Submitted:
08 March 2024
Posted:
11 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
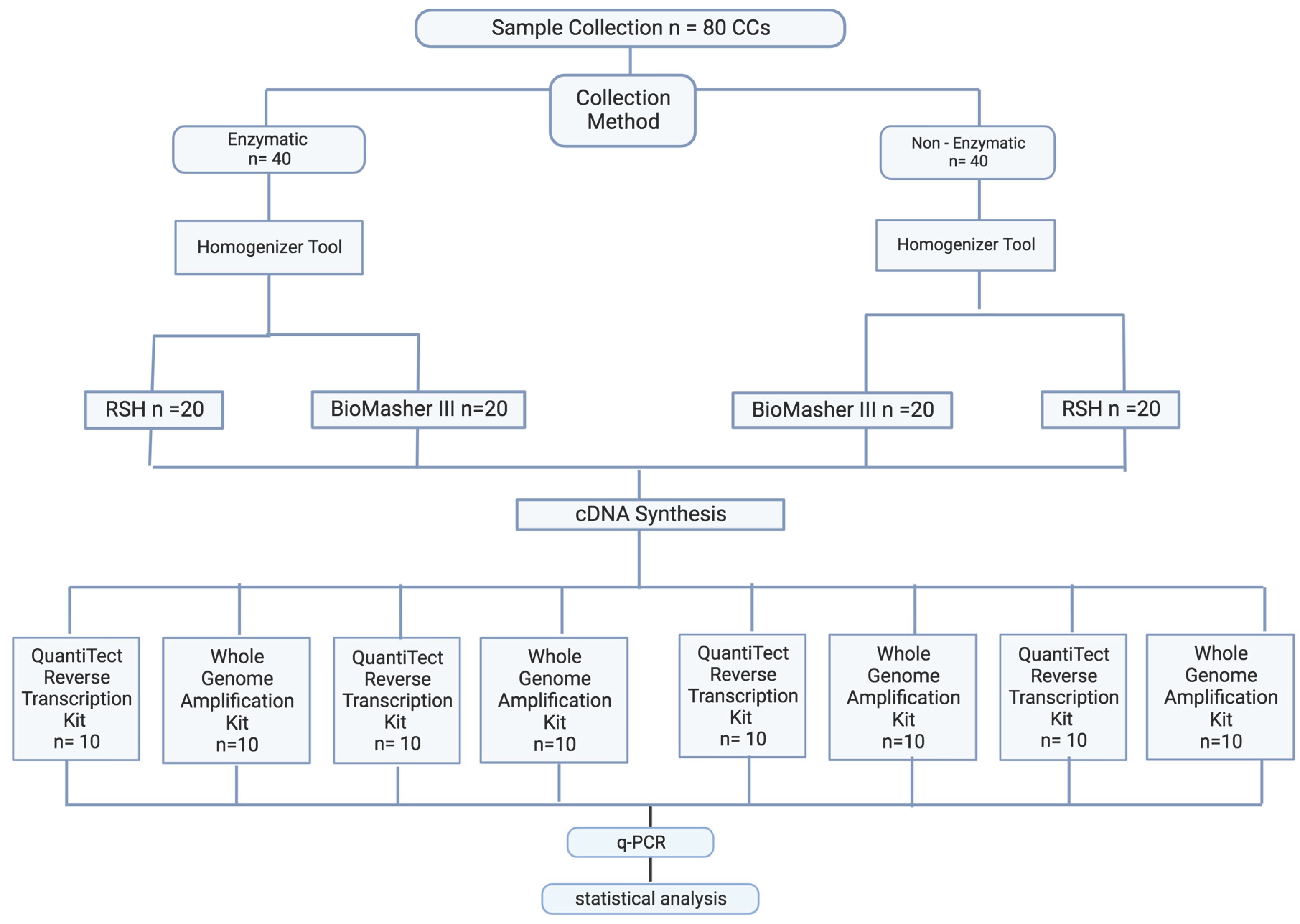
2.1. Clinical Study
2.1.1. Study Design, Recruitment, and Ethical Approval
2.1.2. RNA Extraction Method Optimization
- A.
- Sample collection optimization
- A.
- B. Sample preparation optimization
- A.
- C. RNA purification optimization
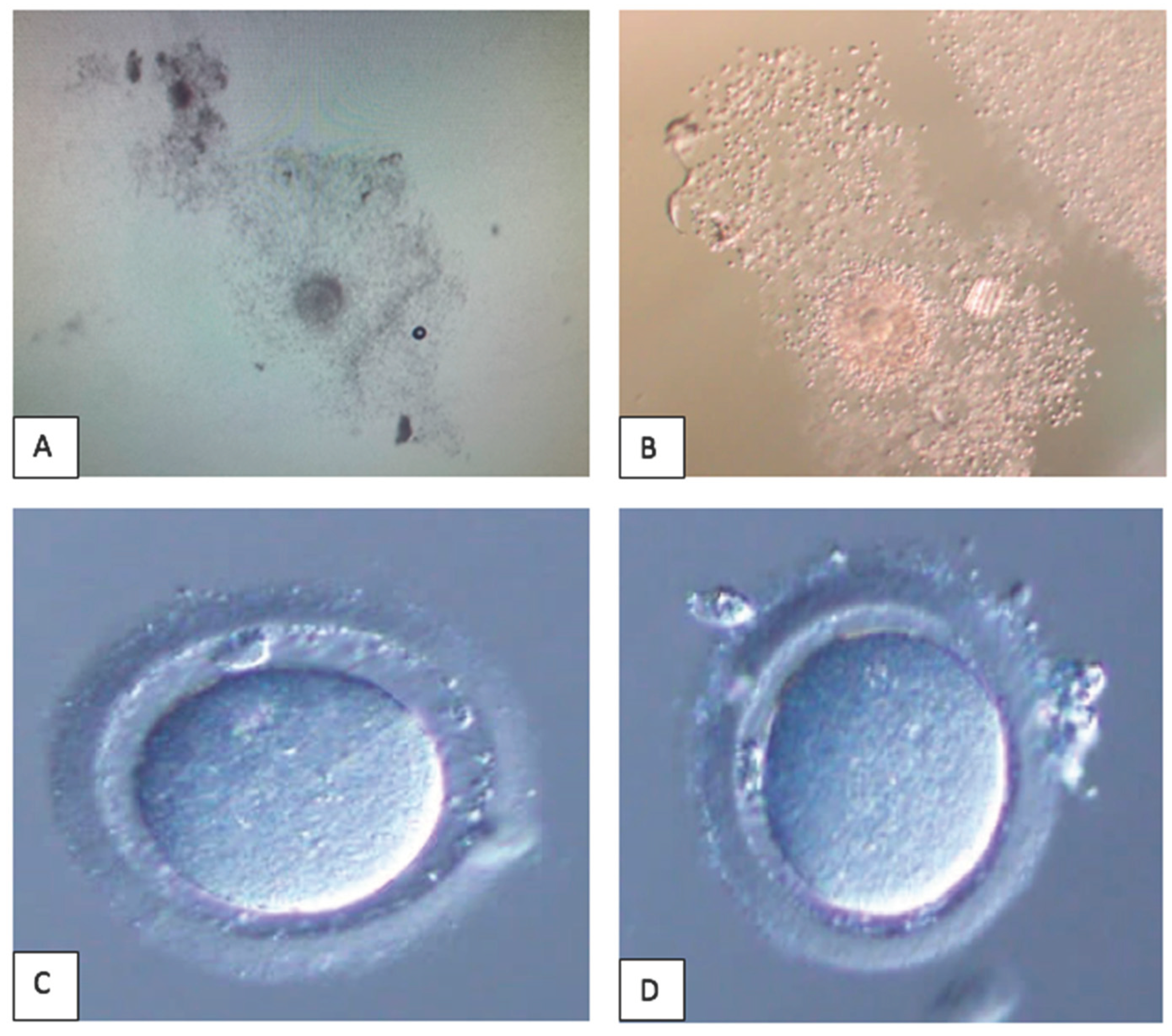
2.1.2.1. Sample Collection Optimization
2.1.2.2 Sample Preparation Optimization
- Rotor–stator homogenizer [20].
- 2.
- BioMasher III
2.1.2.3. RNA Purification Optimization
2.1.2. RNA Concentration and Purity Measurement
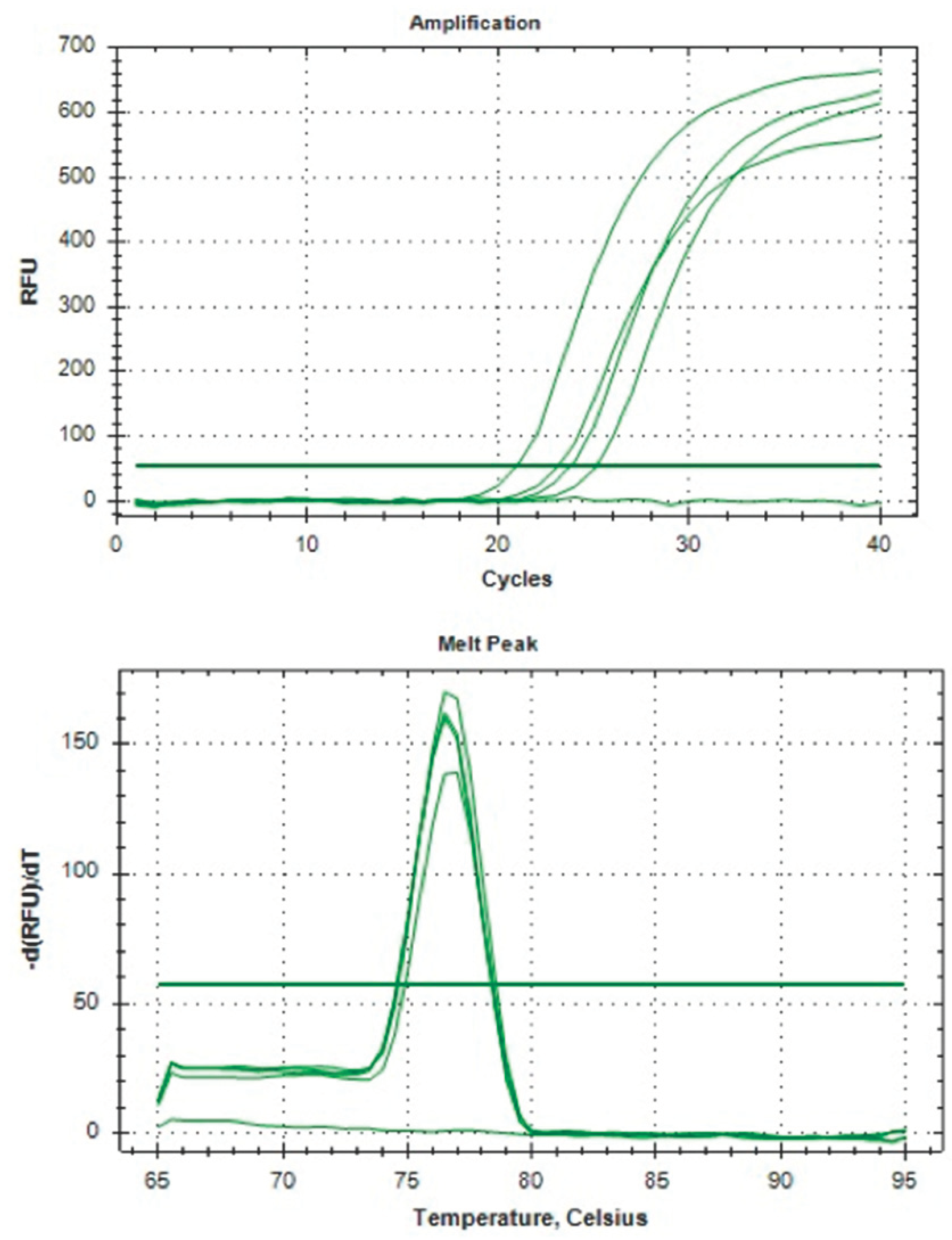
2.1.3. cDNA Synthesis and qPCR
2.2. The Systematic Review
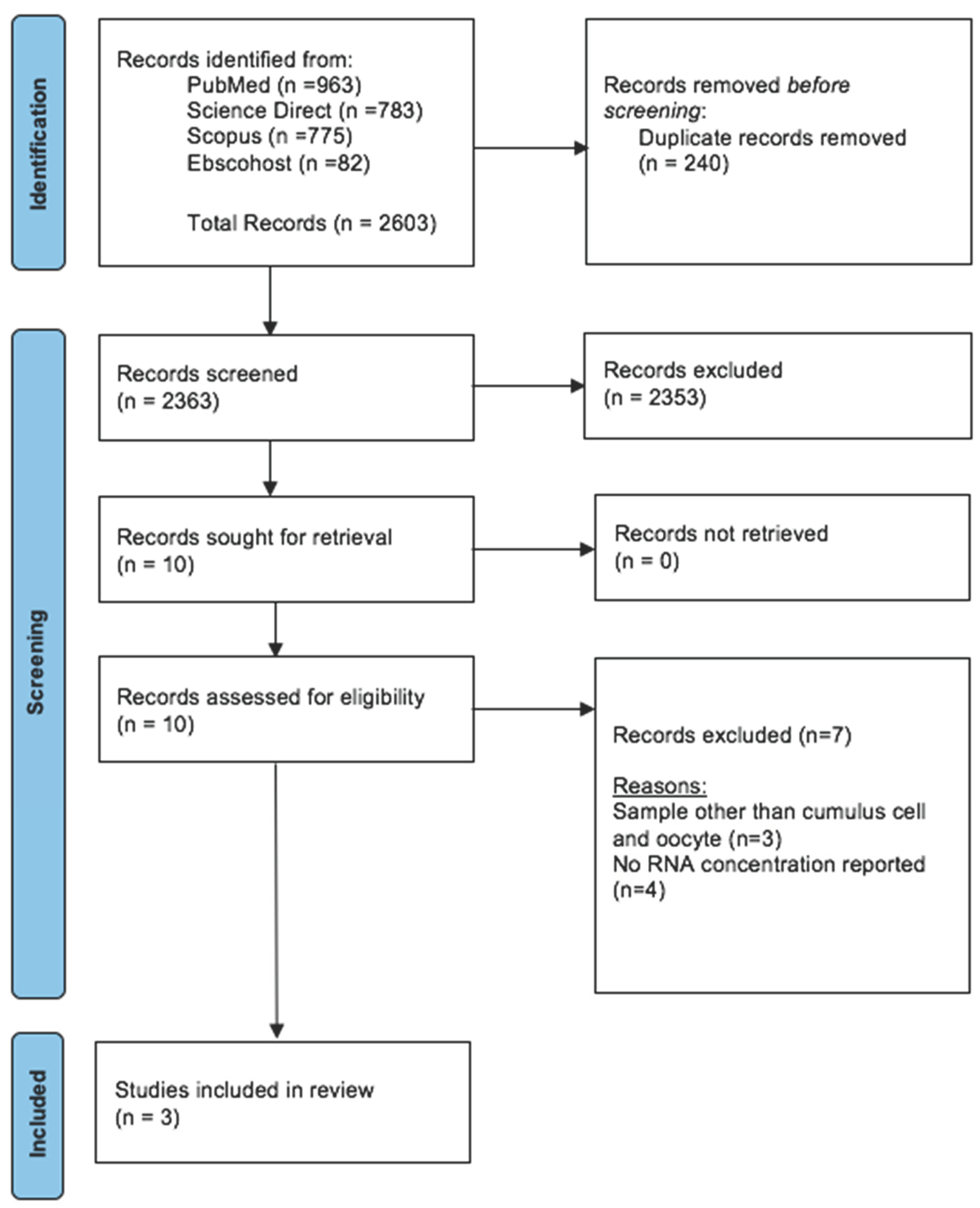
2.2.1. Information Source and Search Strategy
2.2.1.1. Inclusion & Exclusion Criteria
2.2.1.2. Screening of Articles for Eligibility
2.2.2. Study Selection, Data Extraction and Risk of Bias Assessment
3. Results
3.1. Clinical Study
3.2. Systematic Review
3.2.1. Search Sequence and Quality Assessment
3.2.2. Studies Characteristics
3.2.3. Main Outcome
4. Discussion
| Author, Year (References) | Title | Country | Sample size (n) | Organism | Method for RNA extraction | RNA concentration (ng μL-1) | RNA Purity |
|---|---|---|---|---|---|---|---|
| Maisarah et al. (2020) [16] | The challenge of getting a high quality of RNA from oocyte for gene expression study | Malaysia | COC (19) Oocyte (400) |
Mouse | TRIzol | COC = 151.0 Oocyte = 126.7 |
COC = 1.7 Oocyte = 1.68 |
| RNeasy Mini Kit | COC = 3.8 Oocyte = 1.9 |
COC = 1.68 Oocyte = 10.5 |
|||||
| Wiweko et al. (2017) [25] | The quality of RNA isolation from frozen granulosa cells | Indonesia | Oocyte (28) | Human | QIAamp RNA Blood Mini Kit | 250 | 1.85 |
| Pavani et al. (2015) [26] | Optimisation of Total Rna Extraction from Bovine Oocytes and Embryos for Gene Expression Studies and Effects of Cryoprotectants on Total Rna Extraction | Portugal | Oocyte (795) | Bovine | TRIzol | 152.8 | 1.5 |
| Guanidinium thiocyanate | 47 | 1.18 | |||||
| Commercial kit | 31.2 | 2.06 |
4.1. Strengths
4.2. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carvacho, I.; Piesche, M.; Maier, T.J.; Machaca, K. Ion Channel Function During Oocyte Maturation and Fertilization. Front Cell Dev Biol 2018, 6, 63. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhang, T.; Yang, Y.; Wang, C. Mechanisms of Oocyte Maturation and Related Epigenetic Regulation. Front Cell Dev Biol 2021, 9, 654028. [Google Scholar] [CrossRef] [PubMed]
- Turathum, B.; Gao, E.M.; Chian, R.C. The Function of Cumulus Cells in Oocyte Growth and Maturation and in Subsequent Ovulation and Fertilization. Cells 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wells, D. The human oocyte and cumulus cells relationship: new insights from the cumulus cell transcriptome. Mol Hum Reprod 2010, 16, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Assidi, M.; Richard, F.J.; Sirard, M.A. FSH in vitro versus LH in vivo: similar genomic effects on the cumulus. J Ovarian Res 2013, 6, 68. [Google Scholar] [CrossRef] [PubMed]
- van Gijn, J.; Gijselhart, J.P. [Von Willebrand and his factor]. Ned Tijdschr Geneeskd 2011, 155, A2022. [Google Scholar] [PubMed]
- Zhang, C.H.; Liu, X.Y.; Wang, J. Essential Role of Granulosa Cell Glucose and Lipid Metabolism on Oocytes and the Potential Metabolic Imbalance in Polycystic Ovary Syndrome. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Salimov, D.; Lisovskaya, T.; Otsuki, J.; Gzgzyan, A.; Bogolyubova, I.; Bogolyubov, D. Chromatin Morphology in Human Germinal Vesicle Oocytes and Their Competence to Mature in Stimulated Cycles. Cells 2023, 12, 1976. [Google Scholar] [CrossRef]
- Xie, J.; Xu, X.; Liu, S. Intercellular communication in the cumulus-oocyte complex during folliculogenesis: A review. Front Cell Dev Biol 2023, 11, 1087612. [Google Scholar] [CrossRef]
- Sfakianoudis, K.; Maziotis, E.; Karantzali, E.; Kokkini, G.; Grigoriadis, S.; Pantou, A.; Giannelou, P.; Petroutsou, K.; Markomichali, C.; Fakiridou, M.; et al. Molecular Drivers of Developmental Arrest in the Human Preimplantation Embryo: A Systematic Review and Critical Analysis Leading to Mapping Future Research. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef]
- Coticchio, G.; Dal Canto, M.; Mignini Renzini, M.; Guglielmo, M.C.; Brambillasca, F.; Turchi, D.; Novara, P.V.; Fadini, R. Oocyte maturation: gamete-somatic cells interactions, meiotic resumption, cytoskeletal dynamics and cytoplasmic reorganization. Hum Reprod Update 2015, 21, 427–454. [Google Scholar] [CrossRef]
- Nana, M.; Hodson, K.; Lucas, N.; Camporota, L.; Knight, M.; Nelson-Piercy, C. Diagnosis and management of covid-19 in pregnancy. BMJ 2022, 377, e069739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Song, S.; Yang, M.; Yan, L.; Qiao, J. Diminished ovarian reserve causes adverse ART outcomes attributed to effects on oxygen metabolism function in cumulus cells. BMC Genomics 2023, 24, 655. [Google Scholar] [CrossRef]
- Gilchrist, R.B.; Smitz, J. Oocyte in vitro maturation: physiological basis and application to clinical practice. Fertility and sterility 2023, 119, 524–539. [Google Scholar] [CrossRef]
- Ferrari, S.; Lattuada, D.; Paffoni, A.; Brevini, T.A.; Scarduelli, C.; Bolis, G.; Ragni, G.; Gandolfi, F. Procedure for rapid oocyte selection based on quantitative analysis of cumulus cell gene expression. J Assist Reprod Genet 2010, 27, 429–434. [Google Scholar] [CrossRef]
- Maisarah, Y.; Hashida, H.N.; Yusmin, M.Y. The challenge of getting a high quality of RNA from oocyte for gene expression study. Vet Res Forum 2020, 11, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Biase, F.H. Isolation of high-quality total RNA and RNA sequencing of single bovine oocytes. STAR Protoc 2021, 2, 100895. [Google Scholar] [CrossRef]
- Tjahyadi, D.; Susiarno, H.; Chandra, B.A.; Permadi, W.; Djuwantono, T.; Wiweko, B. The effect of oocyte denudation time and intracytoplasmic sperm injection time on embryo quality at assisted reproductive technology clinic - A cross-sectional study. Ann Med Surg (Lond) 2022, 80, 104234. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.C.; Yiap, B.C. DNA, RNA, and protein extraction: the past and the present. J Biomed Biotechnol 2009, 2009, 574398. [Google Scholar] [CrossRef]
- Kaivo-oja, N.; Jeffery, L.A.; Ritvos, O.; Mottershead, D.G. Smad signalling in the ovary. Reprod Biol Endocrinol 2006, 4, 21. [Google Scholar] [CrossRef]
- Maa, Y.-F.; Hsu, C. Liquid-liquid emulsification by rotor/stator homogenization. Journal of Controlled Release 1996, 38, 219–228. [Google Scholar] [CrossRef]
- Iwai, H.; Mori, M.; Tomita, M.; Kono, N.; Arakawa, K. Molecular Evidence of Chemical Disguise by the Socially Parasitic Spiny Ant Polyrhachis lamellidens (Hymenoptera: Formicidae) When Invading a Host Colony. Frontiers in Ecology and Evolution 2022, 10. [Google Scholar] [CrossRef]
- Olson, N.D.; Morrow, J.B. DNA extract characterization process for microbial detection methods development and validation. BMC Res Notes 2012, 5, 668. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Wiweko, B.; Utami, R.; Putri, D.; Muna, N.; Aulia, S.N.; Riayati, O.; Bowolaksono, A. The Quality of RNA Isolation from Frozen Granulosa Cells. Advanced Science Letters 2017, 23, 6717–6719. [Google Scholar] [CrossRef]
- Pavani, K.C.; Baron, E.E.; Faheem, M.; Chaveiro, A.; Da Silva, F.M. Optimisation of Total Rna Extraction from Bovine Oocytes and Embryos for Gene Expression Studies and Effects of Cryoprotectants on Total Rna Extraction. Tsitol Genet 2015, 49, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Moura, B.R.; Gurgel, M.C.; Machado, S.P.; Marques, P.A.; Rolim, J.R.; Lima, M.C.; Salgueiro, L.L. Low concentration of hyaluronidase for oocyte denudation can improve fertilization rates and embryo quality. JBRA Assist Reprod 2017, 21, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, Y.; Takeo, T.; Nakao, S.; Yoshimoto, H.; Hirose, Y.; Sakai, Y.; Horikoshi, Y.; Takeuji, S.; Tsuchiyama, S.; Nakagata, N. Prolonged exposure to hyaluronidase decreases the fertilization and development rates of fresh and cryopreserved mouse oocytes. J Reprod Dev 2014, 60, 454–459. [Google Scholar] [CrossRef]
- Nagyova, E. The Biological Role of Hyaluronan-Rich Oocyte-Cumulus Extracellular Matrix in Female Reproduction. Int J Mol Sci 2018, 19. [Google Scholar] [CrossRef]
- Attanasio, M.; Cirillo, M.; Coccia, M.E.; Castaman, G.; Fatini, C. Factors VIII and Von Willebrand Levels in Women Undergoing Assisted Reproduction: Are Their Levels Associated with Clinical Pregnancy Outcome? Mediterr J Hematol Infect Dis 2020, 12, e2020058. [Google Scholar] [CrossRef] [PubMed]
- Salustri, A.; Camaioni, A.; Tirone, E.; D'Alessandris, C. Hyaluronic acid and proteoglycan accumulation in the cumulus oophorus matrix. Ital J Anat Embryol 1995, 100 Suppl 1, 479–484. [Google Scholar]
- Esbert, M.; Florensa, M.; Riqueros, M.; Teruel, J.; Ballesteros, A. Effect of oocyte denudation timing on clinical outcomes in 1212 oocyte recipients. Fertility and sterility 2013, 100, S482. [Google Scholar] [CrossRef]
- Maldonado Rosas, I.; Anagnostopoulou, C.; Singh, N.; Gugnani, N.; Singh, K.; Desai, D.; Darbandi, M.; Manoharan, M.; Darbandi, S.; Chockalingam, A.; et al. Optimizing embryological aspects of oocyte retrieval, oocyte denudation, and embryo loading for transfer. Panminerva Med 2022, 64, 156–170. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, Y.; Fang, Z.; Hu, S.; Li, Z.; Zhu, L.; Jin, L. Performing ICSI within 4 hours after denudation optimizes clinical outcomes in ICSI cycles. Reprod Biol Endocrinol 2020, 18, 27. [Google Scholar] [CrossRef]
- Nouvel, A.; Laget, J.; Duranton, F.; Leroy, J.; Desmetz, C.; Servais, M.D.; de Preville, N.; Galtier, F.; Nocca, D.; Builles, N.; et al. Optimization of RNA extraction methods from human metabolic tissue samples of the COMET biobank. Sci Rep 2021, 11, 20975. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, S. Mechanical/Physical Methods of Cell Disruption and Tissue Homogenization. Methods Mol Biol 2021, 2261, 563–585. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Attanasio, C.; Pedano, M.S.; Cadenas de Llano-Perula, M. Comparison of human dental tissue RNA extraction methods for RNA sequencing. Arch Oral Biol 2023, 148, 105646. [Google Scholar] [CrossRef]
- Choudhary, S.; Choudhary, R.K. Rapid and Efficient Method of Total RNA Isolation from Milk Fat for Transcriptome Analysis of Mammary Gland. Proceedings of the National Academy of Sciences, India Section B: Biological Sciences 2018, 89, 455–460. [Google Scholar] [CrossRef]
- Ali, N.; Rampazzo, R.C.P.; Costa, A.D.T.; Krieger, M.A. Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. Biomed Res Int 2017, 2017, 9306564. [Google Scholar] [CrossRef]
- Amiri Samani, S.; Naji, M.H. Effect of homogenizer pressure and temperature on physicochemical, oxidative stability, viscosity, droplet size, and sensory properties of Sesame vegetable cream. Food Sci Nutr 2019, 7, 899–906. [Google Scholar] [CrossRef]
- Pagani, S.; Maglio, M.; Sicuro, L.; Fini, M.; Giavaresi, G.; Brogini, S. RNA Extraction from Cartilage: Issues, Methods, Tips. Int J Mol Sci 2023, 24, 2120. [Google Scholar] [CrossRef]
- Yamamoto, T.; Nakashima, K.; Maruta, Y.; Kiriyama, T.; Sasaki, M.; Sugiyama, S.; Suzuki, K.; Fujisaki, H.; Sasaki, J.; Kaku-Ushiki, Y.; et al. Improved RNA extraction method using the BioMasher and BioMasher power-plus. J Vet Med Sci 2012, 74, 1561–1567. [Google Scholar] [CrossRef]
- Jones, G.M.; Cram, D.S.; Song, B.; Magli, M.C.; Gianaroli, L.; Lacham-Kaplan, O.; Findlay, J.K.; Jenkin, G.; Trounson, A.O. Gene expression profiling of human oocytes following in vivo or in vitro maturation. Human reproduction (Oxford, England) 2008, 23, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Wells, D.; Patrizio, P. Gene expression profiling of human oocytes at different maturational stages and after in vitro maturation. Am J Obstet Gynecol 2008, 198, 455–e451. [Google Scholar] [CrossRef]
- Trakunram, K.; Champoochana, N.; Chaniad, P.; Thongsuksai, P.; Raungrut, P. MicroRNA Isolation by Trizol-Based Method and Its Stability in Stored Serum and cDNA Derivatives. Asian Pac J Cancer Prev 2019, 20, 1641–1647. [Google Scholar] [CrossRef]
- Duy, J.; Koehler, J.W.; Honko, A.N.; Minogue, T.D. Optimized microRNA purification from TRIzol-treated plasma. BMC Genomics 2015, 16, 95. [Google Scholar] [CrossRef]
- Beltrame, C.O.; Cortes, M.F.; Bandeira, P.T.; Figueiredo, A.M. Optimization of the RNeasy Mini Kit to obtain high-quality total RNA from sessile cells of Staphylococcus aureus. Braz J Med Biol Res 2015, 48, 1071–1076. [Google Scholar] [CrossRef]
- Tavares, L.; Alves, P.M.; Ferreira, R.B.; Santos, C.N. Comparison of different methods for DNA-free RNA isolation from SK-N-MC neuroblastoma. BMC Res Notes 2011, 4, 3. [Google Scholar] [CrossRef]
- Al-Adsani, A.M.; Barhoush, S.A.; Bastaki, N.K.; Al-Bustan, S.A.; Al-Qattan, K.K. Comparing and Optimizing RNA Extraction from the Pancreas of Diabetic and Healthy Rats for Gene Expression Analyses. Genes (Basel) 2022, 13. [Google Scholar] [CrossRef]
- Ramon-Nunez, L.A.; Martos, L.; Fernandez-Pardo, A.; Oto, J.; Medina, P.; Espana, F.; Navarro, S. Comparison of protocols and RNA carriers for plasma miRNA isolation. Unraveling RNA carrier influence on miRNA isolation. PLoS One 2017, 12, e0187005. [Google Scholar] [CrossRef]
- Wright, K.; de Silva, K.; Purdie, A.C.; Plain, K.M. Comparison of methods for miRNA isolation and quantification from ovine plasma. Sci Rep 2020, 10, 825. [Google Scholar] [CrossRef]
- Ogunbayo, A.E.; Sabiu, S.; Nyaga, M.M. Evaluation of extraction and enrichment methods for recovery of respiratory RNA viruses in a metagenomics approach. J Virol Methods 2023, 314, 114677. [Google Scholar] [CrossRef]
| Parameter | Age (years) |
AMH Level (pmol/L) |
RNA concentration (ng/µl) |
RNA Purification | p- value |
|---|---|---|---|---|---|
| Denudation Technique | |||||
| Recruitment Profile | 37.19 (3.70) | 7.507(4.33) | 6.416(5.15) | 1.712(0.244) | |
| Enzymatic (Hyase) | 36.83 (3.75) | 8.99 (5.35) | 3.83 (1.20) | 1.720 (0.25) | P<0.05* |
| Non-Enzymatic (Non-Hyase) | 37.55 (3.67) | 6.02 (2.17) | 9.00 (6.22) | 1.703 (0.23) | |
| Homogenizer | |||||
| RSH | 37.50 (3.55) | 7.05 (4.05) | 3.68 (1.15) | 1.711(0.23) | P = 0.18* |
| BioMasher III | 36.88 (3.85) | 7.96 (4.59) | 9.15 (6.10) | 1.712 (0.25) | |
| cDNA & qPCR | |||||
| QuantiTect RT | 36.08(3.25) | 7.82(4.12) | 6.82(5.62) | 1.701(0.26) | |
| Whole Genome AK | 38.30 (3.82) | 7.19(4.56) | 6.01(4.68) | 1.722(0.23) | |
| Amplification of HKG | |||||
| Type of cDNA kit | Yes (n, %) |
No (n, %) |
|||
| QuantiTect RT | 20 (50) | 20 (50) | P<0.05# | ||
| Whole Genome AK | 0 | 40 (100) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





