Submitted:
26 March 2024
Posted:
27 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. The Diversity of Higher Plant Genomes and Transcriptomes as a Basis for the Complexity of Transcriptomic Responsiveness
3. Tissue-Specific and Organo-Specific Alterations in Plant Transcriptomes
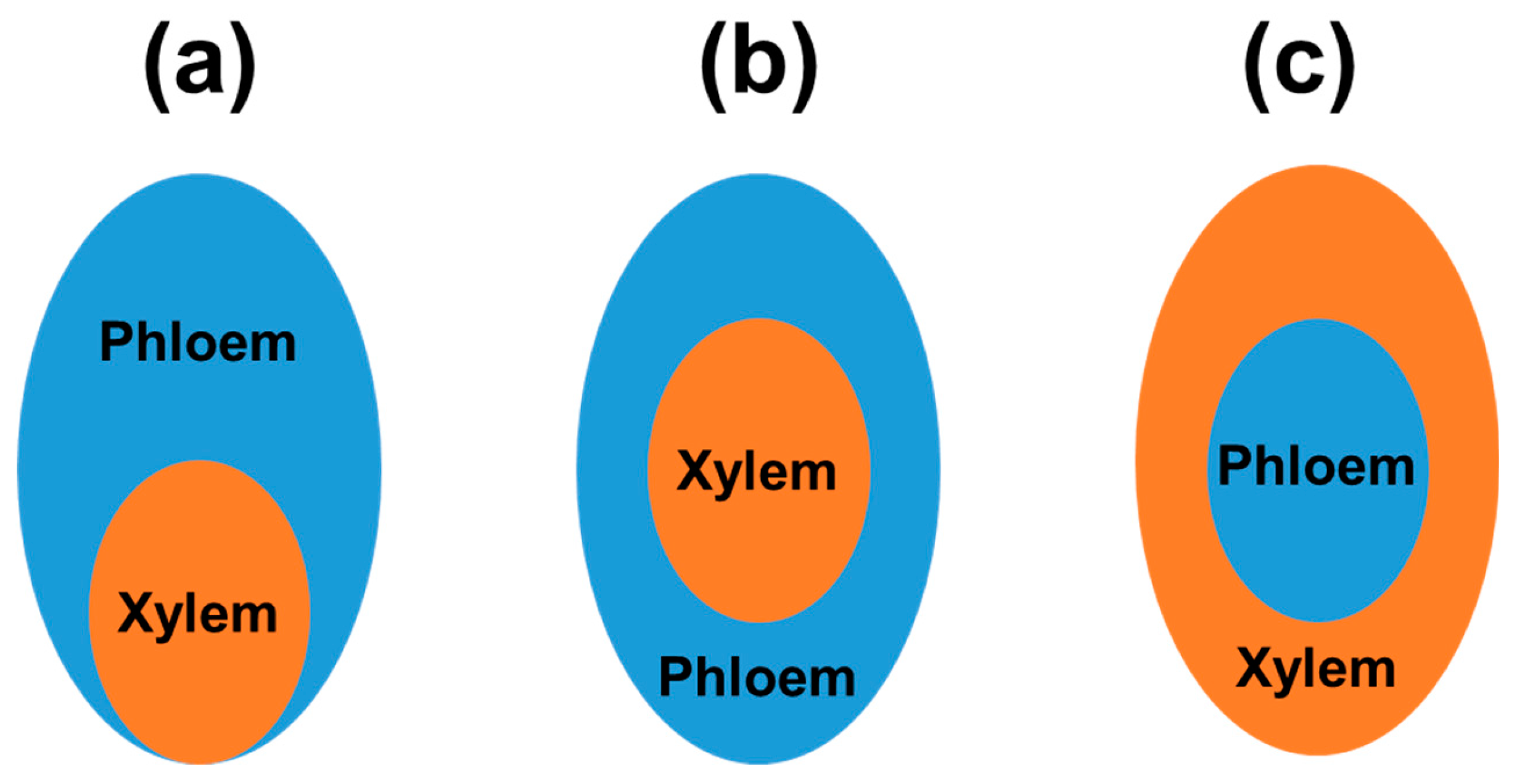
3.1. Xylem and Phloem Transcriptomes as Examples of Tissue-Specific Transcriptomes
3.2. Selected Organo-Specific Transcriptomes
3.2.1. Root and Leaf Transcriptomes
3.2.2. Transcriptomes of Generative Organs
3.2.3. Seed Transcriptomes
3.3. Transcriptomic Analyses of Plant Species with Medicinal Applications
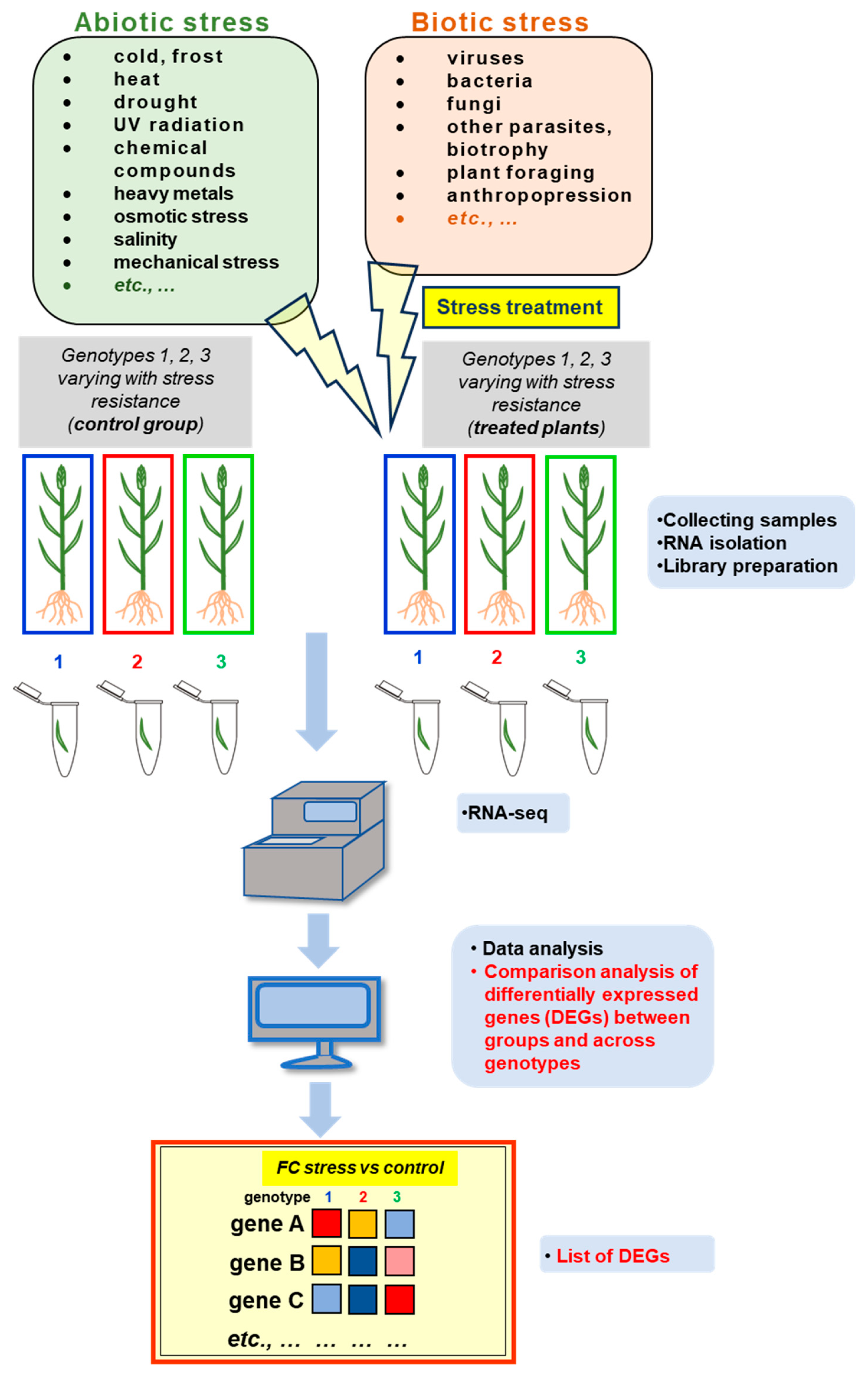
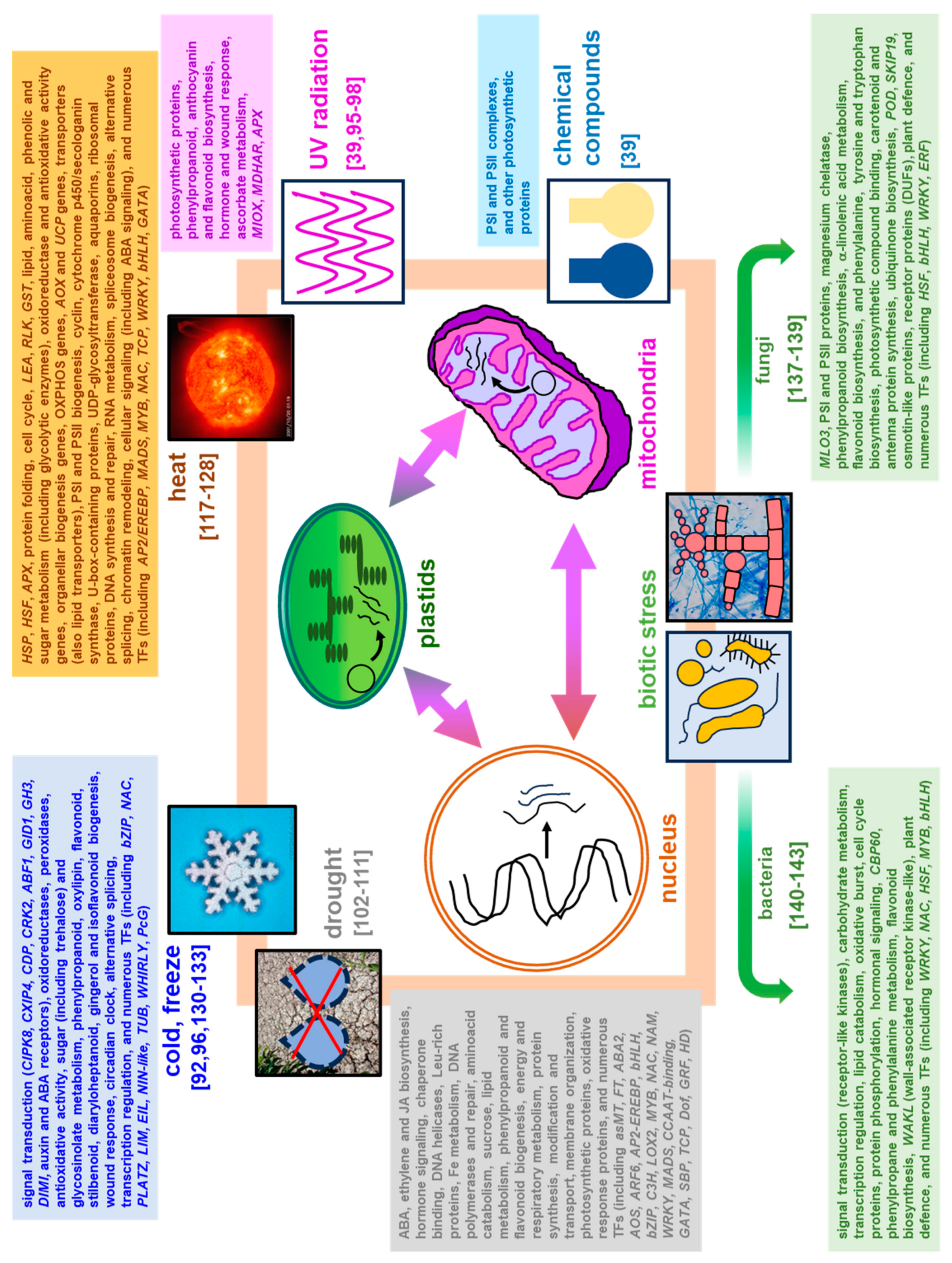
4. Plant Transcriptomic Response under Stress Acclimation
4.1. Alterations in Plant Transcriptomes during Abiotic Stress – the Selected Examples
4.1.1. Chemical Treatment and UV Radiation
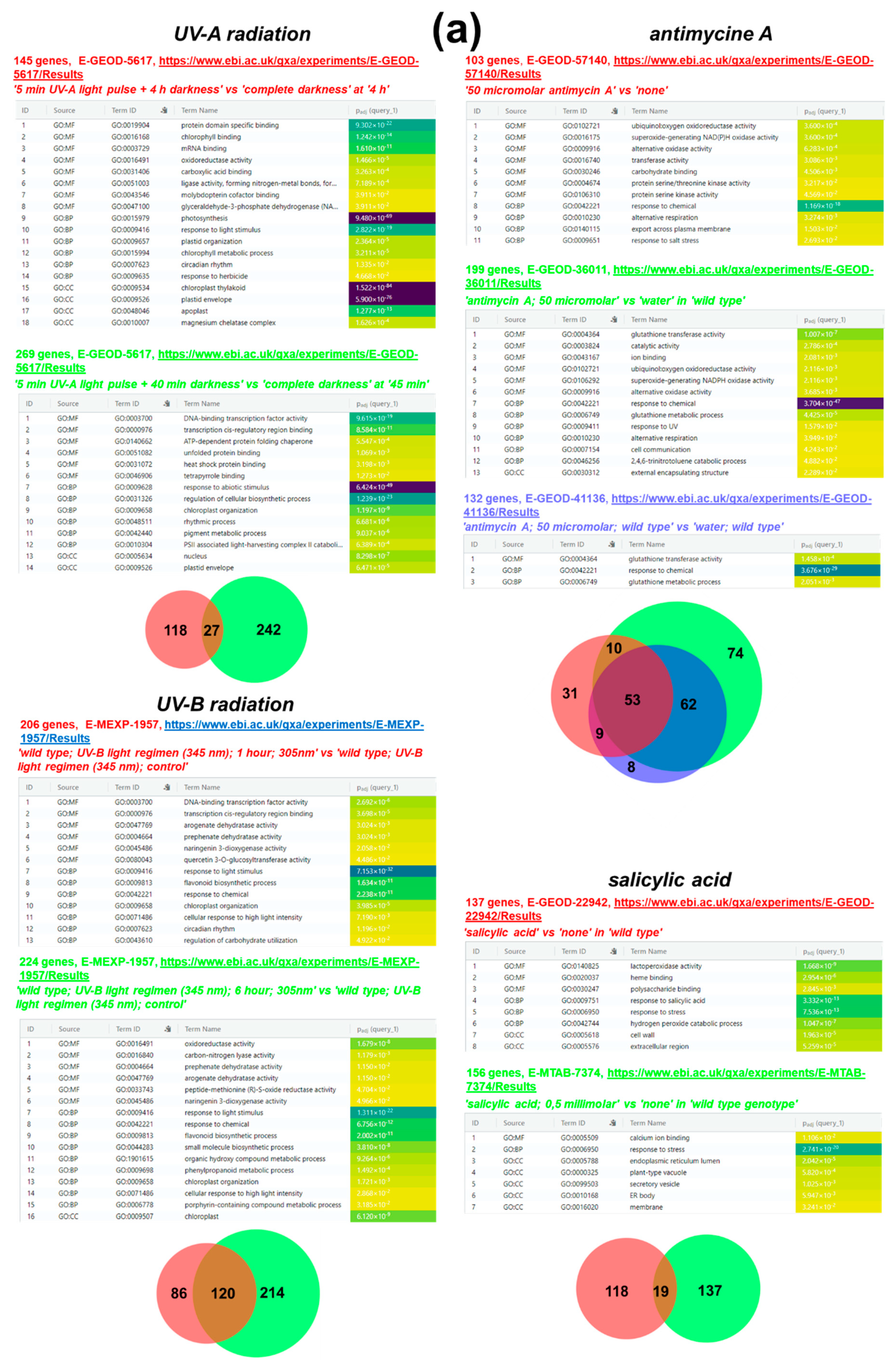

4.1.2. Water Deficiency (drought)
4.1.3. Elevated Temperature (Heat Stress)
4.1.4. Low Temperature
4.2. Plant Transcriptomic Response under Biotic Stress
4.2.1. Fungal Infections
4.2.2. Bacterial Infections
5. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAO | aldehyde oxidase |
| ABA | abscisic acid |
| ABF | ABSCISIC ACID RESPONSIVE ELEMENT-BINDING FACTOR |
| ACADM | medium-chain acyl-CoA dehydrogenase |
| ACO | 1-aminocyclopropane-1-carboxylic oxidase |
| ACOX | acyl-CoA oxidase |
| ACS | 1-aminocyclopropane-1-carboxylic synthase |
| ALA | 5-aminolevulinic acid |
| AOS | allene oxide synthase |
| AOX | alternative oxidase |
| AP | APETALA |
| APX | ascorbate peroxidase |
| ARF | auxin response factor |
| AS | anthocyanidin synthase |
| asMT | acetylserotonin O-methyltransferase |
| ASC | L-ascorbic acid |
| ATHB | small homeodomain-leucine zipper family |
| AUX | auxin receptor |
| bHLH | basic helix–loop–helix |
| BLS | bacterial leaf streak |
| bZIP | basic (region) leucine zipper |
| CAM | crassulacean acid metabolism |
| CAT | catalase |
| CBP | CALMODULIN-BINDING PROTEIN |
| CCOAOMT | caffeoyl-CoA O-methyltransferase |
| CDP | CAAT displacement protein (transcriptional repressor) |
| CCR | cinnamoyl-CoA:NADP reductase |
| C3H | zinc finger transcription factor |
| CHI | chalcone isomerase |
| CHS | chalcone synthase |
| C4H | 4-cinnamate hydroxylase |
| CIPK | CBL-interacting protein kinase |
| 4CL | 4-coumarate: CoA ligase |
| Clp | caseinolytic protease |
| CO | CONSTANS |
| CRK | cysteine-rich receptor-like kinase |
| CRY | cryptochrome |
| CXIP | CAX-interacting protein |
| DEGs | differentially-expressed genes |
| DFR | dihydroflavonol 4-reductase |
| DIMI | DIMINUTO |
| Dof | DNA-binding with one finger |
| DREB | dehydration-responsive element-binding |
| ECHS | short chain enoyl-CoA hydratase |
| EF | early flowering |
| EIL | ethylene insensitive-like |
| ER | endoplasmic reticulum |
| EREBP | ethylene-responsive element binding protein |
| ERF | ethylene response factor |
| ESTs | expressed sequence tags |
| ETI | effector-triggered immunity |
| F3H | flavanone-3-hydroxylase |
| FIE | fertilization-independent endospermia |
| FSL | flavonol synthase |
| FT | FLOWERING LOCUS T |
| FUL | FRUITFUL |
| FUS | fused in sarcoma |
| GABA | g- aminobutyric acid |
| GID | GA-INSENSITIVE DWARF |
| GRF | Growth Regulating Factor |
| HCT | hydroxycinnamoyl transferase |
| HD | homeodomain |
| HLH | helix-loop-helix |
| HR | hypersensitive response |
| HSF | heat shock factor |
| HSP | heat shock protein |
| IFL | INTERFASCICULAR FIBERLESS |
| JA | jasmonic acid |
| KAN | KANADI |
| LEA | late-embryogenesis abundant |
| LHC | light-harvesting complex |
| LEC | little elongation complex |
| LIM | homeobox transcription factor |
| LOX | lipoxygenase |
| MADS | MINICHROMOSOME MAINTENANCE 1/ AGAMOUS/ DEFICIENS/ SERUM RESPONSE FACTOR |
| MAPK | mitogen-assiociated protein kinase |
| MDHAR | monodehydroascorbate reductase |
| MEDEA | motif enrichment in differential elements of accessibility |
| MFP | multifunctional protein |
| MIOX | myo-inositol oxygenase |
| MLO | mildew locus o |
| MYB | myeloblastosis |
| NAC | no apical meristem/ ATAF1/ cup-shaped cotyledon |
| NAM | no apical meristem |
| NCED | 9-cis-epoxycarotenoid dioxygenase |
| NIN | nodule inception |
| OPR | 12-oxophytodienoic acid reductase |
| OSM | osmotin-like protein |
| OXPHOS | oxidative phosphorylation |
| PAAF | 23-dehydroadipyl-CoA hydratase |
| PAL | phenylalanine ammonia lyase |
| PAMP | pathogen-associated molecular pattern |
| PCD | programmed cell death |
| PcG | Polycomb group |
| PEAR | PHLOEM EARLY DNA-BINDING-WITH-ONE-FINGER |
| PEI | Cys3His zinc finger domain-containing protein |
| PHB | PHABULOSA |
| PHV | PHAVOLUTA |
| PHY | phytochrome |
| POD | peroxidase |
| PS | photosystem |
| PTI | PAMP-triggered immunity |
| PLATZ | plant AT-rich sequence and zinc-binding protein |
| REV | REVOLUTA |
| RGA | resistance gene analogue |
| RLK | receptor-like kinase |
| ROS | reactive oxidative species |
| SA | salicylic acid |
| SBP | SQUAMOSA-pROMOTER BINDING PROTEIN |
| SOC | suppressor of overexpression of constans |
| SOD | superoxide dismutase |
| STS | stachyose synthase |
| TCP | bHLH DNA-binding domain |
| TF | transcription factor |
| TUB | transcription factor family |
| UF3GT | UDP-glucose: flavonoid 3-O-glycosyltransferase |
| WAKL20 | wall-associated receptor kinase-like 2 |
| WGD | whole-genome duplication |
| WRKY | transcription factor family |
| VIN | VERNALIZATION INSENSITIVE |
| ZEP | zeaxanthin epoxidase |
| ZIP | leucine zipper |
References
- Kenrick, P.; Crane, P.R. The Origin and Early Diversification of Land Plants: A Cladistic Study; Smithsonian series in comparative evolutionary biology, 1st ed.; Smithsonian Institution Press: Washington, DC, USA, 1997. [Google Scholar]
- Forster, B.P.; Heberle-Bors, E.; Kasha, K.J.; Touraev, A. The Resurgence of Haploids in Higher Plants. Trend Plant Sci. 2007, 12, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.L.; Puttick, M.N.; Clark, J.W.; Edwards, D.; Kenrick, P.; Pressel, S.; Wellman, C.H.; Yang, Z.; Schneider, H.; Donoghue, P.C.J. The Timescale of Early Land Plant Evolution. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, E2274–E2283. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.; Prahalad, S.; Colbert, R. Integrative Genomics. Textbook of Pediatric Rheumatology, 7th ed.; Elsevier: Philadelphia, PA, USA, 2016; pp. 43–53.e3. [Google Scholar]
- Schliesky, S.; Gowik, U.; Weber, A.P.M.; Bräutigam, A. RNA-Seq Assembly – Are We There Yet? Front. Plant Sci. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, P.; Singh, D.; Mathur, S.; Singh, A.; Ranjan, R. Upcoming Progress of Transcriptomics Studies on Plants: An Overview. Front. Plant Sci. 2022, 13, 1030890. [Google Scholar] [CrossRef]
- Chen, C.; Ge, Y.; Lu, L. Opportunities and Challenges in the Application of Single-Cell and Spatial Transcriptomics in Plants. Front. Plant Sci. 2023, 14, 1185377. [Google Scholar] [CrossRef]
- Singh, S.; Parihar, P.; Singh, R.; Singh, V.P.; Prasad, S.M. Heavy Metal Tolerance in Plants: Role of Transcriptomics, Proteomics, Metabolomics, and Ionomics. Front. Plant Sci. 2016, 6, 1143. [Google Scholar] [CrossRef]
- Ahmad, M. Genomics and Transcriptomics to Protect Rice (Oryza sativa. L.) from Abiotic Stressors: -Pathways to Achieving Zero Hunger. Front. Plant Sci. 2022, 13, 1002596. [Google Scholar] [CrossRef]
- Bhat, K.A.; Mahajan, R.; Pakhtoon, M.M.; Urwat, U.; Bashir, Z.; Shah, A.A.; Agrawal, A.; Bhat, B.; Sofi, P.A.; Masi, A.; et al. Low Temperature Stress Tolerance: An Insight Into the Omics Approaches for Legume Crops. Front. Plant Sci. 2022, 13, 888710. [Google Scholar] [CrossRef] [PubMed]
- Kourani, M.; Mohareb, F.; Rezwan, F.I.; Anastasiadi, M.; Hammond, J.P. Genetic and Physiological Responses to Heat Stress in Brassica napus. Front. Plant Sci. 2022, 13, 832147. [Google Scholar] [CrossRef]
- Son, S.; Park, S.R. Plant Translational Reprogramming for Stress Resilience. Front. Plant Sci. 2023, 14, 1151587. [Google Scholar] [CrossRef]
- Tu, M.; Du, C.; Yu, B.; Wang, G.; Deng, Y.; Wang, Y.; Chen, M.; Chang, J.; Yang, G.; He, G.; et al. Current Advances in the Molecular Regulation of Abiotic Stress Tolerance in Sorghum via Transcriptomic, Proteomic, and Metabolomic Approaches. Front. Plant Sci. 2023, 14, 1147328. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Wang, Z.; Li, X.; Ai, Q.; Wong, D.C.J.; Zhang, F.; Yang, J.; Zhang, N.; Si, H. Current Perspectives of lncRNAs in Abiotic and Biotic Stress Tolerance in Plants. Front. Plant Sci. 2024, 14, 1334620. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, C.N.; Robin Buell, C. Tapping the Promise of Genomics in Species with Complex, Nonmodel Genomes. Annu. Rev. Plant Biol. 2013, 64, 89–110. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Soltis, D.E.; Kersey, P.J.; Wegrzyn, J.L.; Leebens-Mack, J.H.; Gostel, M.R.; Liu, X.; Soltis, P.S. Green Plant Genomes: What We Know in an Era of Rapidly Expanding Opportunities. Proc. Natl. Acad. Sci. U.S.A. 2022, 119, e2115640118. [Google Scholar] [CrossRef] [PubMed]
- Pellicer, J.; Hidalgo, O.; Dodsworth, S.; Leitch, I. Genome Size Diversity and Its Impact on the Evolution of Land Plants. Genes 2018, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Margulis, L. Origin of Eukaryotic Cells: Evidence and Research Implications for a Theory of the Origin and Evolution of Microbial, Plant, and Animal Cells on the Precambrian Earth, 1st ed.; Yale Univ. Press: New Haven, U.S.A, 1970. [Google Scholar]
- McFadden, G.I. Primary and Secondary Endosymbiosis and the Origin of Plastids. J. Phycol. 2001, 37, 951–959. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Lagarias, J.C.; Bhattacharya, D. Primary Endosymbiosis and the Evolution of Light and Oxygen Sensing in Photosynthetic Eukaryotes. Front. Ecol. Evol. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Best, C.; Mizrahi, R.; Ostersetzer-Biran, O. Why so Complex? The Intricacy of Genome Structure and Gene Expression, Associated with Angiosperm Mitochondria, May Relate to the Regulation of Embryo Quiescence or Dormancy—Intrinsic Blocks to Early Plant Life. Plants 2020, 9, 598. [Google Scholar] [CrossRef]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The Chloroplast Genome: A Review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Provan, J.; Powell, W.; Hollingsworth, P.M. Chloroplast Microsatellites: New Tools for Studies in Plant Ecology and Evolution. Trends Ecol. Evol. 2001, 16, 142–147. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, A.L.; Aaqil Khan, M.; Muhammad Imran, Q.; Kang, S.-M.; Al-Hosni, K.; Jeong, E.J.; Lee, K.E.; Lee, I.-J. Comparative Analysis of Complete Plastid Genomes from Wild Soybean (Glycine Soja) and Nine Other Glycine Species. PLoS ONE 2017, 12, e0182281. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast Genomes: Diversity, Evolution, and Applications in Genetic Engineering. Genome Biol 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Rurek, M. Participation of Non-Coding RNAs in Plant Organelle Biogenesis. Acta Biochim. Pol. 2016, 63, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Goldschmidt-Clermont, M. Participation of Nuclear Genes in Chloroplast Gene Expression. Biochimie 2000, 82, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Watson, S.J.; Sowden, R.G.; Jarvis, P. Abiotic Stress-Induced Chloroplast Proteome Remodelling: A Mechanistic Overview. J. Exp. Bot. 2018, 69, 2773–2781. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.-S.; Cho, J.-H.; Im, J.-S.; Choi, J.-G.; Park, Y.-E.; Hong, S.-Y.; Kwon, M.; Kang, J.-H.; Park, T.-H. The Complete Mitochondrial Genome Sequences of Potato (Solanum tuberosum L., Solanaceae). Mitochondrial DNA Part B 2017, 2, 781–782. [Google Scholar] [CrossRef] [PubMed]
- Logan, D.C. Plant Mitochondrial Dynamics. Biochim. Biophys. Acta - Molecular Cell Research 2006, 1763, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. Rapid Evolution of Enormous, Multichromosomal Genomes in Flowering Plant Mitochondria with Exceptionally High Mutation Rates. PLoS Biol 2012, 10, e1001241. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Keller, S.R.; Berardi, A.E.; Sanderson, B.J.; Karpovich, J.F.; Taylor, D.R. De novo Transcriptome Assembly and Polymorphism Detection in the Flowering Plant Silene vulgaris (Caryophyllaceae). Mol. Ecol. Res. 2012, 12, 333–343. [Google Scholar] [CrossRef]
- Imadi, S.R.; Kazi, A.G.; Ahanger, M.A.; Gucel, S.; Ahmad, P. Plant Transcriptomics and Responses to Environmental Stress: An Overview. J. Genet. 2015, 94, 525–537. [Google Scholar] [CrossRef]
- Schneider, G.F.; Dekker, C. DNA Sequencing with Nanopores. Nat. Biotechnol. 2012, 30, 326–328. [Google Scholar] [CrossRef] [PubMed]
- Best, C.; Sultan, L.D.; Murik, O.; Ostersetzer, O. Insights into the Mitochondrial Transcriptome Landscapes of Two Brassicales Plant Species, Arabidopsis thaliana (var. Col-0) and Brassica oleracea (var. botrytis). Endocyt. Cell Res. 2020, 30, 16–38. [Google Scholar] [CrossRef]
- Cahoon, A.; Nauss, J.; Stanley, C.; Qureshi, A. Deep Transcriptome Sequencing of Two Green Algae, Chara vulgaris and Chlamydomonas reinhardtii, Provides No Evidence of Organellar RNA Editing. Genes 2017, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H. The Review of Transcriptome Sequencing: Principles, History and Advances. IOP Conf. Ser.: Earth Environ. Sci. 2019, 332, 042003. [Google Scholar] [CrossRef]
- Booth, M.W.; Breed, M.F.; Kendrick, G.A.; Bayer, P.E.; Severn-Ellis, A.A.; Sinclair, E.A. Tissue-Specific Transcriptome Profiles Identify Functional Differences Key to Understanding Whole Plant Response to Life in Variable Salinity. Biology Open 2022, 11, bio059147. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, O.; Xu, Y.; Liew, L.C.; Wang, Y.; Zhu, Y.; Hurgobin, B.; Lewsey, M.G.; Whelan, J. RNA-seq Analysis of Laser Microdissected Arabidopsis Thaliana Leaf Epidermis, Mesophyll and Vasculature Defines Tissue-specific Transcriptional Responses to Multiple Stress Treatments. Plant J. 2021, 107, 938–955. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Z.; Dixon, R.A. On–Off Switches for Secondary Cell Wall Biosynthesis. Mol. Plant 2012, 5, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Růžička, K.; Ursache, R.; Hejátko, J.; Helariutta, Y. Xylem Development – from the Cradle to the Grave. New Phytol. 2015, 207, 519–535. [Google Scholar] [CrossRef] [PubMed]
- Dinneny, J.R.; Yanofsky, M.F. Vascular Patterning: Xylem or Phloem? Curr. Biol. 2004, 14, R112–R114. [Google Scholar] [CrossRef]
- Eshed, Y.; Baum, S.F.; Perea, J.V.; Bowman, J.L. Establishment of Polarity in Lateral Organs of Plants. Curr. Biol. 2001, 11, 1251–1260. [Google Scholar] [CrossRef]
- Eshed, Y.; Izhaki, A.; Baum, S.F.; Floyd, S.K.; Bowman, J.L. Asymmetric Leaf Development and Blade Expansion in Arabidopsis Are Mediated by KANADI and YABBY Activities. Development 2004, 131, 2997–3006. [Google Scholar] [CrossRef] [PubMed]
- Roszak, P.; Heo, J.; Blob, B.; Toyokura, K.; Sugiyama, Y.; De Luis Balaguer, M.A.; Lau, W.W.Y.; Hamey, F.; Cirrone, J.; Madej, E.; et al. Cell-by-Cell Dissection of Phloem Development Links a Maturation Gradient to Cell Specialization. Science 2021, 374, eaba5531. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, F.; Tang, Y.; Yuan, Y.; Guo, Q. Transcriptome Sequencing and Profiling of Expressed Genes in Phloem and Xylem of Ramie (Boehmeria nivea L. Gaud). PLoS ONE 2014, 9, e110623. [Google Scholar] [CrossRef] [PubMed]
- Kalckar, H.M. Galactose Metabolism and Cell “Sociology”: Galactose, One of the Freaks of Evolution, Furnishes a Simple Illustration of the Extravagances of Nature. Science 1965, 150, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Weber, H. Fatty Acid-Derived Signals in Plants. Trend Plant Sci. 2002, 7, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Lashbrook, C.C.; Cho, S.K.; Butler, N.M.; Sharma, P.; Muppirala, U.; Severin, A.J.; Hannapel, D.J. Transcriptional Analysis of Phloem-Associated Cells of Potato. BMC Genomics 2015, 16, 665. [Google Scholar] [CrossRef] [PubMed]
- Mishima, K.; Fujiwara, T.; Iki, T.; Kuroda, K.; Yamashita, K.; Tamura, M.; Fujisawa, Y.; Watanabe, A. Transcriptome Sequencing and Profiling of Expressed Genes in Cambial Zone and Differentiating Xylem of Japanese Cedar (Cryptomeria japonica). BMC Genomics 2014, 15, 5920. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Huang, X.; Deng, J.; Liu, H.; Liu, Y.; Yu, K.; Huang, B. Differential Gene Expression between Leaf and Rhizome in Atractylodes Lancea: A Comparative Transcriptome Analysis. Front. Plant Sci. 2016, 7, 348. [Google Scholar] [CrossRef] [PubMed]
- Mason, P.J.; Furtado, A.; Marquardt, A.; Hodgson-Kratky, K.; Hoang, N.V.; Botha, F.C.; Papa, G.; Mortimer, J.C.; Simmons, B.; Henry, R.J. Variation in Sugarcane Biomass Composition and Enzymatic Saccharification of Leaves, Internodes and Roots. Biotechnol. Biofuels 2020, 13, 201. [Google Scholar] [CrossRef]
- Schmidt-Rohr, K. O2 and Other High-Energy Molecules in Photosynthesis: Why Plants Need Two Photosystems. Life 2021, 11, 1191. [Google Scholar] [CrossRef]
- Denyer, T.; Ma, X.; Klesen, S.; Scacchi, E.; Nieselt, K.; Timmermans, M.C.P. Spatiotemporal Developmental Trajectories in the Arabidopsis Root Revealed Using High-Throughput Single-Cell RNA Sequencing. Dev. Cell 2019, 48, 840–852.e5. [Google Scholar] [CrossRef] [PubMed]
- Shulse, C.N.; Cole, B.J.; Ciobanu, D.; Lin, J.; Yoshinaga, Y.; Gouran, M.; Turco, G.M.; Zhu, Y.; O’Malley, R.C.; Brady, S.M. High-Throughput Single-Cell Transcriptome Profiling of Plant Cell Types. Cell Rep. 2019, 27, 2241–2247.e4. [Google Scholar] [CrossRef] [PubMed]
- Efroni, I.; Mello, A.; Nawy, T.; Ip, P.-L.; Rahni, R.; DelRose, N.; Powers, A.; Satija, R.; Birnbaum, K.D. Root Regeneration Triggers an Embryo-like Sequence Guided by Hormonal Interactions. Cell 2016, 165, 1721–1733. [Google Scholar] [CrossRef] [PubMed]
- Kortz, A.; Hochholdinger, F.; Yu, P. Cell Type-Specific Transcriptomics of Lateral Root Formation and Plasticity. Front. Plant Sci. 2019, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Tenorio Berrío, R.; Verstaen, K.; Vandamme, N.; Pevernagie, J.; Achon, I.; Van Duyse, J.; Van Isterdael, G.; Saeys, Y.; De Veylder, L.; Inzé, D.; et al. Single-Cell Transcriptomics Sheds Light on the Identity and Metabolism of Developing Leaf Cells. Plant Physiol. 2022, 188, 898–918. [Google Scholar] [CrossRef] [PubMed]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.; Penfold, C.A.; Jenkins, D.; et al. High-Resolution Temporal Profiling of Transcripts during Arabidopsis Leaf Senescence Reveals a Distinct Chronology of Processes and Regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef] [PubMed]
- Chrobok, D.; Law, S.R.; Brouwer, B.; Lindén, P.; Ziolkowska, A.; Liebsch, D.; Narsai, R.; Szal, B.; Moritz, T.; Rouhier, N.; et al. Dissecting the Metabolic Role of Mitochondria during Developmental Leaf Senescence. Plant Physiol. 2016, 172, 2132–2153. [Google Scholar] [CrossRef]
- Mason, P.J.; Hoang, N.V.; Botha, F.C.; Furtado, A.; Marquardt, A.; Henry, R.J. Comparison of the Root, Leaf and Internode Transcriptomes in Sugarcane (Saccharum Spp. Hybrids). Curr. Res. Biotech. 2022, 4, 167–178. [Google Scholar] [CrossRef]
- Li, P.; Ponnala, L.; Gandotra, N.; Wang, L.; Si, Y.; Tausta, S.L.; Kebrom, T.H.; Provart, N.; Patel, R.; Myers, C.R.; et al. The Developmental Dynamics of the Maize Leaf Transcriptome. Nat. Genet. 2010, 42, 1060–1067. [Google Scholar] [CrossRef]
- Huang, Y.-J.; Liu, L.-L.; Huang, J.-Q.; Wang, Z.-J.; Chen, F.-F.; Zhang, Q.-X.; Zheng, B.-S.; Chen, M. Use of Transcriptome Sequencing to Understand the Pistillate Flowering in Hickory (Carya cathayensis Sarg.). BMC Genomics 2013, 14, 691. [Google Scholar] [CrossRef]
- Melzer, S.; Lens, F.; Gennen, J.; Vanneste, S.; Rohde, A.; Beeckman, T. Flowering-Time Genes Modulate Meristem Determinacy and Growth Form in Arabidopsis thaliana. Nat. Genet. 2008, 40, 1489–1492. [Google Scholar] [CrossRef] [PubMed]
- Mutasa-Gottgens, E.; Hedden, P. Gibberellin as a Factor in Floral Regulatory Networks. J. Exp. Bot. 2009, 60, 1979–1989. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, A.; Schmid, M. Regulation of Flowering Time: All Roads Lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Doyle, M.R.; Sung, S.; Amasino, R.M. Vernalization: Winter and the Timing of Flowering in Plants. Annu. Rev. Cell Dev. Biol. 2009, 25, 277–299. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Feng, S.; Pan, Y.; Zhong, J.; Chen, Y.; Yuan, C.; Li, H. Transcriptome Analysis and Identification of Genes Associated with Floral Transition and Flower Development in Sugar Apple (Annona squamosa L.). Front. Plant Sci. 2016, 7, 1695. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Hu, Q.; Zhu, Y.; Zhu, L.; Ma, T.; Zeng, H.; Zang, Q.; Li, X.; Lin, X. Comparative Transcriptomic Analysis of the Flower Induction and Development of the Lei Bamboo (Phyllostachys violascens). BMC Bioinformatics 2019, 20, 687. [Google Scholar] [CrossRef] [PubMed]
- Roberts, W.R.; Roalson, E.H. Comparative Transcriptome Analyses of Flower Development in Four Species of Achimenes (Gesneriaceae). BMC Genomics 2017, 18, 240. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yuan, J.; Wu, M.; Han, Z.; Li, L.; Jiang, H.; Jia, Y.; Han, X.; Liu, M.; Sun, D.; et al. Transcriptome and DNA Methylome Reveal Insights into Yield Heterosis in the Curds of Broccoli (Brassica oleracea L var. italica). BMC Plant Biol. 2018, 18, 168. [Google Scholar] [CrossRef] [PubMed]
- Boesewinkel, F.D.; Bouman, F. The Seed: Structure. In Embryology of Angiosperms, 1st ed.; Johri, B.M., Ed.; Springer Berlin Heidelberg: Berlin, Heidelberg, Germany, 1984; pp. 567–610. [Google Scholar]
- Martín-Gómez, J.J.; Gutiérrez del Pozo, D.; Ucchesu, M.; Bacchetta, G.; Cabello Sáenz de Santamaría, F.; Tocino, Á.; Cervantes, E. Seed Morphology in the Vitaceae Based on Geometric Models. Agronomy 2020, 10, 739. [Google Scholar] [CrossRef]
- Peirats-Llobet, M.; Yi, C.; Liew, L.C.; Berkowitz, O.; Narsai, R.; Lewsey, M.G.; Whelan, J. Spatially Resolved Transcriptomic Analysis of the Germinating Barley Grain. Nucl. Acids Res. 2023, 51, 7798–7819. [Google Scholar] [CrossRef]
- Zhu, M.; Taylor, I.W.; Benfey, P.N. Single-Cell Genomics Revolutionizes Plant Development Studies across Scales. Development 2022, 149, dev200179. [Google Scholar] [CrossRef] [PubMed]
- Betts, N.S.; Berkowitz, O.; Liu, R.; Collins, H.M.; Skadhauge, B.; Dockter, C.; Burton, R.A.; Whelan, J.; Fincher, G.B. Isolation of Tissues and Preservation of RNA from Intact, Germinated Barley Grain. Plant J. 2017, 91, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Le, B.H.; Cheng, C.; Bui, A.Q.; Wagmaister, J.A.; Henry, K.F.; Pelletier, J.; Kwong, L.; Belmonte, M.; Kirkbride, R.; Horvath, S.; et al. Global Analysis of Gene Activity during Arabidopsis Seed Development and Identification of Seed-Specific Transcription Factors. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 8063–8070. [Google Scholar] [CrossRef] [PubMed]
- Narsai, R.; Gouil, Q.; Secco, D.; Srivastava, A.; Karpievitch, Y.V.; Liew, L.C.; Lister, R.; Lewsey, M.G.; Whelan, J. Extensive Transcriptomic and Epigenomic Remodelling Occurs during Arabidopsis thaliana Germination. Genome Biol. 2017, 18, 172. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Lu, J.; Xing, J.; Du, M.; Wang, M.; Zhang, L.; Li, Y.; Zhang, C.; Wu, Y. Transcriptome and Metabolome Analyses Revealing the Potential Mechanism of Seed Germination in Polygonatum cyrtonema. Sci. Rep. 2021, 11, 12161. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, N.; Li, W.; Gao, X.; Cha, M.; Qin, L.; Liu, L. Advances in Transcriptomics in the Response to Stress in Plants. Global Med. Genet. 2020, 07, 030–034. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Huang, Z.; Sun, J.; Cui, X.; Liu, Y. Research Progress and Future Development Trends in Medicinal Plant Transcriptomics. Front. Plant Sci. 2021, 12, 691838. [Google Scholar] [CrossRef] [PubMed]
- Choudhri, P.; Rani, M.; Sangwan, R.S.; Kumar, R.; Kumar, A.; Chhokar, V. De Novo Sequencing, Assembly and Characterisation of Aloe Vera Transcriptome and Analysis of Expression Profiles of Genes Related to Saponin and Anthraquinone Metabolism. BMC Genomics 2018, 19, 427. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Singh, G.; Bhandawat, A.; Singh, G.; Parmar, R.; Seth, R.; Sharma, R.K. Spatial Transcriptome Analysis Provides Insights of Key Gene(s) Involved in Steroidal Saponin Biosynthesis in Medicinally Important Herb Trillium govanianum. Sci. Rep. 2017, 7, 45295. [Google Scholar] [CrossRef]
- Liao, W.; Mei, Z.; Miao, L.; Liu, P.; Gao, R. Comparative Transcriptome Analysis of Root, Stem, and Leaf Tissues of Entada Phaseoloides Reveals Potential Genes Involved in Triterpenoid Saponin Biosynthesis. BMC Genomics 2020, 21, 639. [Google Scholar] [CrossRef]
- Yan, J.; Qian, L.; Zhu, W.; Qiu, J.; Lu, Q.; Wang, X.; Wu, Q.; Ruan, S.; Huang, Y. Integrated Analysis of the Transcriptome and Metabolome of Purple and Green Leaves of Tetrastigma hemsleyanum Reveals Gene Expression Patterns Involved in Anthocyanin Biosynthesis. PLoS ONE 2020, 15, e0230154. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Pu, T.; Gui, C.; Zhang, X.; Gong, L. Transcriptome Analysis Reveals Biosynthesis of Important Bioactive Constituents and Mechanism of Stem Formation of Dendrobium huoshanense. Sci. Rep. 2020, 10, 2857. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Peng, D.; Zhu, J.; Zhao, D.; Shi, Y.; Zhang, S.; Ma, K.; Wu, J.; Huang, L. Transcriptome Analysis of Polygonatum cyrtonema Hua: Identification of Genes Involved in Polysaccharide Biosynthesis. Plant Methods 2019, 15, 65. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Tan, H.; Li, S.; Cheung, F.; Wang, N.; Nagamatsu, T.; Feng, Y. Cancer Stem Cells: The Potential Targets of Chinese Medicines and Their Active Compounds. Int. J. Mol. Sci. 2016, 17, 893. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Zhou, C.; Li, T.; Yuan, Z.; Zhou, H.; Tamada, Y.; Zhao, Y.; Wang, J.; Zheng, Q.; Hao, X.; et al. Global Transcriptome Analysis Reveals Dynamic Gene Expression Profiling and Provides Insights into Biosynthesis of Resveratrol and Anthraquinones in a Medicinal Plant Polygonum cuspidatum. Ind. Crops and Prod. 2021, 171, 113919. [Google Scholar] [CrossRef]
- Umar, O.; Ranti, L.; Abdulbaki, A.; Bola, A.; Abdulhamid, A.; Biola, M. Stresses in Plants: Biotic and Abiotic. In Current Trends in Wheat Research; ed. Mahmood-ur-Rahman Ansari, Government College University, Faisalabad: IntechOpen 2021. [CrossRef]
- Zhu, J.-K. Salt and Drought Stress Signal Transduction in Plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Gull, A.; Ahmad Lone, A.; Ul Islam Wani, N. Biotic and Abiotic Stresses in Plants. In Abiotic and Biotic Stress in Plants, 1st ed.; Bosco de Oliveira, A., Ed.; IntechOpen, 2019.
- EMBL-EBI. Expression Atlas. Available online: https://www.ebi.ac.uk/gxa/home (accessed on 9 January 2024). Gene expression across species and biological conditions.
- ELIXIR Recommended Interoperability Resource. g:Profiler. Available online: https://biit.cs.ut.ee/gprofiler/gost (accessed on 9 January 2024).
- Song, Y.; Ma, B.; Guo, Q.; Zhou, L.; Lv, C.; Liu, X.; Wang, J.; Zhou, X.; Zhang, C. UV-B Induces the Expression of Flavonoid Biosynthetic Pathways in Blueberry (Vaccinium corymbosum) Calli. Front. Plant Sci. 2022, 13, 1079087. [Google Scholar] [CrossRef]
- Dong, Y.; Gupta, S.; Wargent, J.J.; Putterill, J.; Macknight, R.C.; Gechev, T.S.; Mueller-Roeber, B.; Dijkwel, P.P. Comparative Transcriptomics of Multi-Stress Responses in Pachycladon cheesemanii and Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 11323. [Google Scholar] [CrossRef]
- Lorence, A.; Chevone, B.I.; Mendes, P.; Nessler, C.L. Myo -Inositol Oxygenase Offers a Possible Entry Point into Plant Ascorbate Biosynthesis. Plant Physiol. 2004, 134, 1200–1205. [Google Scholar] [CrossRef]
- Zhou, H.; Yu, L.; Liu, S.; Zhu, A.; Yang, Y.; Chen, C.; Yang, A.; Liu, L.; Yu, F. Transcriptome Comparison Analyses in UV-B Induced AsA Accumulation of Lactuca sativa L. BMC Genomics 2023, 24, 61. [Google Scholar] [CrossRef]
- DeepVenn - Create Area-Proportional Venn Diagrams Using the Deep Learning Framework Tensorflow. Available online: http://www.deepvenn.com/ (accessed on 8 01 2024).
- Verma, S.; Nizam, S.; Verma, P.K. Biotic and Abiotic Stress Signaling in Plants. In Stress Signaling in Plants: Genomics and Proteomics Perspective, Volume 1, 1st ed.; Sarwat, M., Ahmad, A., Abdin, M., Eds.; Springer New York: New York, NY, U.S.A, 2013; pp. 25–49. [Google Scholar]
- Shanker, A.K.; Maheswari, M.; Yadav, S.K.; Desai, S.; Bhanu, D.; Attal, N.B.; Venkateswarlu, B. Drought Stress Responses in Crops. Funct. Integr. Genomics 2014, 14, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhou, Y.; Zhai, H.; He, S.; Zhao, N.; Liu, Q. Transcriptome Profiling Reveals Insights into the Molecular Mechanism of Drought Tolerance in Sweetpotato. J. Integr. Agri. 2019, 18, 9–23. [Google Scholar] [CrossRef]
- Kumar, M.; Chauhan, A.S.; Kumar, M.; Yusuf, M.A.; Sanyal, I.; Chauhan, P.S. Transcriptome Sequencing of Chickpea (Cicer arietinum L.) Genotypes for Identification of Drought-Responsive Genes Under Drought Stress Condition. Plant Mol. Biol. Rep. 2019, 37, 186–203. [Google Scholar] [CrossRef]
- Li, X.; Liu, L.; Sun, S.; Li, Y.; Jia, L.; Ye, S.; Yu, Y.; Dossa, K.; Luan, Y. Leaf-Transcriptome Profiles of Phoebe bournei Provide Insights into Temporal Drought Stress Responses. Front. Plant Sci. 2022, 13, 1010314. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Gunapati, S.; Kianian, S.F.; Singh, S.P. Comparative Analysis of Transcriptome in Two Wheat Genotypes with Contrasting Levels of Drought Tolerance. Protoplasma 2018, 255, 1487–1504. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Kumar, S.; Mohapatra, T. Biochemical, Physiological and Molecular Responses of Rice to Terminal Drought Stress: Transcriptome Profiling of Leaf and Root Reveals the Key Stress-Responsive Genes. J. Plant Biochem. Biotechnol. 2023. [Google Scholar] [CrossRef]
- Miao, Z.; Xu, W.; Li, D.; Hu, X.; Liu, J.; Zhang, R.; Tong, Z.; Dong, J.; Su, Z.; Zhang, L.; et al. De Novo Transcriptome Analysis of Medicago falcata Reveals Novel Insights about the Mechanisms Underlying Abiotic Stress-Responsive Pathway. BMC Genomics 2015, 16, 818. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira-Busatto, L.A.; De Almeida, R.M.C.; Weber, R.L.M.; Favero, D.; Bredemeier, C.; Da Silva Giordano, C.P.; Bodanese-Zanettini, M.H. The Soybean Transcriptogram Allows a Wide Genome-to-Single-Gene Analysis That Evinces Time-Dependent Drought Response. Plant Mol. Biol. Rep. 2022, 40, 1–27. [Google Scholar] [CrossRef]
- Xu, X.; Legay, S.; Sergeant, K.; Zorzan, S.; Leclercq, C.C.; Charton, S.; Giarola, V.; Liu, X.; Challabathula, D.; Renaut, J.; et al. Molecular Insights into Plant Desiccation Tolerance: Transcriptomics, Proteomics and Targeted Metabolite Profiling in Craterostigma plantagineum. Plant J. 2021, 107, 377–398. [Google Scholar] [CrossRef]
- Fang, S.; Zhao, P.; Tan, Z.; Peng, Y.; Xu, L.; Jin, Y.; Wei, F.; Guo, L.; Yao, X. Combining Physio-Biochemical Characterization and Transcriptome Analysis Reveal the Responses to Varying Degrees of Drought Stress in Brassica napus L. Int. J. Mol. Sci. 2022, 23, 8555. [Google Scholar] [CrossRef]
- Yang, Y.; Xia, J.; Fang, X.; Jia, H.; Wang, X.; Lin, Y.; Liu, S.; Ge, M.; Pu, Y.; Fang, J.; et al. Drought Stress in ‘Shine Muscat’ Grapevine: Consequences and a Novel Mitigation Strategy–5-Aminolevulinic Acid. Front. Plant Sci. 2023, 14, 1129114. [Google Scholar] [CrossRef] [PubMed]
- Young, L.W.; Wilen, R.W.; Bonham-Smith, P.C. High Temperature Stress of Brassica napus during Flowering Reduces Micro- and Megagametophyte Fertility, Induces Fruit Abortion, and Disrupts Seed Production. J. Exp. Bot. 2004, 55, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Taohua, Z.; Gui, W.; Xu, L.; Li, J.; Ding, Y. Five Pectinase Gene Expressions Highly Responding to Heat Stress in Rice Floral Organs Revealed by RNA-Seq Analysis. Biochem. Biophys. Res. Commun. 2015, 463, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Bita, C.E.; Gerats, T. Plant Tolerance to High Temperature in a Changing Environment: Scientific Fundamentals and Production of Heat Stress-Tolerant Crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Lu, X.; Zeng, H.; Zhang, Y.; Zhu, J. Heat Stress Induction of miR398 Triggers a Regulatory Loop That Is Critical for Thermotolerance in Arabidopsis. Plant J. 2013, 74, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Asseng, S.; Foster, I.; Turner, N.C. The Impact of Temperature Variability on Wheat Yields. Global Change Biology 2011, 17, 997–1012. [Google Scholar] [CrossRef]
- Xing, X.; Ding, Y.; Jin, J.; Song, A.; Chen, S.; Chen, F.; Fang, W.; Jiang, J. Physiological and Transcripts Analyses Reveal the Mechanism by Which Melatonin Alleviates Heat Stress in Chrysanthemum Seedlings. Front. Plant Sci. 2021, 12, 673236. [Google Scholar] [CrossRef] [PubMed]
- Frank, G.; Pressman, E.; Ophir, R.; Althan, L.; Shaked, R.; Freedman, M.; Shen, S.; Firon, N. Transcriptional Profiling of Maturing Tomato (Solanum Lycopersicum L.) Microspores Reveals the Involvement of Heat Shock Proteins, ROS Scavengers, Hormones, and Sugars in the Heat Stress Response. J. Exp.Bot. 2009, 60, 3891–3908. [Google Scholar] [CrossRef]
- Valdés-López, O.; Batek, J.; Gomez-Hernandez, N.; Nguyen, C.T.; Isidra-Arellano, M.C.; Zhang, N.; Joshi, T.; Xu, D.; Hixson, K.K.; Weitz, K.K.; et al. Soybean Roots Grown under Heat Stress Show Global Changes in Their Transcriptional and Proteomic Profiles. Front. Plant Sci. 2016, 7, 517. [Google Scholar] [CrossRef]
- Sewelam, N.; Brilhaus, D.; Bräutigam, A.; Alseekh, S.; Fernie, A.R.; Maurino, V.G. Molecular Plant Responses to Combined Abiotic Stresses Put a Spotlight on Unknown and Abundant Genes. J. Exp. Bot. 2020, 71, 5098–5112. [Google Scholar] [CrossRef]
- Rurek, M.; Czołpińska, M.; Pawłowski, T.; Krzesiński, W.; Spiżewski, T. Cold and Heat Stress Diversely Alter Both Cauliflower Respiration and Distinct Mitochondrial Proteins Including OXPHOS Components and Matrix Enzymes. Int. J. Mol. Sci. 2018, 19, 877. [Google Scholar] [CrossRef]
- Rashid, F.A.A.; Crisp, P.A.; Zhang, Y.; Berkowitz, O.; Pogson, B.J.; Day, D.A.; Masle, J.; Dewar, R.C.; Whelan, J.; Atkin, O.K.; et al. Molecular and Physiological Responses during Thermal Acclimation of Leaf Photosynthesis and Respiration in Rice. Plant Cell Environ. 2020, 43, 594–610. [Google Scholar] [CrossRef] [PubMed]
- Mikołajczak, K.; Kuczyńska, A.; Krajewski, P.; Kempa, M.; Nuc, M. Transcriptome Profiling Disclosed the Effect of Single and Combined Drought and Heat Stress on Reprogramming of Genes Expression in Barley Flag Leaf. Front. Plant Sci. 2023, 13, 1096685. [Google Scholar] [CrossRef]
- Mahalingam, R.; Duhan, N.; Kaundal, R.; Smertenko, A.; Nazarov, T.; Bregitzer, P. Heat and Drought Induced Transcriptomic Changes in Barley Varieties with Contrasting Stress Response Phenotypes. Front. Plant Sci. 2022, 13, 1066421. [Google Scholar] [CrossRef]
- Martin, R.C.; Kronmiller, B.A.; Dombrowski, J.E. Transcriptome Analysis of Lolium temulentum Exposed to a Combination of Drought and Heat Stress. Plants 2021, 10, 2247. [Google Scholar] [CrossRef] [PubMed]
- Vicente, R.; Bolger, A.M.; Martínez-Carrasco, R.; Pérez, P.; Gutiérrez, E.; Usadel, B.; Morcuende, R. De Novo Transcriptome Analysis of Durum Wheat Flag Leaves Provides New Insights Into the Regulatory Response to Elevated CO2 and High Temperature. Front. Plant Sci. 2019, 10, 1605. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, H. Heat Stress Regulates the Expression of Genes at Transcriptional and Post-Transcriptional Levels, Revealed by RNA-Seq in Brachypodium distachyon. Front. Plant Sci. 2017, 7, 2067. [Google Scholar] [CrossRef]
- Obaid, A.Y.; Sabir, J.S.M.; Atef, A.; Liu, X.; Edris, S.; El-Domyati, F.M.; Mutwakil, M.Z.; Gadalla, N.O.; Hajrah, N.H.; Al-Kordy, M.A.; et al. Analysis of Transcriptional Response to Heat Stress in Rhazya stricta. BMC Plant Biol. 2016, 16, 252. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Cold Stress Regulation of Gene Expression in Plants. Trend Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Xu, W.; Li, R.; Zhang, N.; Ma, F.; Jiao, Y.; Wang, Z. Transcriptome Profiling of Vitis amurensis, an Extremely Cold-Tolerant Chinese Wild Vitis Species, Reveals Candidate Genes and Events That Potentially Connected to Cold Stress. Plant Mol. Biol. 2014, 86, 527–541. [Google Scholar] [CrossRef]
- Cheng, Z.; Lei, N.; Li, S.; Liao, W.; Shen, J.; Peng, M. The Regulatory Effects of MeTCP4 on Cold Stress Tolerance in Arabidopsis Thaliana: A Transcriptome Analysis. Plant Physiol. Biochem. 2019, 138, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Li, X.; Li, T.; Yu, D.; Han, B. Genome-Wide Transcriptome Profiling Provides Overwintering Mechanism of Agropyron Mongolicum. BMC Plant Biol. 2017, 17, 138. [Google Scholar] [CrossRef] [PubMed]
- Naydenov, N.G.; Khanam, S.; Siniauskaya, M.; Nakamura, C. Profiling of Mitochondrial Transcriptome in Germinating Wheat Embryos and Seedlings Subjected to Cold, Salinity and Osmotic Stresses. Genes Genet. Syst. 2010, 85, 31–42. [Google Scholar] [CrossRef] [PubMed]
- McDowell, J.M.; Dangl, J.L. Signal Transduction in the Plant Immune Response. Trend Biochem. Sci. 2000, 25, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Tsuda, K.; Wang, L.; Coller, J.; Watanabe, Y.; Glazebrook, J.; Katagiri, F. Network Modeling Reveals Prevalent Negative Regulatory Relationships between Signaling Sectors in Arabidopsis Immune Signaling. PLoS Pathog. 2010, 6, e1001011. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C.M. Networking by Small-Molecule Hormones in Plant Immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.-L.; Chen, B.-H.; Chen, X.-J.; Guo, Y.-Y.; Yang, H.-L.; Li, X.-Z.; Wang, G.-Y. Transcriptome Profiling of Pumpkin (Cucurbita moschata Duch.) Leaves Infected with Powdery Mildew. PLoS ONE 2018, 13, e0190175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, Y.; Wang, C.; Liu, M.; Li, H.; Fu, Y.; Wang, Y.; Nie, Y.; Liu, X.; Ji, W. Large-Scale Transcriptome Comparison Reveals Distinct Gene Activations in Wheat Responding to Stripe Rust and Powdery Mildew. BMC Genomics 2014, 15, 898. [Google Scholar] [CrossRef]
- Kumar, A.; Kanak, K.R.; Arunachalam, A.; Dass, R.S.; Lakshmi, P.T.V. Comparative Transcriptome Profiling and Weighted Gene Co-Expression Network Analysis to Identify Core Genes in Maize (Zea mays L.) Silks Infected by Multiple Fungi. Front. Plant Sci. 2022, 13, 985396. [Google Scholar] [CrossRef]
- Kottapalli, K.R.; Rakwal, R.; Satoh, K.; Shibato, J.; Kottapalli, P.; Iwahashi, H.; Kikuchi, S. Transcriptional Profiling of Indica Rice Cultivar IET8585 (Ajaya) Infected with Bacterial Leaf Blight Pathogen Xanthomonas oryzae pv oryzae. Plant Physiol. Biochem. 2007, 45, 834–850. [Google Scholar] [CrossRef]
- Yokotani, N.; Hasegawa, Y.; Sato, M.; Hirakawa, H.; Kouzai, Y.; Nishizawa, Y.; Yamamoto, E.; Naito, Y.; Isobe, S. Transcriptome Analysis of Clavibacter michiganensis subsp. michiganensis-Infected Tomatoes: A Role of Salicylic Acid in the Host Response. BMC Plant Biol. 2021, 21, 476. [Google Scholar] [CrossRef]
- Lu, L.; Yang, D.; Tang, D.; Li, S.; Chen, Z. Transcriptome Analysis of Different Rice Cultivars Provides Novel Insights into the Rice Response to Bacterial Leaf Streak Infection. Funct. Integr. Genomics 2020, 20, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Li, Z.; Liu, X.; Yang, W.; Wang, Y. Comparative Transcriptome Analysis Reveals Potential Genes Conferring Resistance or Susceptibility to Bacterial Canker in Tomato. Horticulturae 2023, 9, 242. [Google Scholar] [CrossRef]
- Rivero, R.M.; Mittler, R.; Blumwald, E.; Zandalinas, S.I. Developing Climate-resilient Crops: Improving Plant Tolerance to Stress Combination. Plant J. 2022, 109, 373–389. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Prusty, M.R.; Pandey, M.K.; Singh, P.K.; Bohra, A.; Guo, B.; Varshney, R.K. Application of CRISPR/Cas9-Mediated Gene Editing for Abiotic Stress Management in Crop Plants. Front. Plant Sci. 2023, 14, 1157678. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




