1. Introduction
Klebsiella pneumoniae is a facultative anaerobe, rod-shaped, encapsulated, non-motile, gram-negative and lactose-fermenting bacterium, normally within the normal flora of the mouth, skin, and intestines (1). Nonetheless, virulence factors that result in variety of infections such as septicemia, lobar pneumonia and urinary tract infections can also cause harmful and pathological changes in the host. The last 20 years have seen a significant increase in the public health risk posed by ESBL-producing bacteria (2). It is clear from the information given that bacteria that acquire and develop ESBL-type resistance are typically members of the Enterobacteriaceae family, of which K. pneumoniae and E. coli are the most significant (2). Extended Spectrum Beta-Lactamases (ESBL) are enzymes that can hydrolyze and break down the beta-lactam ring of Penicillins and Cephalosporins (1, 2). This means that the inactivation of these particular antimicrobial drugs will lead to bacterial resistance and the ineffectiveness of the host defense.
The importance of ESBLs stems from the fact that they represent a growing public health risk, which is connected to the overuse and misuse of antibiotics. Firstly, ESBL-producing bacteria are resistant to many common antibiotics, making infections difficult to treat. Consequently, patients with ESBL infections may have delays in receiving the most effective treatment while doctors try to determine which antibiotics the infecting bacteria are susceptible to. This leads to increased mortality rates. It has been shown that patients with ESBL-producing bacterial infections have a 57% chance of death compared to an infection with a non-ESBL-producing strain (3). Secondly, the spread of ESBLs is also a risk to public and community health, as patients infected in hospitals may well have prolonged stays in healthcare environments which can become centers for the spread of ESBL-producing bacteria (4). It is challenging to control the spread of these bacteria in hospital environments as the patients are already highly susceptible to infection.
However, many K. pneumoniae strains are resistant to Carbapenem antibiotics, which are the most effective drugs available for the treatment of serious infections, and this type of resistance is becoming more common in hospitals and also in non-hospitalized patients (5). Therefore, if the spread of ESBLs is not effectively managed, it is very likely that there will be a new wave of antibiotic resistance that will pose a significant worldwide health risk. Furthermore, as the likelihood of sepsis and other severe conditions resulting from bacterial infection increases, the burden on healthcare providers and systems could become overwhelming. This scenario perfectly illustrates the wider social and economic importance of combating ESBLs and other antibiotic resistance mechanisms (6, 7).
Several studies have been carried out to estimate the prevalence of various forms of antibiotic resistance in many parts of Nigeria (8). The emergence of antimicrobial resistance in numerous microbes poses a serious threat to the healthcare system now more than ever. Several studies indicate that antibiotic resistance is rising and that new forms of acquired resistance are emerging (8, 9). The investigation of ESBL enzymes in clinical specimens is gaining attention due to the failure of therapy with antibiotics, resulting in poor patient outcomes and the search for better antibiotics, which impose a significant economic burden on healthcare. Since the treatment of ESBL-producing organisms is limited to a few "last resort" antibiotics, such as Carbapenems and Tigecycline, which are more toxic and costly compared to conventional antibiotics, it is essential to understand the prevalence and the mechanism of spread of ESBL enzymes to identify best practice for the treatment of these organisms and to explore novel therapeutic options to optimize patient's outcome and reduce the economic healthcare burden (8). The study's goal is to find out how common it is for urine samples from non-hospitalized patients with suspected UTIs to contain K. pneumoniae encoding genes for the extended-spectrum beta-lactamases (ESBL) enzymes CTX-M, TEM, and SHV-1. It is hoped that this study can provide further insight to clinicians in choosing the first-line antibiotics, hence to achieve better prognosis for the patients and at the same time, could contribute to the ongoing strategies to ensure the relevance of antibiotic therapy and to reduce the burden of antibiotic resistance in Nigeria.
2. Methodology
2.1. Study Area
2.1.1. Study Population
All K. pneumoniae urine cultures processed at a diagnostic centre (Lagos, Nigeria) between May and August 2023 were analyzed in this retrospective study. All urine cultures analyzed in this study were from only outpatients who employed the services of the diagnostic centre and were confirmed to have UTI. In cases where multiple urine cultures from the same patient were positive for the same organism, only the first episode was reviewed and recorded. All the urine cultures including all age groups and sexes in which Klebsiella was implicated in the confirmed UTI cases were analysed.
2.1.2. Characterization and Identification of Bacterial Isolates
To re-identify Klebsiella, the isolates were sub-cultured on MacConkey. The colony morphology revealed that the bacterial isolates were large, mucoid, convex, smooth, lactose fermenting, and translucent. Gram-staining revealed uniformly stained Gram-negative rods with parallel or bulging sides and slightly pointed or rounded ends; the bacterial isolates were non-sporing and non-motile in the preparation of hanging drops; and the biochemical reactions revealed that the bacteria was lactose fermenter, catalase test positive, O/F (oxidation/fermentation) test showing glucose fermentation, motility, and gas production, indole test negative, citrate used, urease test positive, lactose fermenter.
2.1.3. Antibiotic Susceptibility Test of Klebsiella
This test was conducted using the Kirby-Bauer disk diffusion method, as modified by the Clinical and Laboratory Standards Institute (CLSI, 2018) (10). Antimicrobial susceptibility testing was conducted using Mueller-Hinton agar (MHA) media and antibiotic discs. Amoxicillin/Clavulanic acid (AUG) (30µg), Tetracycline (30µg), Gentamicin (10µg), Erythromycin (30µg), Colistin (30µg), Ceftazidime (CAZ) (30μg), Ceftriaxone (CTR) (30μg), Ciprofloxacin (5µg), and Imipenem (10µg) are among the commercial antibiotics used. The organisms were categorized as sensitive, resistant, or intermediate based on the measurement of the zone of inhibitions' diameter.
Ceftazidime and Ceftriaxone discs spaced 20 mm from edge to edge from an Amoxicillin-clavulanic acid disc were placed on inoculated MHA plates, and the discs were incubated for 24 hours at 37 °C to test for ESBL production. An expanded zone of inhibition between the Amoxicillin-Clavulanic acid disc and any of the Cephalosporin discs was considered proof of an ESBL (10). The formation of a "keyhole" with Amoxicillin-Clavulanic acid or either of the antibiotics indicated a positive confirmatory screening test.
2.1.4. Identification of Resistance Genes
The existence of resistance genes was examined in each isolate. This analysis was conducted using recently cultured bacterial cells. Using the Boiling method (30), the isolates' DNA was extracted for genetic analysis. Polymerase chain reaction was used to target the blaSHV, blaCTX-M, and blaTEM genes using specific oligonucleotide primers, as indicated in
Table 1. One Taq Quick Load Master Mix was used along with standard buffer. Ethidium bromide staining was used to identify the amplified DNA bands after the PCR products were separated on a 1.5% agarose gel. The sizes of the PCR product were accessed using a molecular marker.
3. Results
Patient information such as age and gender were retrieved from laboratory records at the centre. A total of 41 isolates which fulfill the inclusion criteria were screened for the study population. 16 (34.14%) were from the male population and 25 (60.97%) were from the female population. The majority of the patients suffering from UTI clustered in the age group of 21-30 years (17/41 i.e. 41.46%) i.e. sexually active age. All 41 of the isolates were reconfirmed as Klebsiella isolates based on the biochemical characterization.
3.1. Antibiotic Resistance Pattern
Resistance to Erythromycin was the highest with 100% of the isolates being resistant to the antibiotics. Similarly, marked resistance was noted against Ciprofloxacin, Colistin, Amoxicillin-Clavulanic acid and Tetracycline with 40 (97.56%), 34 (82.93%), 30 (73.17%) and 28 (68.29%) of the isolates respectively being resistant to the antibiotics (
Table 2).
Using the conventional disk diffusion method, 41 K. pneumoniae isolates were screened for the production of ESBL in this study. The isolates were all resistant to third-generation Cephalosporins; the percentages of isolates resistant to Ceftazidime and Ceftriaxone were 100% and 80.49%, respectively. The double disk synergy method was used to test all confirmed cases of K. pneumoniae for ESBL. Following testing of the chosen bacterial samples, 8 (19.51%) of the K. pneumoniae isolates were found to be ESBL positive and 33 (80.48%) to be ESBL negative using the combination disc method.
3.2. Resistance Genes Detection:
PCR and gel electrophoresis were used to check for the presence of genes encoding ESBL production in each of the 41 isolates. Using blaTEM, blaSHV, and blaCTX-M specific primers, PCR was used to screen all isolates. In Klebsiella isolates, the frequency of the SHV, TEM, and CTX-M genes was 31 (75.61%) (Figure 10), 4 (9.76%) (
Figure 2), and 10 (24.39%) (
Figure 2), respectively. Additionally, four isolates (9.67%) showed evidence of the SHV, TEM, and CTX-M genes. Nevertheless, the CTX-M and SHV genes were present in 8 (19.51%) of the isolates.
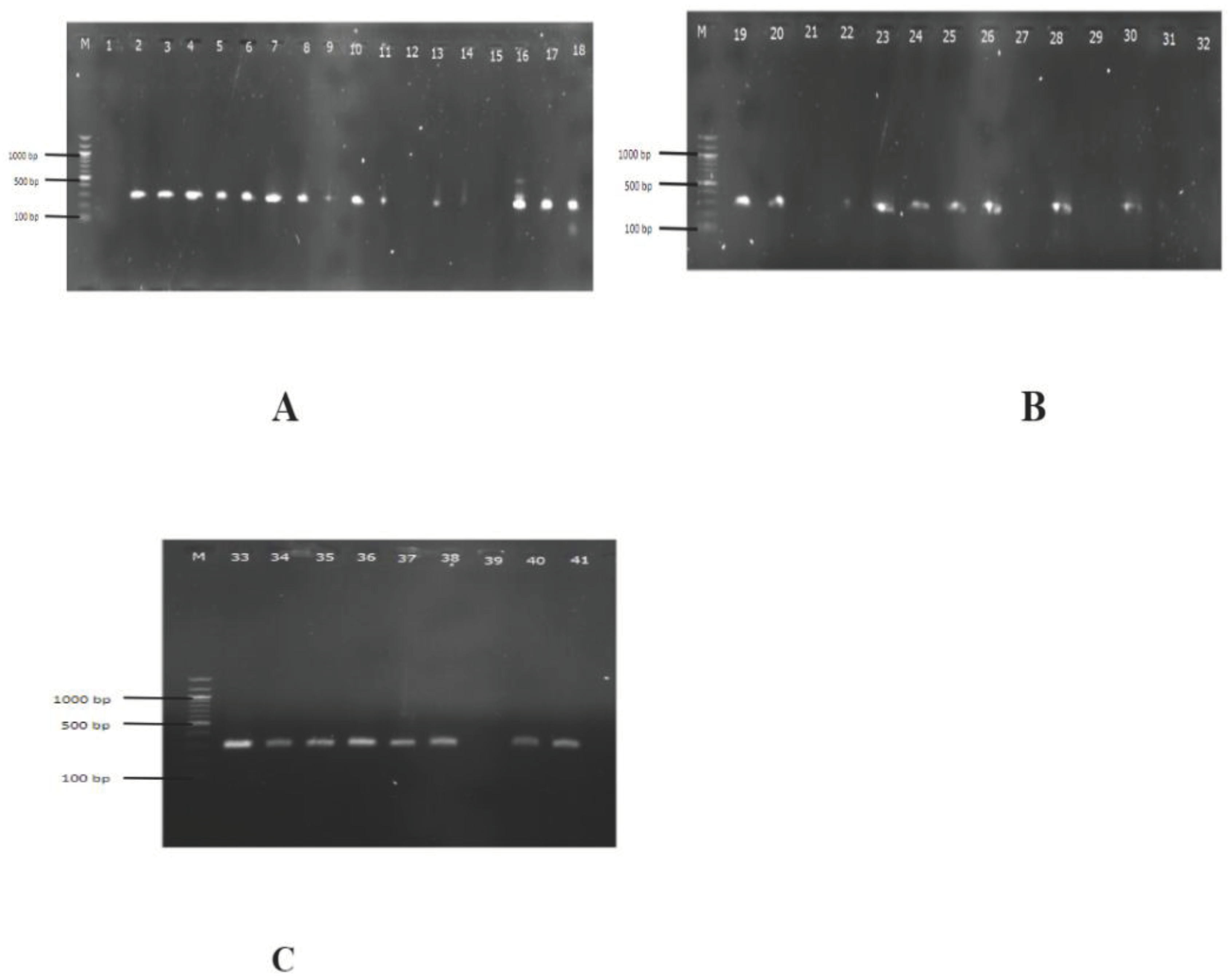
Figure 1.
Gel pictures of amplified products; SHV gene: 293 bps. *M: DNA ladder of 100 bps.
Figure 1.
Gel pictures of amplified products; SHV gene: 293 bps. *M: DNA ladder of 100 bps.
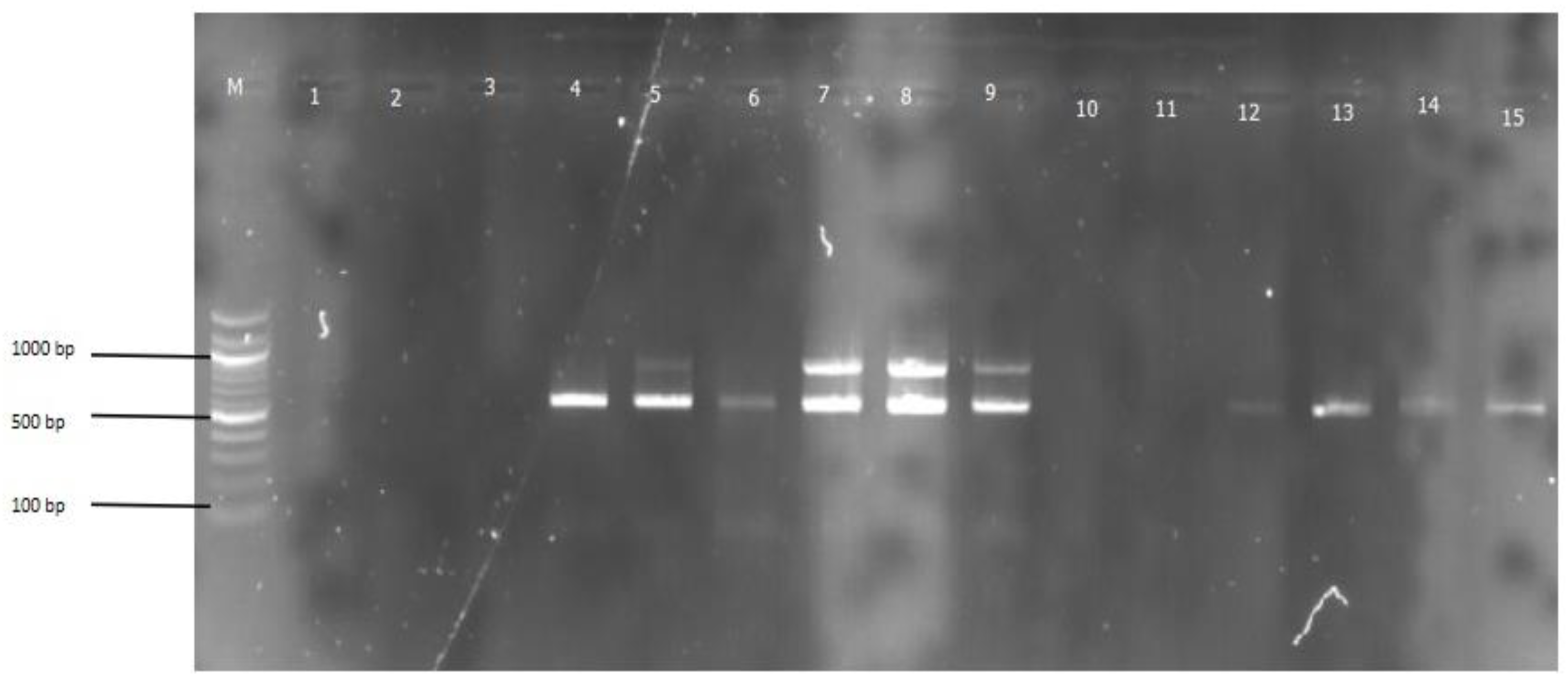
Figure 2.
Gel pictures of amplified products; TEM gene: 860 bps. (b) CTX-M gene: 588 bps. *M: DNA ladder of 100 bps.
Figure 2.
Gel pictures of amplified products; TEM gene: 860 bps. (b) CTX-M gene: 588 bps. *M: DNA ladder of 100 bps.
4. Discussion
The use of antimicrobial drugs in human treatment has led to an increase in antimicrobial resistance in animals, humans, and the environment. UTIs are frequently caused by K. pneumoniae, both in inpatient and outpatient settings. Bacteria known as Extended Spectrum Beta-Lactamase (ESBL)-producing Klebsiella pneumoniae have evolved resistance to a variety of beta-lactam antibiotics, such as Cephalosporins and Penicillins. Urinary tract infections are among the many infections that these organisms can cause. Concerns regarding the spread of K. pneumoniae infections that produce ESBLs have escalated due to the substantial health risks associated with these infections. Hospitals and the general public are the primary sources of ESBL-producing organisms (14, 15, 16).
Examining the frequency of Klebsiella pneumoniae encoding genes for SHV, TEM, and CTX-M extended-spectrum beta-lactamases (ESBL) enzymes extracted from urine samples of out-of-hospital patients with suspected UTIs was the study's main objective. The current study found that 19.51% (8/41) of K. pneumoniae cases produced ESBL. The results of the present study are in line with a report on comparable research conducted in Enugu, Nigeria (17), where the prevalence of ESBL Klebsiella was found to be 10.99%. However, the prevalence was lower than that of a similar study conducted in Bahir Dar City, Northwest Ethiopia (18), where the prevalence was found to be 10.26.3%. Additionally, the prevalence of ESBL Klebsiella in our data was marginally higher than that of previous research on community-onset UTIs in China, where the rate was 6.3% (19). The reported findings of various scientific investigations on the production of ESBL in Nigeria range widely, from 7.5% to 82.3% (20).
Antibiotics have been reported to be successful in treating varieties of infections for many years, but a report revealed that the development of antimicrobial resistance in hospitals and the community is undermining the progress made possible by antibiotics. The African Center for Disease Control estimates that drug resistance results in 700,000 deaths annually (21); if this trend continues, it is predicted that antimicrobial resistance could cause over 100 million deaths by the year 2050. The overuse of antibiotics due to environmental factors and societal norms has led to an increase in antibiotic resistance (21).
Antimicrobial resistance has been accepted as a major public health issue in patient care and it is more predominant in developing countries like Nigeria. In this study, resistance to Erythromycin and Ceftazidime were the highest with 100% of the isolates being resistant to the antibiotics, followed by Ciprofloxacin with 97.56%, Colistin with 82.93%, Ceftriaxone with 80.49% and Amoxicillin-Clavulanate with 73.17%. The least resistance was to Gentamicin and Imipenem, with 24 (58.54%) of the isolates being resistant to each. Over the past 30 years, there have been reports of an increase in the resistance of community-acquired organisms to several important antibiotics in various countries, including Nigeria. This could be attributed to over-prescription of antibiotics, the use of inferior or fake medications, use of over-the-counter medications without stringent regulations, and patient dosage inadequacies. Therefore, the amount of MDR strains in the community has been greatly influenced by these factors. These organisms are still sources of antibiotic-resistant genes, and the day is not far off when these newly discovered pathogen-causing organisms pose a threat to medicine (22).
The prevalence rates of the genes included in this study were as follows: β-lactamase SHV (75.61%), β-lactamase CTX-M (24.39%), and β-lactamase TEM (9.76%). Dehshiri et al. (2018) found that the prevalence of the SHV, TEM, and CTX-M genes was 85.5%, 16.1%, and 27.4%, respectively. Meanwhile, Nasehi et al. (23), reported gene isolation rates of 18% and 26% for β-lactamase TEM and β-lactamase SHV. Moreover, a comparable study conducted in Enugu, Nigeria, revealed that the frequencies of the SHV, TEM, and CTX-M genes were higher—90%, 100%, and 40%, respectively (17). Consistent with the current investigation, the majority of studies found that K. pneumoniae isolates containing β-lactamase were derived from SHV group derivatives.
Because the SHV genes are implicated in the resistance of expanded-spectrum cephalosporins (typically third generation cephalosporins), β-lactamase inhibitors/β-lactam combinations, and monobactams, their prevalence in our study and other reports is of great clinical importance.
9.67% of the isolates had all SHV, TEM and CTX- M genes while 8 (19.51%) of the isolates had both the CTX-M and SHV genes. The treatment of UTI becomes more challenging because the isolates will probably be resistant to more antibiotics and the majority of third-generation cephalosporins due to the presence of multiple ESBL resistance genes, which could result in retained resistance to beta-lactamases (24).
Our study demonstrated a lack of correlation between the phenotypic test for ESBL production and the molecular detection of ESBL genes, which underscores the necessity of integrating an enhanced ESBL detection technique into standard susceptibility protocols. The disparity between the phenotypic and genotypic approaches' ability to identify ESBL-positive isolates may be due to the lower sensitivity of the phenotypic methods as well as the impact of environmental factors on the incidence of resistance. It might also be influenced by the phenotypic techniques used to identify ESBL in bacterial isolates. Certain ESBLs might not accumulate to a level where disk diffusion tests can identify them, which would mean that the infected patients would not respond to treatment (25, 26, 27). There is, thus, a need that the performance of these phenotypic tests to be assessed regularly because the introduction and prevalence of new enzymes could alter it. A wrong diagnosis of antibiotic resistance can result in the wrong antibiotic being prescribed, which can then favor the emergence of new resistance genes. Phenotypic tests for ESBL detection are limited to determining whether an ESBL is produced; they are unable to identify the ESBL genes whose expression is concealed or disguised. Thus, it is recommended that the genotypic approach be used as the primary technique for identifying ESBL-producing strains of Klebsiella and other Enterobacteriaceae.
The statistics show that β-lactamase genes SHV, CTXM and TEM are increasing and worrying. The general population can make a significant contribution by acting to stop infections, reduce the need for antibiotics, and only take them as directed by a licensed healthcare provider. According to reports, one significant risk factor for antibiotic resistance is the public's misuse of antibiotics. Nigeria is a developing nation, thus the misuse of antibiotics tends to rise as a result of a lack of knowledge and communication about the use of antibiotic medications, particularly in rural areas. (28) The majority of people either self-medicate or buy antibiotics from an unapproved website or source. Additionally, some people stop taking their antibiotics before finishing the prescribed prescription. Certain Nigerian communities demonstrated a lack of knowledge and a disapproving attitude toward the usage of antibiotics (29).
5. Conclusion
This investigation demonstrates the high prevalence, wide range of patterns, and coexistence of ESBL genes in K. pneumoniae isolates from outpatient UTI patients. The ESBL testing methodology is critical to the identification of ESBL production. Even though there are many phenotypic tests available for ESBL detection, testing directly for the presence of ESBL genes yields more accurate results. Molecular detection techniques are the only ones that allow for the definitive identification of ESBL genes. The majority of K. pneumoniae isolates that produced ESBL had the SHV gene identified in them.
Funding
This research received no external funding and was self-sponsored by all the authors.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ashurst, J.V. and Dawson, A. (2023). Klebsiella pneumoniae. In: Stat Pearls. Treasure Island (FL): Stat Pearls Publishing. Available from: https://www.ncbi.nlm.nih.gov/books/NBK519004.
- Brolund A. (2014). Overview of ESBL-producing Enterobacteriaceae from a Nordic perspectives. Infection Ecology and Epidemiology, 4:10.3402/iee.v4.24555. [CrossRef]
- Husna, A., Rahman, M.M., Badruzzaman, A.T.M., Sikder, M.H., Islam, M.R., Rahman, M.T., Alam, J. and Ashour, H.M. (2023). Extended-spectrum β-lactamases (ESBL): Challenges and opportunities. Biomedicines, 11(11):2937. [CrossRef]
- Tufa, T.B., Fuchs, A., Tufa, T.B., Stötter, L., Kaasch, A.J., Feldt, T., Häussinger, D. and Mackenzie, C.R. (2020). High rate of extended-spectrum beta-lactamase-producing gram-negative infections and associated mortality in Ethiopia: A systematic review and meta-analysis. Antimicrobial Resistance Infectious Control, 9:128. [CrossRef]
- Li, Y., Kumar, S., Zhang, L., Wu, H., and Wu, H. (2023). Characteristics of antibiotic resistance mechanisms and genes of Klebsiella pneumoniae. Open medicine (Warsaw, Poland), 18(1):0707. [CrossRef]
- Muteeb, G., Rehman, M.T., Shahwan, M. and Aatif, M. (2023). Origin of antibiotics and antibiotic resistance, and their impacts on drug development: A narrative review. Pharmaceuticals, 16:1615. [CrossRef]
- Serwecińska, L. (2020). Antimicrobials and antibiotic-resistant bacteria: A risk to the environment and public health. Water, 12:333–341. [CrossRef]
- Antimicrobial Resistance Collaborators. (2022). Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. The Lancet, 399(10325):629–655. [CrossRef]
- Enyi, E.O. and Ekpunobi, N.F. (2022). Secondary metabolites from endophytic fungi of Moringa oleifera: antimicrobial and antioxidant properties. Journal of Microbiology Exp. 10(5):150‒154. [CrossRef]
- Clinical and Laboratory Standards Institute (2013). Performance standards for antimicrobial disk susceptibility tests, 23rd informational supplement. CLSI document M100-S23. Wayne, PA. Clinical and Laboratory Standards Institute, 34(1):1–230.
- Schlesinger, J., Navon-Venezia, S., Chmelnitsky, I., Hammer-Münz, O., Leavitt, A., Gold, H. S., Schwaber, M.J., and Carmeli, Y. (2005). Extended-spectrum beta-lactamases among Enterobacter isolates obtained in Tel Aviv, Israel. Antimicrobial Agents and Chemotherapy, 49(3):1150–1156. [CrossRef]
- Bali, E., Açık, L. and Sultan, N. (2010). Phenotypic and molecular characterization of SHV, TEM, CTX-M and extended-spectrum β-lactamase produced by Escherichia coli, Acinobacter baumannii and Klebsiella isolates in a Turkish hospital. African Journal of Microbiology Research, 4:650–654.
- Afzali, H., Firoozeh, F., Amiri, A., Moniri, R., and Zibaei, M. (2015). Characterization of CTX-M-Type extend-spectrum β-lactamase producing Klebsiella sp. in Kashan, Iran. Jundishapur Journal of Microbiology, 8(10):e27967. [CrossRef]
- Okoye, E.L., Kemakolam, C., Ugwuoji, E.T. and Ogbonna, I. (2022). Multidrug resistance tracing by plasmid profile analysis and the curing of bacteria from different clinical specimens. Adv. Gut Microbiology Research, 3170342. [CrossRef]
- Ekpunobi, N., Akinsuyi, O., Ariri, T. and Ogunmola, T. (2023). The re-emergence of Monkeypox in Nigeria. Challenges, 14:22–29. [CrossRef]
- Ekpunobi, N.F., Markjonathan, I., Olanrewaju, O. and Olanihun, D. (2020). Idiosyncrasies of COVID-19: A Review. Iranian Journal of Medical Microbiology, 14(3):290–296. [CrossRef]
- Chinedu, A.C., Tochukwu, E.J., Anibueze, E.G. and Nympha, E.O. (2022). Prevalence of ESBL genes in Klebsiella pneumoniae from individuals with community-acquired urinary tract infection in rural communities of Enugu State, Nigeria. GSC Biological and Pharmaceutical Sciences, 19(3):294–303. [CrossRef]
- Ameshe, A., Engda, T., and Gizachew, M. (2022). Antimicrobial resistance patterns, extended-spectrum beta-lactamase production, and associated risk factors of Klebsiella species among UTI-suspected patients at Bahir Dar City, Northwest Ethiopia. International Journal of Microbiology, 826545. [CrossRef]
- Quan, J., Dai, H., Liao, W., Zhao, D., Shi, Q., Zhang, L., Shi, K., Akova, M. and Yu, Y. (2021). Etiology and prevalence of ESBLs in adult community-onset urinary tract infections in East China: A prospective multicenter study. Journal of Infection, 83(2):175-181. [CrossRef]
- Tanko, N., Bolaji, R.O., Olayinka, A.T. and Olayinka, B.O. (2020). A systematic review on the prevalence of extended-spectrum beta lactamase-producing Gram-negative bacteria in Nigeria. Journal of Global Antimicrobial Resistance, 22:488–496. [CrossRef]
- Chukwu, E.E., Oladele, D.A., Awoderu, O.B., Afocha, E.E., Lawal, R.G., Abdus-Salam, I., Ogunsola, F.T., and Audu, R.A. (2020). A national survey of public awareness of antimicrobial resistance in Nigeria. Antimicrobial resistance and infection control, 9(1):72. [CrossRef]
- Ekpunobi, N. F. and Adeleye, I. A. (2020). Phenotypic characterization of biofilm formation and efflux pump activity in multidrug-resistant Staphylococcus species isolated from asymptomatic students. Journal of Microbiology and Experimentation, 8(6):223–229. [CrossRef]
- Nasehi, L., Shahcheraghi, F., Nikbin, V., Shoeib, N. (2010). PER, CTX-M, TEM and SHV Beta-lactamases in clinical isolates of Klebsiella pneumoniae isolated from Tehran, Iran. Iranian Journal of Basic Medical Sciences, 13(3):111–118.
- Gundran, R.S., Cardenio, P.A., Villanueva, M.A., Sison, F.B., Benigno, C.C., Kreausukon, K., Pichpol, D., and Punyapornwithaya, V. (2019). Prevalence and distribution of blaCTX-M, blaSHV, blaTEM genes in extended- spectrum β- lactamase- producing Escherichia coli isolates from broiler farms in the Philippines. BMC veterinary research, 15(1):227. [CrossRef]
- Sharma, J., Sharma, M. and Ray, P. (2010). Detection of TEM and SHV genes in Escherichia coli and Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian Journal of Medical Research, 132:332–336.
- Bajpai, T., Pandey, M., Varma, M. and Bhatambare, G.S. (2017). Prevalence of TEM, SHV, and CTX-M Beta-Lactamase genes in the urinary isolates of a tertiary care hospital. Avicenna journal of medicine, 7(1):12–16. [CrossRef]
- Tamma, P.D. and Humphries, R.M. (2021). Testing for ESBL production is necessary for ceftriaxone-non-susceptible Enterobacterailes: perfect should not be the enemy of progress. JAC Antimicrobial Resistance, 3(2):dlab019.
- Ghebremedhin, B., Olugbosi, M. O., Raji, A., Layer, F., Bakare, R., König, B., and König, W. (2009). Emergence of a community-associated methicillin-resistant Staphylococcus aureus strain with a unique resistance profile in Southwest Nigeria. Journal of Clinical Microbiology, 47(9):2975–2980. [CrossRef]
- Pitout, J., Nordmann, P., Laupland, K. B., and Poirel, L. (2005). Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. Journal of Antimicrobial Chemotherapy, 56(1):52–59. [CrossRef]
- Egwuatu, T. O., Ishola, O. D. and Oladele, O. E. (2021).the distribution of extended-spectrum beta-lactamase genes in fomites, healthcare workers, and patients from two hospitals in Lagos state, Nigeria. Ife Journal of Science 23(2):015-024. [CrossRef]
Table 1.
List of Primers used in the study.
Table 1.
List of Primers used in the study.
Table 2.
Antimicrobial sensitivity patterns of Klebsiella isolates in urine samples of UTI patients.
Table 2.
Antimicrobial sensitivity patterns of Klebsiella isolates in urine samples of UTI patients.
| ANTIBIOTICS |
Susceptible |
Intermediate |
Resistant |
| Ciprofloxacin |
0 |
1 (2.44%) |
40 (97.56%) |
| Tetracycline |
12 (29.00%) |
1 (2.44%) |
28 (68.28%) |
| Imipenem |
5 (12.20%) |
12 (29.27%) |
24 (58.54%) |
| Amoxicillin/Clavulanic acid |
4 (9.76%) |
7 (17.07%) |
30 (73.17%) |
| Erythromycin |
0 (0.00%) |
0 (0.00%) |
41 (100%) |
| Gentamicin |
6 (14.60%) |
11 (26.80%) |
24 (58.54%) |
| Colistin |
7 (17.07%) |
0 (0.00%) |
34 (82.93%) |
| Ceftriaxone |
8 (19.51%) |
0(0.00%) |
33 (80.49%) |
| Ceftazidime |
0 (0.00%) |
0 (0.00%) |
41 (100%) |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).