Submitted:
02 May 2023
Posted:
04 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
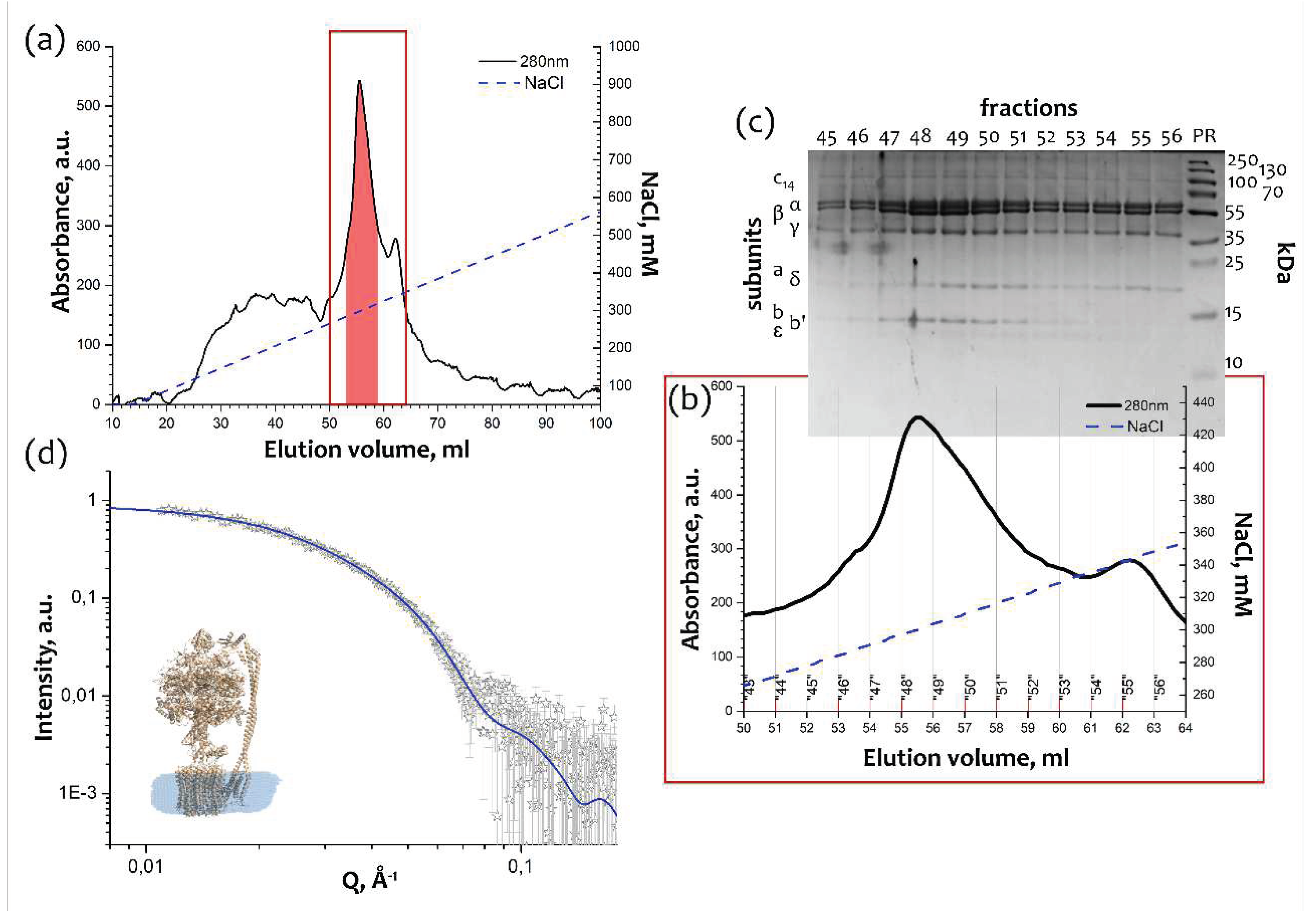
2.1. Purification of the monomeric FOF1 ATP-synthase from thylakoid membranes
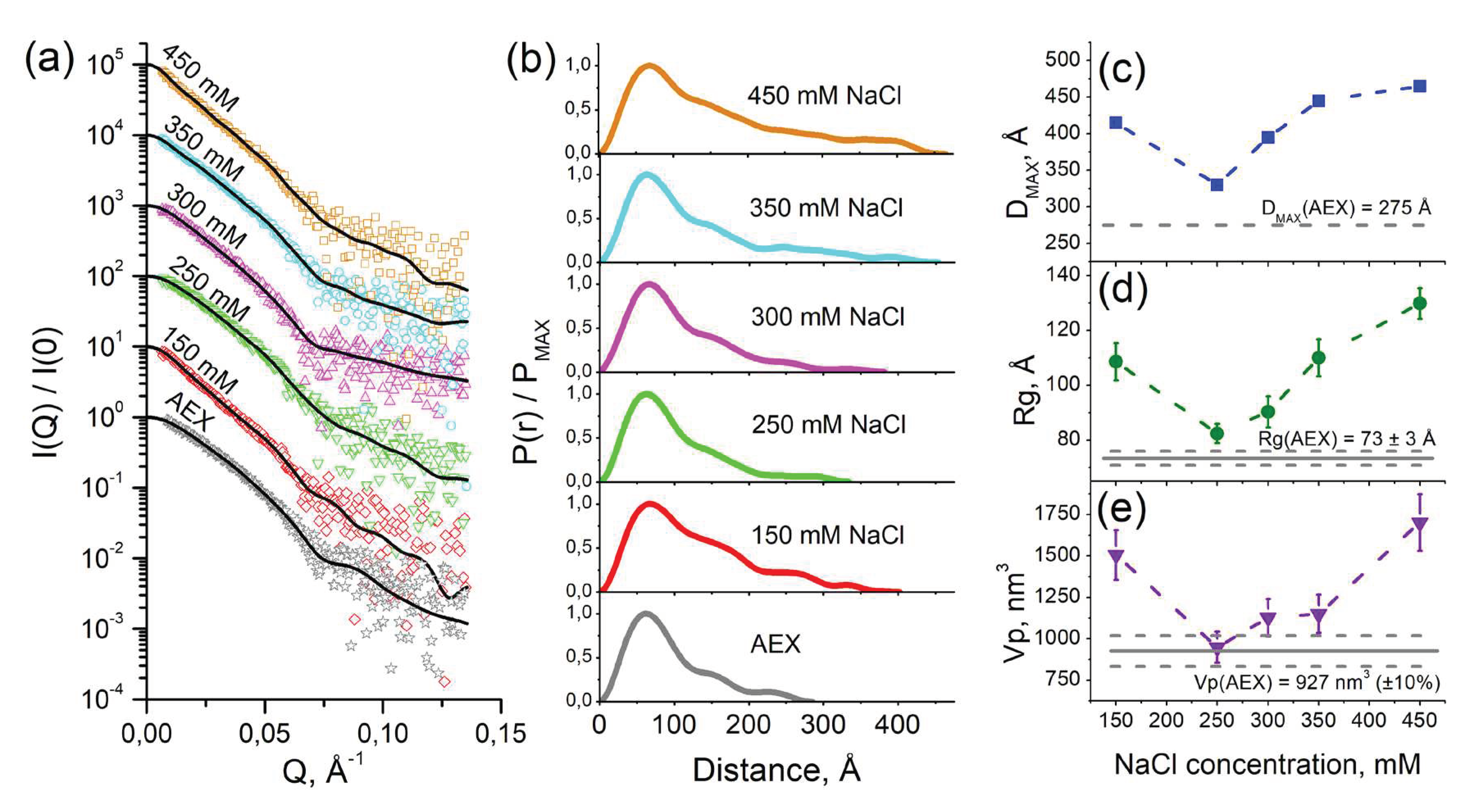
2.2. SAS studies of cFOF1 dimerization at different NaCl concentrations
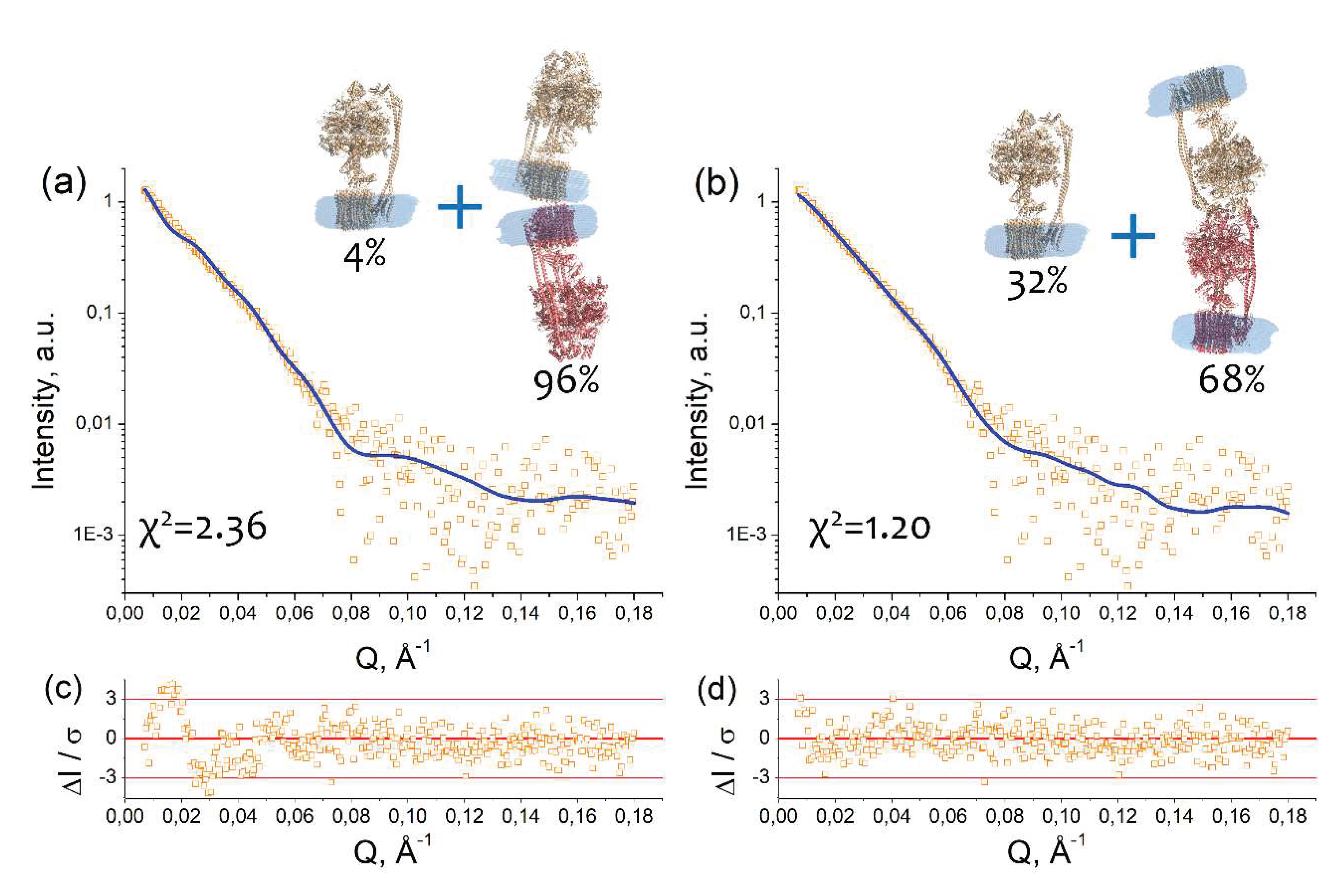
2.3. Models of an I-shaped cFOF1 dimer
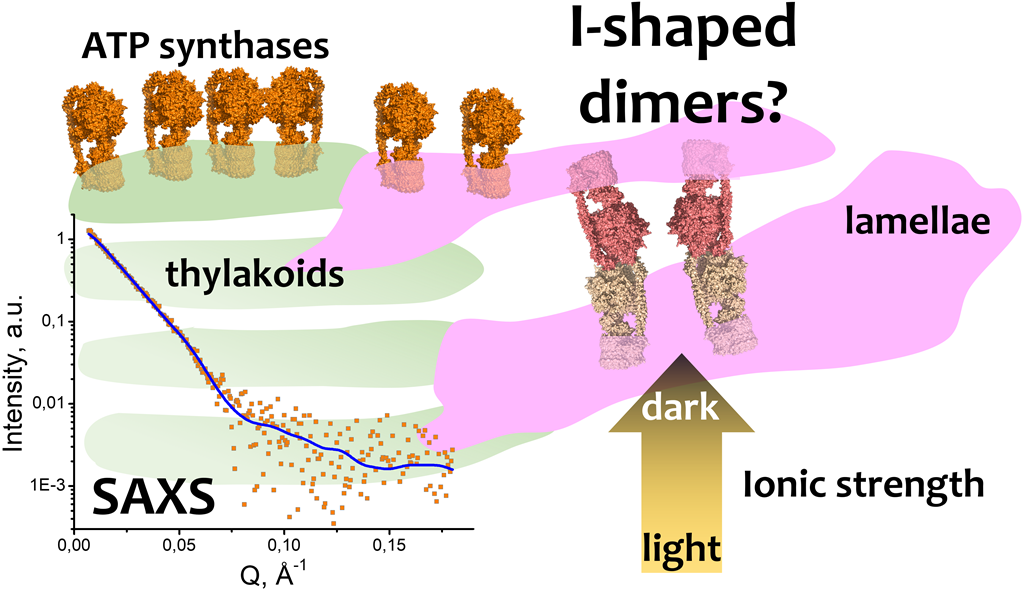
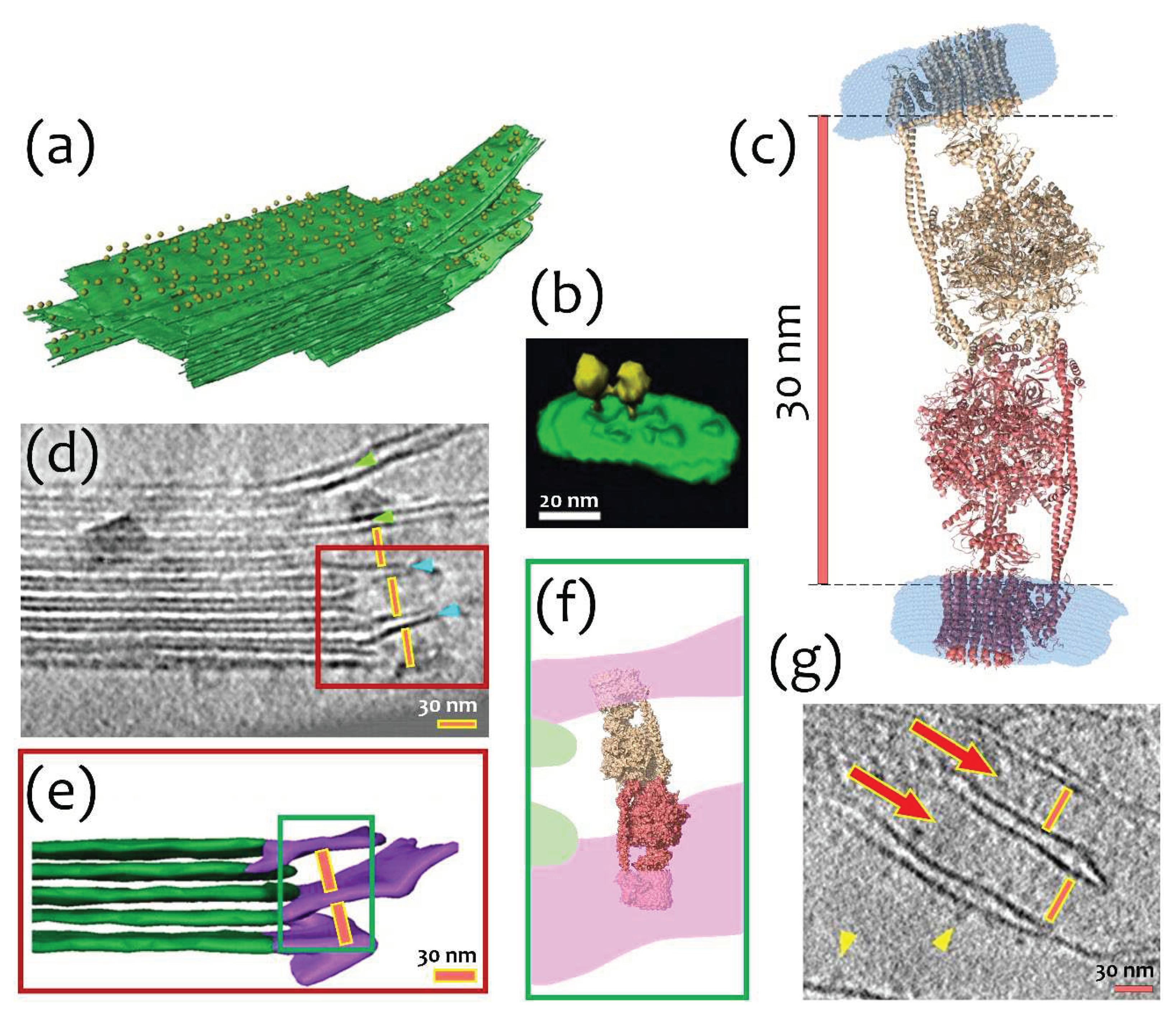
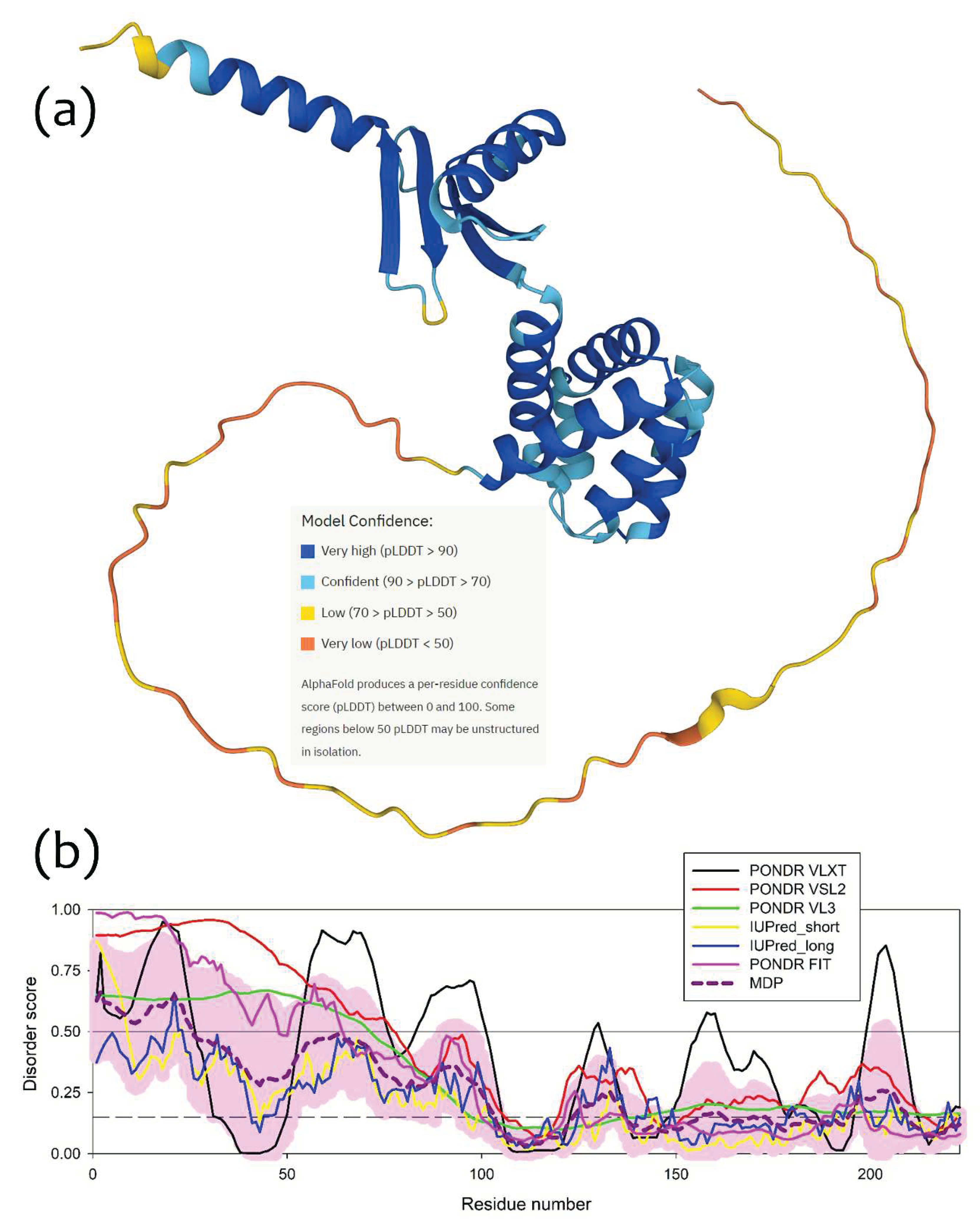
2.4. A possible physiological role of I-shaped cFOF1 dimers
3. Discussion
4. Materials and Methods
4.1. cFOF1 isolation and purification
4.2. Blue Native Polyacrylamide Gel Electrophoresis
4.3. Small-angle scattering measurements
4.4. Small-angle scattering data treatment
4.5. Macromolecular docking
4.6. Dynamic Scanning Fluorimetry
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vlasov, A.V.; Osipov, S.D.; Bondarev, N.A.; Uversky, V.N.; Borshchevskiy, V.I.; Yanyushin, M.F.; Manukhov, I.V.; Rogachev, A.V.; Vlasova, A.D.; Ilyinsky, N.S.; et al. ATP Synthase FOF1 Structure, Function, and Structure-Based Drug Design. Cellular and Molecular Life Sciences 2022, 79, 1–27. [Google Scholar] [CrossRef]
- Kühlbrandt, W. Structure and Mechanisms of F-Type ATP Synthases. Annu Rev Biochem 2019, 88, 515–549. [Google Scholar] [CrossRef]
- Daum, B.; Nicastro, D.; Austin, J.; Richard McIntosh, J.; Kühlbrandt, W. Arrangement of Photosystem II and ATP Synthase in Chloroplast Membranes of Spinach and Pea. Plant Cell 2010, 22, 1299–1312. [Google Scholar] [CrossRef]
- Robinson, S.P.; Downton, W.J.S. Potassium, Sodium, and Chloride Content of Isolated Intact Chloroplasts in Relation to Ionic Compartmentation in Leaves. Arch Biochem Biophys 1984, 228, 197–206. [Google Scholar] [CrossRef]
- Chow, W.; Wagner, A.; Hope, A. Light-Dependent Redistribution of Ions in Isolated Spinach Chloroplasts. Functional Plant Biology 1976, 3, 853–861. [Google Scholar] [CrossRef]
- Nobel, P.S. Light-Induced Changes in the Ionic Content of Chloroplasts in Pisum Sativum. Biochimica et Biophysica Acta (BBA) - Bioenergetics 1969, 172, 134–143. [Google Scholar] [CrossRef]
- Vlasov, A.V. New Structural Insights in Chloroplast F1FO-ATP Synthases - RWTH Publications, RWTH Aachen University, 2021.
- Vlasov, A. V.; Kovalev, K. V.; Marx, S.-H.; Round, E.S.; Gushchin, I.Yu.; Polovinkin, V.A.; Tsoy, N.M.; Okhrimenko, I.S.; Borshchevskiy, V.I.; Büldt, G.D.; et al. Unusual Features of the C-Ring of F1FO ATP Synthases. Sci Rep 2019, 9, 18547. [Google Scholar] [CrossRef] [PubMed]
- Balakrishna, A.M.; Seelert, H.; Marx, S.-H.; Dencher, N.A.; Grüber, G. Crystallographic Structure of the Turbine C-Ring from Spinach Chloroplast F-ATP Synthase. Biosci Rep 2014, 34, e00102. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Vonck, J.; Mills, D.J.; Meier, T.; Kühlbrandt, W. Structure, Mechanism, and Regulation of the Chloroplast ATP Synthase. Science 2018, 360, eaat4318. [Google Scholar] [CrossRef]
- Pérez, J.; Koutsioubas, A. Memprot: A Program to Model the Detergent Corona around a Membrane Protein Based on SEC-SAXS Data. Acta Crystallogr D Biol Crystallogr 2015, 71, 86–93. [Google Scholar] [CrossRef]
- Ryzhykau, Y.L.; Vlasov, A. V.; Orekhov, P.S.; Rulev, M.I.; Rogachev, A. V.; Vlasova, A.D.; Kazantsev, A.S.; Verteletskiy, D.P.; Skoi, V. V.; Brennich, M.E.; et al. Ambiguities in and Completeness of SAS Data Analysis of Membrane Proteins: The Case of the Sensory Rhodopsin II–Transducer Complex. Acta Crystallogr D Struct Biol 2021, 77, 1386–1400. [Google Scholar] [CrossRef]
- Ryzhykau, Y.L.; Orekhov, P.S.; Rulev, M.I.; Vlasov, A. v.; Melnikov, I.A.; Volkov, D.A.; Nikolaev, M.Yu.; Zabelskii, D. v.; Murugova, T.N.; Chupin, V. v.; et al. Molecular Model of a Sensor of Two-Component Signaling System. Sci Rep 2021, 11, 10774. [Google Scholar] [CrossRef] [PubMed]
- Molodenskiy, D.S.; Svergun, D.I.; Mertens, H.D.T. MPBuilder: A PyMOL Plugin for Building and Refinement of Solubilized Membrane Proteins Against Small Angle X-Ray Scattering Data. J Mol Biol 2021, 433, 166888. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Tao, H.; He, J.; Huang, S.Y. The HDOCK Server for Integrated Protein–Protein Docking. Nature Protocols 2020 15:5 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J Mol Biol 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Minges, A.; Groth, G. Structure and Supramolecular Architecture of Chloroplast ATP Synthase. Adv Bot Res 2020, 96, 27–74. [Google Scholar] [CrossRef]
- Rexroth, S.; Meyer Zu Tittingdorf, J.M.W.; Schwaßmann, H.J.; Krause, F.; Seelert, H.; Dencher, N.A. Dimeric H+-ATP Synthase in the Chloroplast of Chlamydomonas Reinhardtii. Biochim Biophys Acta Bioenerg 2004, 1658, 202–211. [Google Scholar] [CrossRef]
- Schwaßmann, H.J.; Rexroth, S.; Seelert, H.; Dencher, N.A. Metabolism Controls Dimerization of the Chloroplast FoF1 ATP Synthase in Chlamydomonas Reinhardtii. FEBS Lett 2007, 581, 1391–1396. [Google Scholar] [CrossRef]
- Disfani, F.M.; Hsu, W.-L.; Mizianty, M.J.; Oldfield, C.J.; Xue, B.; Dunker, A.K.; Uversky, V.N.; Kurgan, L. MoRFpred, a Computational Tool for Sequence-Based Prediction and Characterization of Short Disorder-to-Order Transitioning Binding Regions in Proteins. Bioinformatics 2012, 28, i75–83. [Google Scholar] [CrossRef]
- Cheng, Y.; Oldfield, C.J.; Meng, J.; Romero, P.; Uversky, V.N.; Dunker, A.K. Mining Alpha-Helix-Forming Molecular Recognition Features with Cross Species Sequence Alignments. Biochemistry 2007, 46, 13468–13477. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Romero, P.; Uversky, V.N.; Dunker, A.K. Coupled Folding and Binding with Alpha-Helix-Forming Molecular Recognition Elements. Biochemistry 2005, 44, 12454–12470. [Google Scholar] [CrossRef]
- Garner, E.; Romero, P.; Dunker, A.; Brown, C.; Obradovic, Z. Predicting Binding Regions within Disordered Proteins. Genome Inform Ser Workshop Genome Inform 1999, 10, 41–50. [Google Scholar] [PubMed]
- DeForte, S.; Uversky, V.N. Resolving the Ambiguity: Making Sense of Intrinsic Disorder When PDB Structures Disagree. Protein Sci 2016, 25, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, T.; Romero, P.R.; Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Intrinsic Disorder in the Protein Data Bank. J Biomol Struct Dyn 2007, 24, 325–342. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Kryshtafovych, A.; Schwede, T.; Topf, M.; Fidelis, K.; Moult, J. Critical Assessment of Methods of Protein Structure Prediction (CASP)-Round XIV. Proteins 2021, 89, 1607–1617. [Google Scholar] [CrossRef]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence Complexity of Disordered Protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-Dependent Prediction of Protein Intrinsic Disorder. BMC Bioinformatics 2006, 7, 208. [Google Scholar] [CrossRef]
- Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Optimizing Long Intrinsic Disorder Predictors with Protein Evolutionary Information. J Bioinform Comput Biol 2005, 3, 35–60. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A Meta-Predictor of Intrinsically Disordered Amino Acids. Biochim Biophys Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Mészáros, B.; Erdos, G.; Dosztányi, Z. IUPred2A: Context-Dependent Prediction of Protein Disorder as a Function of Redox State and Protein Binding. Nucleic Acids Res 2018, 46, W329–W337. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Katuwawala, A.; Wang, K.; Wu, Z.; Ghadermarzi, S.; Gao, J.; Kurgan, L. FlDPnn: Accurate Intrinsic Disorder Prediction with Putative Propensities of Disorder Functions. Nat Commun 2021, 12, 4438. [Google Scholar] [CrossRef] [PubMed]
- Dayhoff, G.W.; Uversky, V.N. Rapid Prediction and Analysis of Protein Intrinsic Disorder. Protein Sci 2022, 31, e4496. [Google Scholar] [CrossRef] [PubMed]
- Caffrey, M.; Cherezov, V. Crystallizing Membrane Proteins Using Lipidic Mesophases. Nat Protoc 2009, 4, 706–731. [Google Scholar] [CrossRef]
- Faham, S.; Bowie, J.U. Bicelle Crystallization: A New Method for Crystallizing Membrane Proteins Yields a Monomeric Bacteriorhodopsin Structure. J Mol Biol 2002, 316, 1–6. [Google Scholar] [CrossRef]
- Murugova, T.N.; Ivankov, O.I.; Ryzhykau, Y.L.; Soloviov, D. V.; Kovalev, K. V.; Skachkova, D. V.; Round, A.; Baeken, C.; Ishchenko, A. V.; Volkov, O.A.; et al. Mechanisms of Membrane Protein Crystallization in ‘Bicelles. ’ Scientific Reports 2022 12:1 2022, 12, 1–17. [Google Scholar] [CrossRef]
- Morgan, J.L.W.; McNamara, J.T.; Zimmer, J. Mechanism of Activation of Bacterial Cellulose Synthase by Cyclic Di-GMP. Nature Structural & Molecular Biology 2014, 21, 489–496. [Google Scholar] [CrossRef]
- Noinaj, N.; Kuszak, A.J.; Gumbart, J.C.; Lukacik, P.; Chang, H.; Easley, N.C.; Lithgow, T.; Buchanan, S.K. Structural Insight into the Biogenesis of β-Barrel Membrane Proteins. Nature 2013, 501, 385–390. [Google Scholar] [CrossRef]
- Lin, D.Y.W.; Huang, S.; Chen, J. Crystal Structures of a Polypeptide Processing and Secretion Transporter. Nature 2015, 523, 425–430. [Google Scholar] [CrossRef]
- Stock, D.; Leslie, A.G.W.; Walker, J.E. Molecular Architecture of the Rotary Motor in ATP Synthase. Science (1979) 1999, 286, 1700–1705. [Google Scholar] [CrossRef] [PubMed]
- Dautant, A.; Velours, J.; Giraud, M.F. Crystal Structure of the Mg·ADP-Inhibited State of the Yeast F 1c10-ATP Synthase. Journal of Biological Chemistry 2010, 285, 29502–29510. [Google Scholar] [CrossRef]
- Watt, I.N.; Montgomery, M.G.; Runswick, M.J.; Leslie, A.G.W.; Walker, J.E. Bioenergetic Cost of Making an Adenosine Triphosphate Molecule in Animal Mitochondria. Proc Natl Acad Sci USA 2010, 107, 16823–16827. [Google Scholar] [CrossRef] [PubMed]
- Giraud, M.F.; Paumard, P.; Sanchez, C.; Brèthes, D.; Velours, J.; Dautant, A. Rotor Architecture in the Yeast and Bovine F 1-c-Ring Complexes of F-ATP Synthase. J Struct Biol 2012, 177, 490–497. [Google Scholar] [CrossRef]
- Morales-Rios, E.; Montgomery, M.G.; Leslie, A.G.W.; Walker, J.E. Structure of ATP Synthase from Paracoccus Denitrificans Determined by X-Ray Crystallography at 4.0 Å Resolution. Proc Natl Acad Sci USA 2015, 112, 13231–13236. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Parey, K.; Bublitz, M.; Mills, D.J.; Zickermann, V.; Vonck, J.; Kühlbrandt, W.; Meier, T. Structure of a Complete ATP Synthase Dimer Reveals the Molecular Basis of Inner Mitochondrial Membrane Morphology. Mol Cell 2016, 63, 445–456. [Google Scholar] [CrossRef]
- Zabelskii, D.V.; Vlasov, A.V.; Ryzhykau, Y.L.; Murugova, T.N.; Brennich, M.; Soloviov, D.V.; Ivankov, O.I.; Borshchevskiy, V.I.; Mishin, A.V.; Rogachev, A.V.; et al. Ambiguities and Completeness of SAS Data Analysis: Investigations of Apoferritin by SAXS/SANS EID and SEC-SAXS Methods. In Proceedings of the Journal of Physics: Conference Series; 2018; Vol. 994.
- Böttcher, B.; Gräber, P. The Structure of the H+-ATP Synthase from Chloroplasts and Its Subcomplexes as Revealed by Electron Microscopy. Biochim Biophys Acta Bioenerg 2000, 1458, 404–416. [Google Scholar] [CrossRef]
- Vlasov, A.V.; Kovalev, Y.S.; Utrobin, P.K.; Ryzhykau, Y.L.; Frolov, F.V.; Zinovev, E.V.; Rogachev, A.V.; Kuklin, A.I.; Gordeliy, V.I. Photo-Voltage of Highly-Oriented Bacteriorhodopsin in Purple Membranes: Possibilities for Bio Solar Cells. Optoelectronics and Advanced Materials, Rapid Communications 2017, 11. [Google Scholar]
- Daum, B.; Nicastro, D.; Austin, J.; Richard McIntosh, J.; Kühlbrandt, W. Arrangement of Photosystem II and ATP Synthase in Chloroplast Membranes of Spinach and Pea. Plant Cell 2010, 22, 1299–1312. [Google Scholar] [CrossRef]
- Engel, B.D.; Schaffer, M.; Cuellar, L.K.; Villa, E.; Plitzko, J.M.; Baumeister, W. Native Architecture of the Chlamydomonas Chloroplast Revealed by in Situ Cryo-Electron Tomography. Elife 2015, 2015. [Google Scholar] [CrossRef]
- Schägger, H.; von Jagow, G. Blue Native Electrophoresis for Isolation of Membrane Protein Complexes in Enzymatically Active Form. Anal Biochem 1991, 199, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Džinić, T.; Hartwig, S.; Lehr, S.; Dencher, N.A. Oxygen and Differentiation Status Modulate the Effect of X-Ray Irradiation on Physiology and Mitochondrial Proteome of Human Neuroblastoma Cells. 2016, 122, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Kuklin, A.I.; Ivankov, O.I.; Rogachev, A. V.; Soloviov, D. V.; Islamov, A.K.; Skoi, V. V.; Kovalev, Y.S.; Vlasov, A. V.; Ryzhykau, Y.L.; Soloviev, A.G.; et al. Small-Angle Neutron Scattering at the Pulsed Reactor IBR-2: Current Status and Prospects. Crystallography Reports 2021, 66, 231–241. [Google Scholar] [CrossRef]
- Kuklin, A.I.; Islamov, A.K.; Gordeliy, V.I. Scientific Reviews: Two-Detector System for Small-Angle Neutron Scattering Instrument. Neutron News 2005, 16, 16–18. [Google Scholar] [CrossRef]
- Kuklin, A.I.; Soloviov, D. V; Rogachev, A. V; Utrobin, P.K.; Kovalev, Y.S.; Balasoiu, M.; Ivankov, O.I.; Sirotin, A.P.; Murugova, T.N.; Petukhova, T.B.; et al. New Opportunities Provided by Modernized Small-Angle Neutron Scattering Two-Detector System Instrument (YuMO). J Phys Conf Ser 2011, 291, 012013. [Google Scholar] [CrossRef]
- Covington, A.K.; Paabo, M.; Robinson, R.A.; Bates, R.G. Use of the Glass Electrode in Deuterium Oxide and the Relation between the Standardized PD (PaD) Scale and the Operational PH in Heavy Water. Anal Chem 1968, 40, 700–706. [Google Scholar] [CrossRef]
- Tsoraev, G. V.; Protasova, E.A.; Klimanova, E.A.; Ryzhykau, Y.L.; Kuklin, A.I.; Semenov, Y.S.; Ge, B.; Li, W.; Qin, S.; Friedrich, T.; et al. Anti-Stokes Fluorescence Excitation Reveals Conformational Mobility of the C-Phycocyanin Chromophores. Structural Dynamics 2022, 9, 054701. [Google Scholar] [CrossRef]
- Nyam-Osor, M.; Soloviov, D. V; Kovalev, Y.S.; Zhigunov, A.; Rogachev, A. V; Ivankov, O.I.; Erhan, R. V; Kuklin, A.I. Silver Behenate and Silver Stearate Powders for Calibration of SAS Instruments. J Phys Conf Ser 2012, 351, 012024. [Google Scholar] [CrossRef]
- Soloviev, A.G.; Solovjeva, T.M.; Ivankov, O.I.; Soloviov, D. V.; Rogachev, A. V.; Kuklin, A.I. SAS Program for Two-Detector System: Seamless Curve from Both Detectors. J Phys Conf Ser 2017, 848, 012020. [Google Scholar] [CrossRef]
- Manalastas-Cantos, K.; Konarev, P. V.; Hajizadeh, N.R.; Kikhney, A.G.; Petoukhov, M. V.; Molodenskiy, D.S.; Panjkovich, A.; Mertens, H.D.T.; Gruzinov, A.; Borges, C.; et al. ATSAS 3.0: Expanded Functionality and New Tools for Small-Angle Scattering Data Analysis. urn:issn:1600-5767 2021, 54, 343–355. [Google Scholar] [CrossRef]
- Franke, D.; Petoukhov, M. V.; Konarev, P. V.; Panjkovich, A.; Tuukkanen, A.; Mertens, H.D.T.; Kikhney, A.G.; Hajizadeh, N.R.; Franklin, J.M.; Jeffries, C.M.; et al. ATSAS 2.8: A Comprehensive Data Analysis Suite for Small-Angle Scattering from Macromolecular Solutions. J Appl Crystallogr 2017, 50, 1212–1225. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An Open-Source Molecular Graphics Tool. CCP4 Newsletter on protein crystallography 2002, 40, 82–92. [Google Scholar]
- Lomize, M.A.; Pogozheva, I.D.; Joo, H.; Mosberg, H.I.; Lomize, A.L. OPM Database and PPM Web Server: Resources for Positioning of Proteins in Membranes. Nucleic Acids Res 2012, 40, D370–D376. [Google Scholar] [CrossRef] [PubMed]
- Konarev, P. V.; Volkov, V. V.; Sokolova, A. V.; Koch, M.H.J.; Svergun, D.I. PRIMUS: A Windows PC-Based System for Small-Angle Scattering Data Analysis. urn:issn:0021-8898 2003, 36, 1277–1282. [Google Scholar] [CrossRef]
- Svergun, D.I. ; IUCr Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. urn:issn:0021-8898 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Ferrè, F.; Clote, P. DiANNA: A Web Server for Disulfide Connectivity Prediction. Nucleic Acids Res 2005, 33, W230–W232. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




