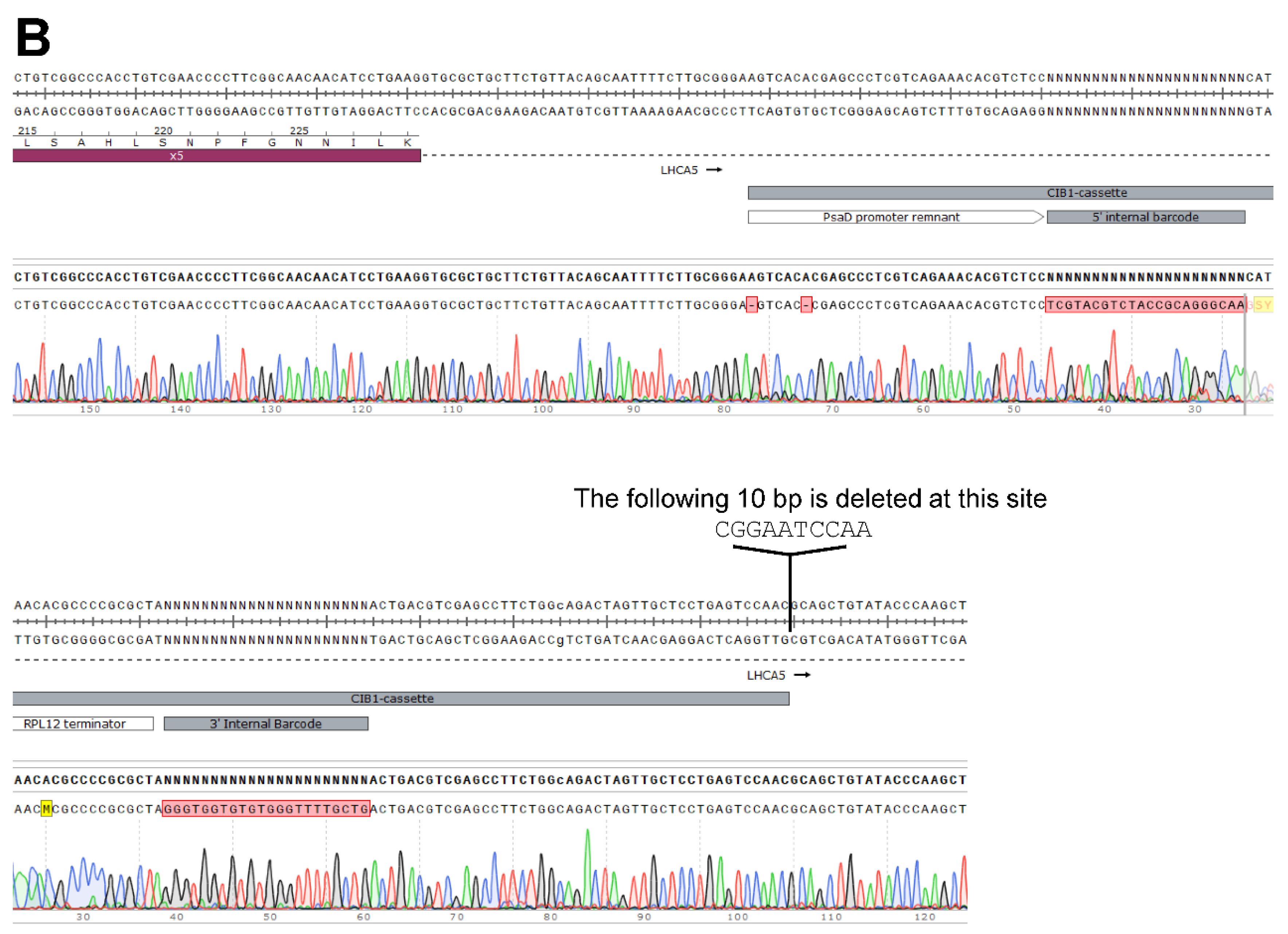
Generation of the LHCA5 Complemented Strain
- Lhca5_SGP1
- CTACAAGAACTTCGGCTCGG
- Lhca5_SGP2
- CGGTCAACAATCGAGGTTTT
- CIB_5′ GCACCAATCATGTCAAGCCT
- CIB_3′ GACGTTACAGCACACCCTTG
- If-EcoRI_ga5_F-up
- GCTTGATATCGAATTGCATCCCCACATTCAAGAGC
- If-EcoRI_ga5_R-up
- CGGGCTGCAGGAATTGTGGGCGTGCGGGTTCGTTT
- Up_a5_F
- CGCCATCATTTGCACTTTCAAGCCCTCCCTCGCCG
- Dwn_ga5_R
- AGGACGCTGGCAAGAAGGACGCTGCCAAGAAGGAC
- seq_pga5_1_F
- CCGGTGCATACGGCAAGGTTCGGTCGGGGGCTGGG
- seq_pga5_1_R
- CCCAGCCCCCGACCGAACCTTGCCGTATGCACCGG
- seq_pga5_2_F
- GTCCCTGGGGGTCCGTCGGGTTTCGGGAGCCCATC
- seq_pga5_3_F
- CCAAACCTTAAAAACGCCACTCATGTCAGAGGCTC
- seq_pga5_4_F
- AGCTCCCTGCCTTTGTAACTTTAACTCTGGTGCTT
- seq_pga5_5_F
- GTACCGCCAGTCGGAGCTGCAGCACGCTCGCTGGG
- seq_pga5_6_F
- AGACCAAGGAGATCAAGAACGGCCGCCTGGCCATG
- seq_pga5_7_F
- CATCCCCCTGACCTGCCTGTGGCCCGGCAGCCAGT
- seq_pga5_8_F
- ACGGGTGTGTAACCCAAAACCTCGATTGTTGACCG
Thylakoid Isolation and Chemical Crosslinking
Thylakoid Solubilization and Sucrose Density Gradient Centrifugation
SDS-PAGE
Immunoblotting
In-Gel digestion
LC-MS/MS analysis
MS Data Analysis
Structural Modelling
Supplementary Materials
References
- Whatley, F.R.; Arnon, D.I.; Tagawa, K. Separation of Light and Dark Reactions in Electron Transfer during Photosynthesis. Proc. Natl. Acad. Sci. 1963, 49, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P. Coupling of Phosphorylation to Electron and Hydrogen Transfer by a Chemi-Osmotic type of Mechanism. Nature 1961, 191, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shem, A.; Frolow, F.; Nelson, N. Crystal structure of plant photosystem I. Nature 2003, 426, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Amunts, A.; Drory, O.; Nelson, N. The structure of a plant photosystem I supercomplex at 3.4 Å resolution. Nature 2007, 447, 58–63. [Google Scholar] [CrossRef]
- Jordan, P.; Fromme, P.; Witt, H.T.; Klukas, O.; Saenger, W.; Krauß, N. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 2001, 411, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Nelson, N.; Ben-Shem, A. The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 2004, 5, 971–982. [Google Scholar] [CrossRef] [PubMed]
- Mazor, Y.; Borovikova, A.; Caspy, I.; Nelson, N. Structure of the plant photosystem I supercomplex at 2.6 Å resolution. Nat. Plants 2017, 3, 17014. [Google Scholar] [CrossRef] [PubMed]
- Mazor, Y.; Borovikova, A.; Nelson, N.; Tel Aviv University; Israel The structure of plant photosystem I super-complex at 2. 8 Å resolution. eLife 2015, 4, e07433. [Google Scholar] [CrossRef]
- Qin, X.; Suga, M.; Kuang, T.; Shen, J.-R. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 2015, 348, 989–995. [Google Scholar] [CrossRef]
- Suga, M.; Ozawa, S.-I.; Yoshida-Motomura, K.; Akita, F.; Miyazaki, N.; Takahashi, Y. Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I. Nat. Plants 2019, 5, 626–636. [Google Scholar] [CrossRef]
- Kubota-Kawai, H.; Burton-Smith, R.N.; Tokutsu, R.; Song, C.; Akimoto, S.; Yokono, M.; Ueno, Y.; Kim, E.; Watanabe, A.; Murata, K.; et al. Ten antenna proteins are associated with the core in the supramolecular organization of the photosystem I supercomplex in Chlamydomonas reinhardtii. J. Biol. Chem. 2019, 294, 4304–4314. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, S.-I.; Bald, T.; Onishi, T.; Xue, H.; Matsumura, T.; Kubo, R.; Takahashi, H.; Hippler, M.; Takahashi, Y. Configuration of Ten Light-Harvesting Chlorophyll a/b Complex I Subunits in Chlamydomonas reinhardtii Photosystem I. Plant Physiol. 2018, 178, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Ma, J.; Pan, X.; Zhao, X.; Chang, W.; Liu, Z.; Zhang, X.; Li, M. Antenna arrangement and energy transfer pathways of a green algal photosystem-I–LHCI supercomplex. Nat. Plants 2019, 5, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Pi, X.; Wang, W.; Han, G.; Zhu, L.; Liu, M.; Cheng, L.; Shen, J.-R.; Kuang, T.; Sui, S.-F. Structure of a green algal photosystem I in complex with a large number of light-harvesting complex I subunits. Nat. Plants 2019, 5, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Crepin, A.; Kučerová, Z.; Kosta, A.; Durand, E.; Caffarri, S. Isolation and characterization of a large photosystem I–light-harvesting complex II supercomplex with an additional Lhca1–a4 dimer in Arabidopsis. Plant J. 2019, 102, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Rochaix, J.-D. Regulation and Dynamics of the Light-Harvesting System. Annu. Rev. Plant Biol. 2014, 65, 287–309. [Google Scholar] [CrossRef]
- Murata, N. Control of excitation transfer in photosynthesis. I. Light-induced change of chlorophyll a fluorescence in Porphyridium cruentum. Biochim. Biophys. Acta 1969, 172, 242–251. [Google Scholar] [PubMed]
- Bonaventura, C.; Myers, J. Fluorescence and oxygen evolution from Chlorella pyrenoidosa. Biochim. et Biophys. Acta (BBA) - Bioenerg. 1969, 189, 366–383. [Google Scholar] [CrossRef]
- Lemeille, S.; Rochaix, J.-D. State transitions at the crossroad of thylakoid signalling pathways. Photosynth. Res. 2010, 106, 33–46. [Google Scholar] [CrossRef]
- Rochaix, J.-D.; Lemeille, S.; Shapiguzov, A.; Samol, I.; Fucile, G.; Willig, A.; Goldschmidt-Clermont, M. Protein kinases and phosphatases involved in the acclimation of the photosynthetic apparatus to a changing light environment. Philos. Trans. R. Soc. B: Biol. Sci. 2012, 367, 3466–3474. [Google Scholar] [CrossRef]
- Cariti, F.; Chazaux, M.; Lefebvre-Legendre, L.; Longoni, P.; Ghysels, B.; Johnson, X.; Goldschmidt-Clermont, M. Regulation of Light Harvesting in Chlamydomonas reinhardtii Two Protein Phosphatases Are Involved in State Transitions. Plant Physiol. 2020, 183, 1749–1764. [Google Scholar] [CrossRef] [PubMed]
- Depège, N.; Bellafiore, S.; Rochaix, J.-D. Role of Chloroplast Protein Kinase Stt7 in LHCII Phosphorylation and State Transition in Chlamydomonas. Science 2003, 299, 1572–1575. [Google Scholar] [CrossRef] [PubMed]
- Bellafiore, S.; Barneche, F.; Peltier, G.; Rochaix, J.-D. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 2005, 433, 892–895. [Google Scholar] [CrossRef]
- Pan, X.; Ma, J.; Su, X.; Cao, P.; Chang, W.; Liu, Z.; Zhang, X.; Li, M. Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II. Science 2018, 360, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Tokutsu, R.; Li, A.; Takizawa, K.; Song, C.; Murata, K.; Yamasaki, T.; Liu, Z.; Minagawa, J.; Li, M. Structural basis of LhcbM5-mediated state transitions in green algae. Nat. Plants 2021, 7, 1119–1131. [Google Scholar] [CrossRef]
- Huang, Z.; Shen, L.; Wang, W.; Mao, Z.; Yi, X.; Kuang, T.; Shen, J.-R.; Zhang, X.; Han, G. Structure of photosystem I-LHCI-LHCII from the green alga Chlamydomonas reinhardtii in State 2. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Bergner, S.V.; Scholz, M.; Trompelt, K.; Barth, J.; Gäbelein, P.; Steinbeck, J.; Xue, H.; Clowez, S.; Fucile, G.; Goldschmidt-Clermont, M.; et al. STATE TRANSITION7-Dependent Phosphorylation Is Modulated by Changing Environmental Conditions, and Its Absence Triggers Remodeling of Photosynthetic Protein Complexes. Plant Physiol. 2015, 168, 615–634. [Google Scholar] [CrossRef]
- Naschberger, A.; Mosebach, L.; Tobiasson, V.; Kuhlgert, S.; Scholz, M.; Perez-Boerema, A.; Ho, T.T.H.; Vidal-Meireles, A.; Takahashi, Y.; Hippler, M.; et al. Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex. Nat. Plants 2022, 8, 1191–1201. [Google Scholar] [CrossRef] [PubMed]
- Caspy, I.; Schwartz, T.; Bayro-Kaiser, V.; Fadeeva, M.; Kessel, A.; Ben-Tal, N.; Nelson, N. Dimeric and high-resolution structures of Chlamydomonas Photosystem I from a temperature-sensitive Photosystem II mutant. Commun. Biol. 2021, 4, 1–10. [Google Scholar] [CrossRef]
- Iwai, M.; Takizawa, K.; Tokutsu, R.; Okamuro, A.; Takahashi, Y.; Minagawa, J. Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 2010, 464, 1210–1213. [Google Scholar] [CrossRef]
- Terashima, M.; Petroutsos, D.; Hüdig, M.; Tolstygina, I.; Trompelt, K.; Gäbelein, P.; Fufezan, C.; Kudla, J.; Weinl, S.; Finazzi, G.; et al. Calcium-dependent regulation of cyclic photosynthetic electron transfer by a CAS, ANR1, and PGRL1 complex. Proc. Natl. Acad. Sci. 2012, 109, 17717–17722. [Google Scholar] [CrossRef] [PubMed]
- Steinbeck, J.; Ross, I.L.; Rothnagel, R.; Gäbelein, P.; Schulze, S.; Giles, N.; Ali, R.; Drysdale, R.; Sierecki, E.; Gambin, Y.; et al. Structure of a PSI–LHCI–cyt b 6 f supercomplex in Chlamydomonas reinhardtii promoting cyclic electron flow under anaerobic conditions. Proc. Natl. Acad. Sci. 2018, 115, 10517–10522. [Google Scholar] [CrossRef] [PubMed]
- Munekage, Y.; Hashimoto, M.; Miyake, C.; Tomizawa, K.-I.; Endo, T.; Tasaka, M.; Shikanai, T. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 2004, 429, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Dang, K.-V.; Plet, J.; Tolleter, D.; Jokel, M.; Cuiné, S.; Carrier, P.; Auroy, P.; Richaud, P.; Johnson, X.; Alric, J.; et al. Combined Increases in Mitochondrial Cooperation and Oxygen Photoreduction Compensate for Deficiency in Cyclic Electron Flow in Chlamydomonas reinhardtii. Plant Cell 2014, 26, 3036–3050. [Google Scholar] [CrossRef] [PubMed]
- Johnson, X.; Steinbeck, J.; Dent, R.M.; Takahashi, H.; Richaud, P.; Ozawa, S.-I.; Houille-Vernes, L.; Petroutsos, D.; Rappaport, F.; Grossman, A.R.; et al. Proton Gradient Regulation 5-Mediated Cyclic Electron Flow under ATP- or Redox-Limited Conditions: A Study of ƊATPase pgr5 and ƊrbcL pgr5 Mutants in the Green Alga Chlamydomonas reinhardtii. Plant Physiol. 2014, 165, 438–452. [Google Scholar] [CrossRef] [PubMed]
- Kukuczka, B.; Magneschi, L.; Petroutsos, D.; Steinbeck, J.; Bald, T.; Powikrowska, M.; Fufezan, C.; Finazzi, G.; Hippler, M. Proton Gradient Regulation5-Like1-Mediated Cyclic Electron Flow Is Crucial for Acclimation to Anoxia and Complementary to Nonphotochemical Quenching in Stress Adaptation. Plant Physiol. 2014, 165, 1604–1617. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-P.; Björkman, O.; Shih, C.; Grossman, A.R.; Rosenquist, M.; Jansson, S.; Niyogi, K.K. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 2000, 403, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Peers, G.; Truong, T.B.; Ostendorf, E.; Busch, A.; Elrad, D.; Grossman, A.R.; Hippler, M.; Niyogi, K.K. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 2009, 462, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Liguori, N.; Roy, L.M.; Opacic, M.; Durand, G.; Croce, R. Regulation of Light Harvesting in the Green Alga Chlamydomonas reinhardtii: The C-Terminus of LHCSR Is the Knob of a Dimmer Switch. J. Am. Chem. Soc. 2013, 135, 18339–18342. [Google Scholar] [CrossRef]
- Bonente, G.; Ballottari, M.; Truong, T.B.; Morosinotto, T.; Ahn, T.K.; Fleming, G.R.; Niyogi, K.K.; Bassi, R. Analysis of LhcSR3, a Protein Essential for Feedback De-Excitation in the Green Alga Chlamydomonas reinhardtii. PLOS Biol. 2011, 9, e1000577. [Google Scholar] [CrossRef]
- Ballottari, M.; Truong, T.B.; De Re, E.; Erickson, E.; Stella, G.R.; Fleming, G.R.; Bassi, R.; Niyogi, K.K. Identification of pH-sensing Sites in the Light Harvesting Complex Stress-related 3 Protein Essential for Triggering Non-photochemical Quenching in Chlamydomonas reinhardtii. J. Biol. Chem. 2016, 291, 7334–7346. [Google Scholar] [CrossRef] [PubMed]
- Tokutsu, R.; Minagawa, J. Energy-dissipative supercomplex of photosystem II associated with LHCSR3 in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. 2013, 110, 10016–10021. [Google Scholar] [CrossRef] [PubMed]
- Allorent, G.; Tokutsu, R.; Roach, T.; Peers, G.; Cardol, P.; Girard-Bascou, J.; Seigneurin-Berny, D.; Petroutsos, D.; Kuntz, M.; Breyton, C.; et al. A Dual Strategy to Cope with High Light in Chlamydomonas reinhardtii. Plant Cell 2013, 25, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Tokutsu, R.; Bergner, S.V.; Scholz, M.; Minagawa, J.; Hippler, M. PHOTOSYSTEM II SUBUNIT R Is Required for Efficient Binding of LIGHT-HARVESTING COMPLEX STRESS-RELATED PROTEIN3 to Photosystem II-Light-Harvesting Supercomplexes in Chlamydomonas reinhardtii. Plant Physiol. 2015, 167, 1566–1578. [Google Scholar] [CrossRef] [PubMed]
- Lemeille, S.; Turkina, M.V.; Vener, A.V.; Rochaix, J.-D. Stt7-dependent Phosphorylation during State Transitions in the Green Alga Chlamydomonas reinhardtii. Mol. Cell. Proteom. 2010, 9, 1281–1295. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Bullock, J.M.A.; Schwab, J.; Thalassinos, K.; Topf, M. The Importance of Non-accessible Crosslinks and Solvent Accessible Surface Distance in Modeling Proteins with Restraints From Crosslinking Mass Spectrometry. Mol. Cell. Proteom. 2016, 15, 2491–2500. [Google Scholar] [CrossRef]
- Cohen, S.; Schneidman-Duhovny, D. A deep learning model for predicting optimal distance range in crosslinking mass spectrometry data. Proteomics 2023, 23, e2200341. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting Subcellular Localization of Proteins Based on their N-terminal Amino Acid Sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef]
- Camargo, F.V.A.; Perozeni, F.; Valbuena, G.d.l.C.; Zuliani, L.; Sardar, S.; Cerullo, G.; D’andrea, C.; Ballottari, M. The Role of Acidic Residues in the C Terminal Tail of the LHCSR3 Protein of Chlamydomonas reinhardtii in Non-Photochemical Quenching. J. Phys. Chem. Lett. 2021, 12, 6895–6900. [Google Scholar] [CrossRef] [PubMed]
- Naumann, B.; Stauber, E.J.; Busch, A.; Sommer, F.; Hippler, M. N-terminal Processing of Lhca3 Is a Key Step in Remodeling of the Photosystem I-Light-harvesting Complex Under Iron Deficiency in Chlamydomonas reinhardtii. J. Biol. Chem. 2005, 280, 20431–20441. [Google Scholar] [CrossRef] [PubMed]
- Schiphorst, C.; Achterberg, L.; Gómez, R.; Koehorst, R.; Bassi, R.; van Amerongen, H.; Dall’osto, L.; Wientjes, E. The role of light-harvesting complex I in excitation energy transfer from LHCII to photosystem I in Arabidopsis. Plant Physiol. 2021, 188, 2241–2252. [Google Scholar] [CrossRef]
- Bressan, M.; Bassi, R.; Dall’osto, L. Loss of LHCI system affects LHCII re-distribution between thylakoid domains upon state transitions. Photosynth. Res. 2017, 135, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Benson, S.L.; Maheswaran, P.; Ware, M.A.; Hunter, C.N.; Horton, P.; Jansson, S.; Ruban, A.V.; Johnson, M.P. An intact light harvesting complex I antenna system is required for complete state transitions in Arabidopsis. Nat. Plants 2015, 1, 15176. [Google Scholar] [CrossRef]
- Bos, P.; Oosterwijk, A.; Koehorst, R.; Bader, A.; Philippi, J.; van Amerongen, H.; Wientjes, E. Digitonin-sensitive LHCII enlarges the antenna of Photosystem I in stroma lamellae of Arabidopsis thaliana after far-red and blue-light treatment. Biochim. et Biophys. Acta (BBA) - Bioenerg. 2019, 1860, 651–658. [Google Scholar] [CrossRef]
- Yadav, K.S.; Semchonok, D.A.; Nosek, L.; Kouřil, R.; Fucile, G.; Boekema, E.J.; Eichacker, L.A. Supercomplexes of plant photosystem I with cytochrome b6f, light-harvesting complex II and NDH. Biochim. et Biophys. Acta (BBA) - Bioenerg. 2017, 1858, 12–20. [Google Scholar] [CrossRef]
- Pinnola, A.; Cazzaniga, S.; Alboresi, A.; Nevo, R.; Levin-Zaidman, S.; Reich, Z.; Bassi, R. Light-Harvesting Complex Stress-Related Proteins Catalyze Excess Energy Dissipation in Both Photosystems of Physcomitrella patens. Plant Cell 2015, 27, 3213–3227. [Google Scholar] [CrossRef]
- Girolomoni, L.; Cazzaniga, S.; Pinnola, A.; Perozeni, F.; Ballottari, M.; Bassi, R. LHCSR3 is a nonphotochemical quencher of both photosystems in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. 2019, 116, 4212–4217. [Google Scholar] [CrossRef]
- Yamano, T.; Iguchi, H.; Fukuzawa, H. Rapid transformation of Chlamydomonas reinhardtii without cell-wall removal. J. Biosci. Bioeng. 2013, 115, 691–694. [Google Scholar] [CrossRef]
- Chua, N.H.; Matlin, K.; Bennoun, P. A chlorophyll-protein complex lacking in photosystem I mutants of Chlamydomonas reinhardtii. J. Cell Biol. 1975, 67, 361–377. [Google Scholar] [CrossRef]
- Tokutsu, R.; Kato, N.; Bui, K.H.; Ishikawa, T.; Minagawa, J. Revisiting the Supramolecular Organization of Photosystem II in Chlamydomonas reinhardtii. J. Biol. Chem. 2012, 287, 31574–31581. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Naumann, B.; Busch, A.; Allmer, J.; Ostendorf, E.; Zeller, M.; Kirchhoff, H.; Hippler, M. Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 2007, 7, 3964–3979. [Google Scholar] [CrossRef]
- Hippler, M.; Klein, J.; Fink, A.; Allinger, T.; Hoerth, P. Towards functional proteomics of membrane protein complexes: analysis of thylakoid membranes from Chlamydomonas reinhardtii. Plant J. 2001, 28, 595–606. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Mendes, M.L.; Fischer, L.; A Chen, Z.; Barbon, M.; O‘Reilly, F.J.; Giese, S.H.; Bohlke-Schneider, M.; Belsom, A.; Dau, T.; Combe, C.W.; et al. An integrated workflow for crosslinking mass spectrometry. Mol. Syst. Biol. 2019, 15, e8994. [Google Scholar] [CrossRef]
- Yılmaz, S.u.; Busch, F.; Nagaraj, N.; Cox, J. Accurate and Automated High-Coverage Identification of Chemically Cross-Linked Peptides with MaxLynx. Anal. Chem. 2022, 94, 1608–1617. [Google Scholar] [CrossRef]
- Götze, M.; Pettelkau, J.; Fritzsche, R.; Ihling, C.H.; Schäfer, M.; Sinz, A. Automated Assignment of MS/MS Cleavable Cross-Links in Protein 3D-Structure Analysis. J. Am. Soc. Mass Spectrom. 2014, 26, 83–97. [Google Scholar] [CrossRef]
- Sarpe, V.; Rafiei, A.; Hepburn, M.; Ostan, N.; Schryvers, A.B.; Schriemer, D.C. High Sensitivity Crosslink Detection Coupled With Integrative Structure Modeling in the Mass Spec Studio. Mol. Cell. Proteom. 2016, 15, 3071–3080. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Elias, J.E.; Gygi, S.P. Target-Decoy Search Strategy for Mass Spectrometry-Based Proteomics. In Proteome Bioinformatics; Hubbard, S., Jones, A., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 604, pp. 55–71. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2020, 30, 70–82. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for Structure Building and Analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]

| Protein 1 ID |
Protein 1 Name |
Phytozome ID 1 |
Protein 2 ID |
Protein 2 Name |
Phytozome ID 2 |
Peptide 1 | Protein Position 1 | Peptide 2 | Protein Position 2 | Search Engines |
|---|---|---|---|---|---|---|---|---|---|---|
| A8IKC8 | LHCA2 | Cre12.g508750 | P13352 | PSAH | Cre07.g330250 | RYEIYKK | K130 | YPDNQAKFFTQATDIISR | K68 | Xi; MSS; MQ |
| A8IKC8 | LHCA2 | Cre12.g508750 | P13352 | PSAH | Cre07.g330250 | YEIYKK | K130 | YPDNQAKFFTQATDIISR | K68 | Xi; MSS; MQ |
| A8IKC8 | LHCA2 | Cre12.g508750 | P13352 | PSAH | Cre07.g330250 | KTGETGFLSFAPFDPMGMK | K131 | YPDNQAKFFTQATDIISR | K68 | MQ; Xi |
| A8IKC8 | LHCA2 | Cre12.g508750 | P09144 | PSAB | CreCp.g802312 | ESVPYFPWNEPWNKV | K245 | LFPKFSQGLAQDPTTR | K8 | Mx; Xi; MSS; MQ |
| A8J26, A8J27, A8J28, Q8S3T9 |
LHCBM4, LHCBM8, LHCBM6, LHCBM9 |
Cre06.g283950, Cre06.g284250, Cre06.g285250, Cre06.g284200 |
Q05093 | LHCA1 | Cre06.g283050 | AAAPKSSGVEFYGPNR | K31 | SGELKLK | K174 | MQ; Xi |
| P09144 | PSAB | CreCp.g802312 | A8IKC8 | LHCA2 | Cre12.g508750 | LFPKFSQGLAQDPTTR | K8 | TLNPGKESVPYFPWNEPWNKV | K231 | MSS; MQ; Xi |
| P14225 | PSAK | Cre17.g724300 | Q75VY9 | LHCA3 | Cre11.g467573 | FGLAPTVKK | K58 | SIAKVDR | K37 | MSS; MQ; Xi |
| P14225 | PSAK | Cre17.g724300 | Q75VY9 | LHCA3 | Cre11.g467573 | NTTAGLKLVDSK | K66 | SIAKVDR | K37 | MSS; MQ; Xi |
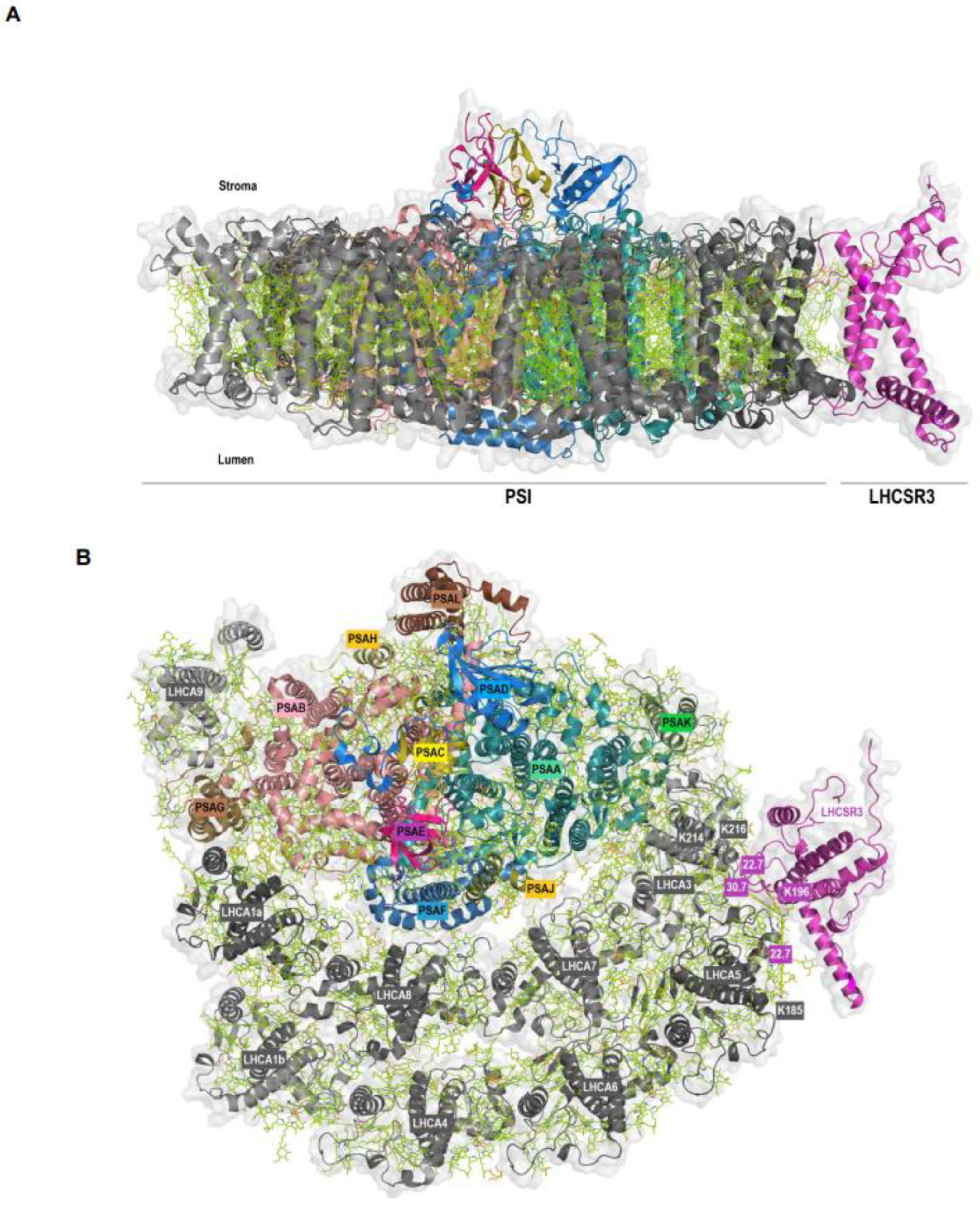
| P93663 | LHCSR3.2 | Cre08.g367400 | Q75VY8 | LHCA5 | Cre10.g425900 | VMQTKELNNGR | K196 | ELQTKEIK | K185 | MQ; Xi |
| P93663 | LHCSR3.2 | Cre08.g367400 | Q75VY9 | LHCA3 | Cre11.g467573 | VMQTKELNNGR | K196 | ELKLKEIK | K214 | MQ |
| P93663 | LHCSR3.2 | Cre08.g367400 | Q75VY9 | LHCA3 | Cre11.g467573 | VMQTKELNNGR | K196 | ELKLKEIK | K216 | MQ |
| Q05093 | LHCA1 | Cre06.g283050 | Q8S3T9, A8J270, A8J287, A8J264 |
LHCBM9, LHCBM8, LHCBM6, LHCBM4 |
Cre06.g285250, Cre06.g284200, Cre06.g283950, Cre06.g284250 |
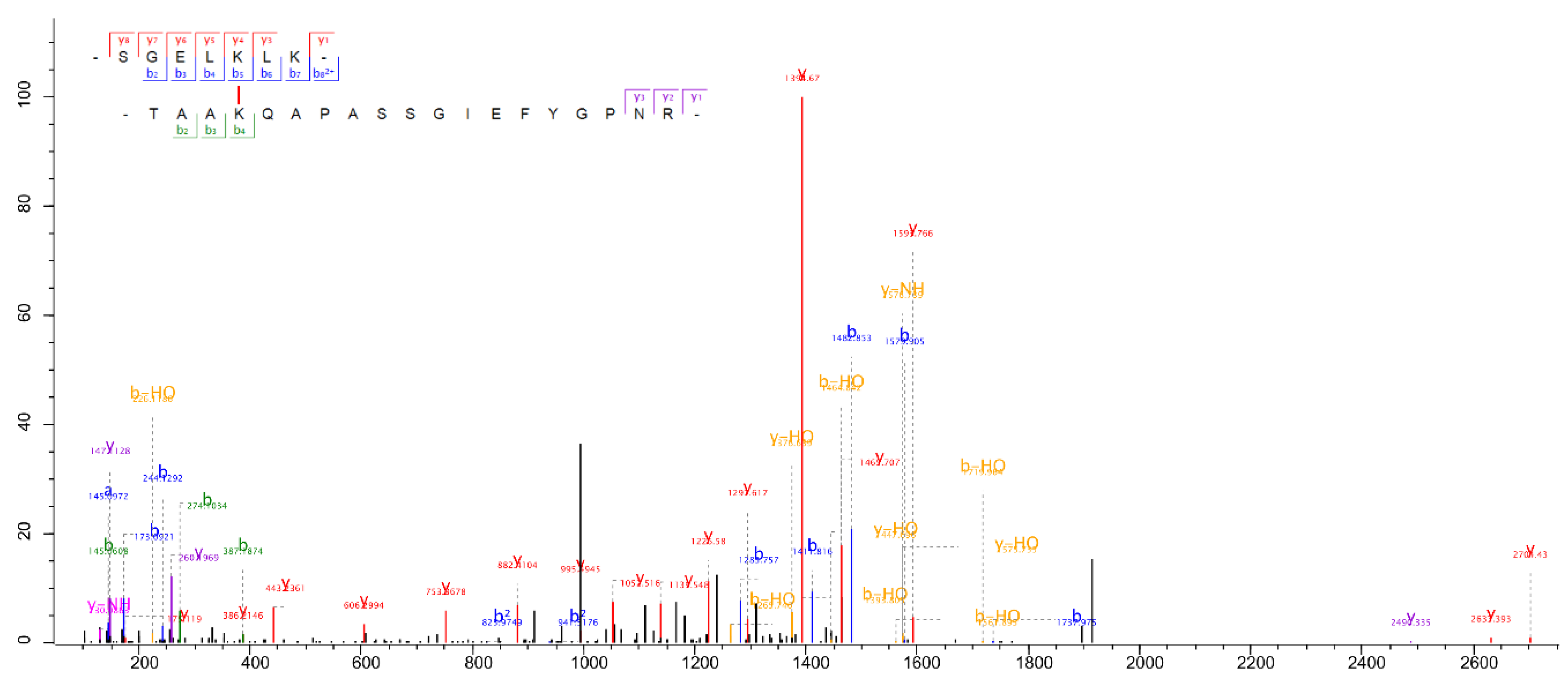
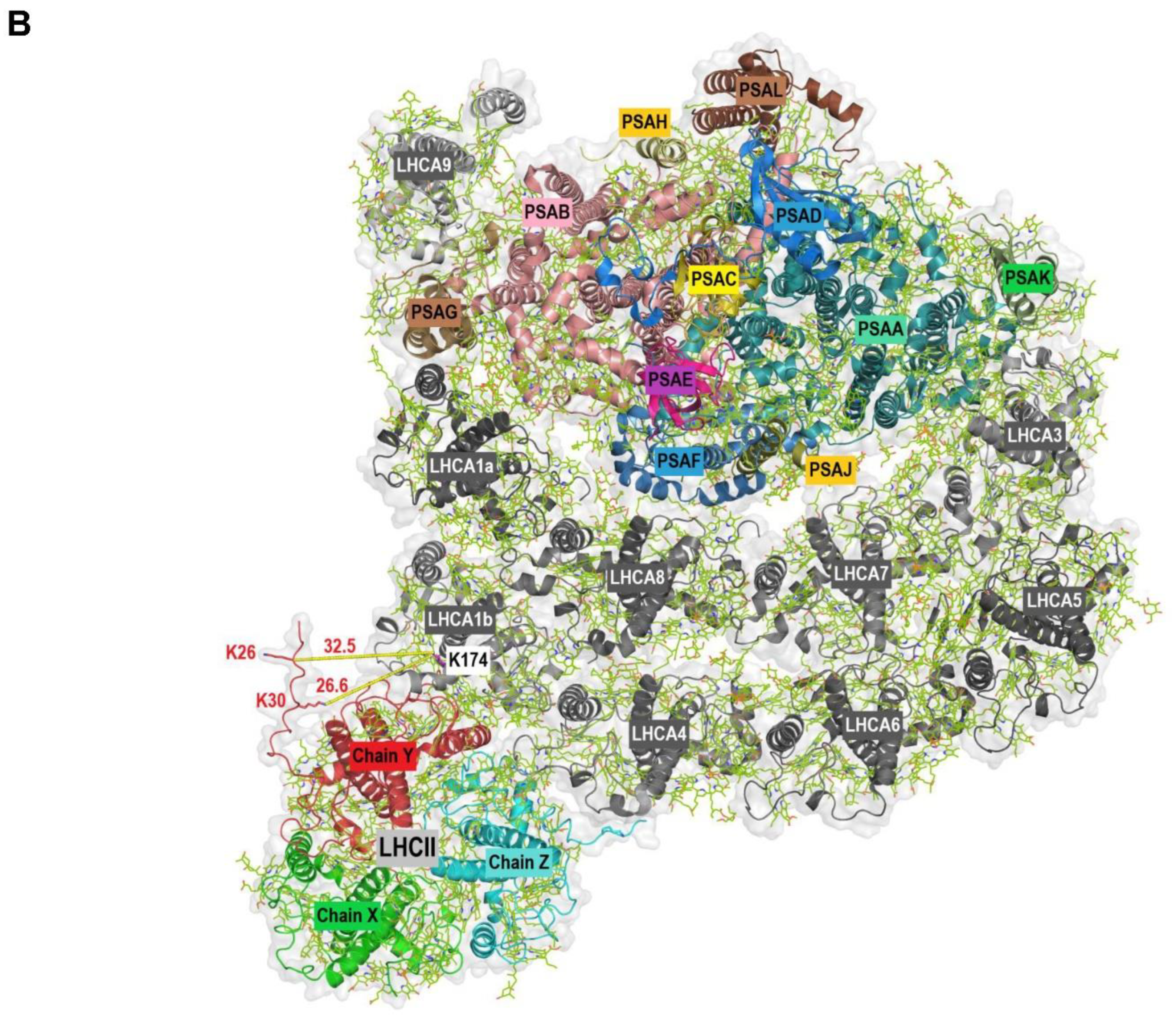
SGELKLK | K174 | TAAKAAAPK | K25 | Xi; Mx; MQ |
| Q05093 | LHCA1 | Cre06.g283050 | Q93VE0 | LHCBM1 | Cre01.g066917 | SGELKLK | K174 | TVKPASK | K29 | MSS; MQ; Xi |
| Q05093 | LHCA1 | Cre06.g283050 | Q93WL4 | LHCBM3 | Cre04.g232104 | SGELKLK | K174 | TAAKQAPASSGIEFYGPNR | K30 | Xi; MQ |
| Q05093, Q75VZ0, A8IKC8, Q75VY9 |
LHCA1, LHCA4, LHCA1- LIKE, LHCA2, LHCA3 |
Cre06.g283050, Cre10.g452050, Cre06.g283050, Cre12.g508750, Cre11.g467573 |
Q42687 | ATPD | Cre11.g467569 | LKELKNGR | K179 | LSALIMNPVVESDKK | K89 | Xi; MQ |
| Q75VY6 | LHCA6 | Cre06.g278213 | Q75VY8 | LHCA5 | Cre10.g425900 | ASSRPLWLPGSTPPAHLK | S28 | GNKVPNPEMGYPGGIFDPFGFSK | K156 | Xi; Mx |
| Q75VY7 | LHCA8 | Cre06.g272650 | Q75VZ0 | LHCA4 | Cre10.g452050 | EADKWADWK | K184 | LKWYAQAELMNAR | K94 | Xi; MSS; Mx; MQ |
| Q75VY9 | LHCA3 | Cre11.g467573 | Q75VY8 | LHCA5 | Cre10.g425900 | ELKLKEIK | K216 | VPNPEMGYPGGIFDPFGFSKGNLK | K176 | MQ; Xi |
| Q93WL4 | LHCBM3 | Cre04.g232104 | Q05093 | LHCA1 | Cre06.g283050 | GTGKTAAK | K26 | SGELKLK | K174 | Xi; MQ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





