Submitted:
29 February 2024
Posted:
06 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
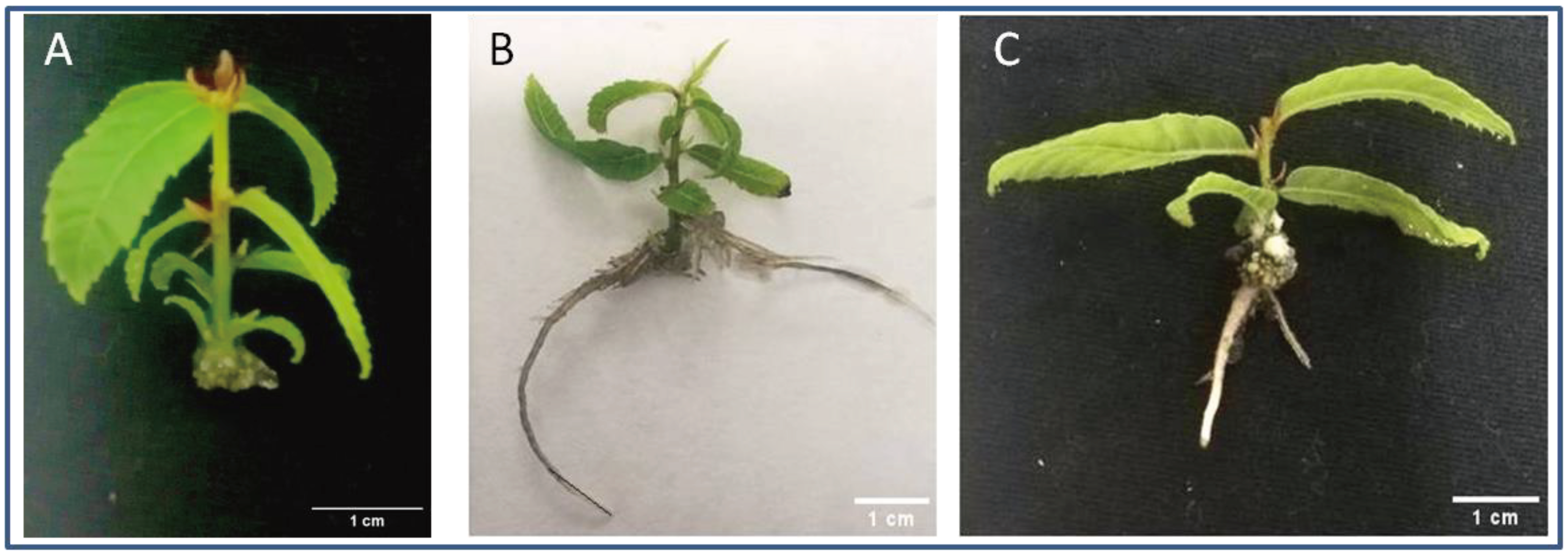
2.1. Plant Material, Wounding and Rooting Treatments
2.2. RNA Extraction, Library Preparation and Bioinformatics Pipeline
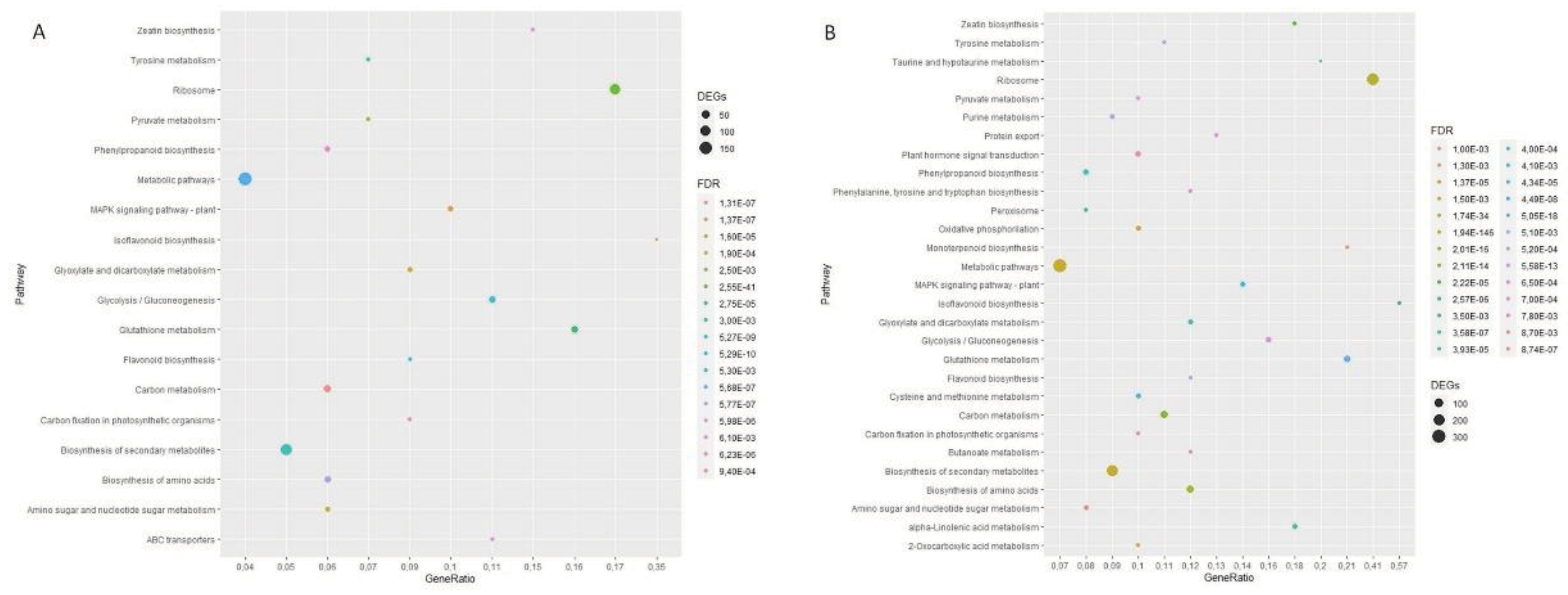
2.3. Differential Gene Expression and Pathway Enrichment Analysis
2.4. Gene Expression Analysis
3. Results
3.1. Auxin Treatment Optimization
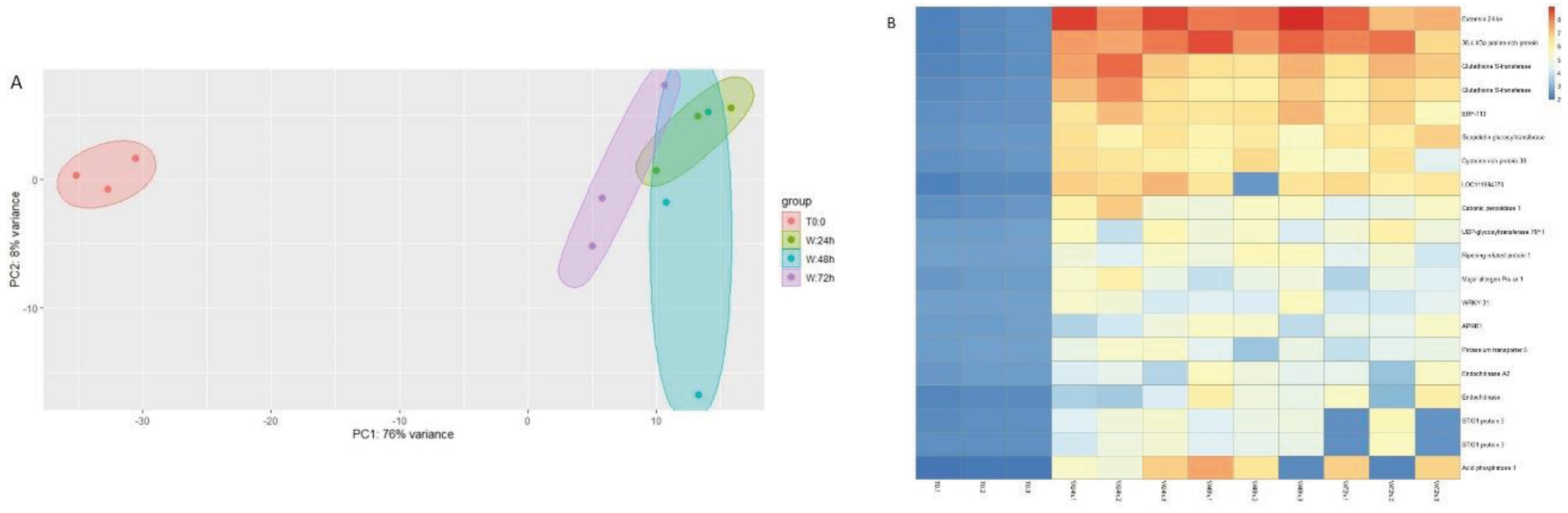
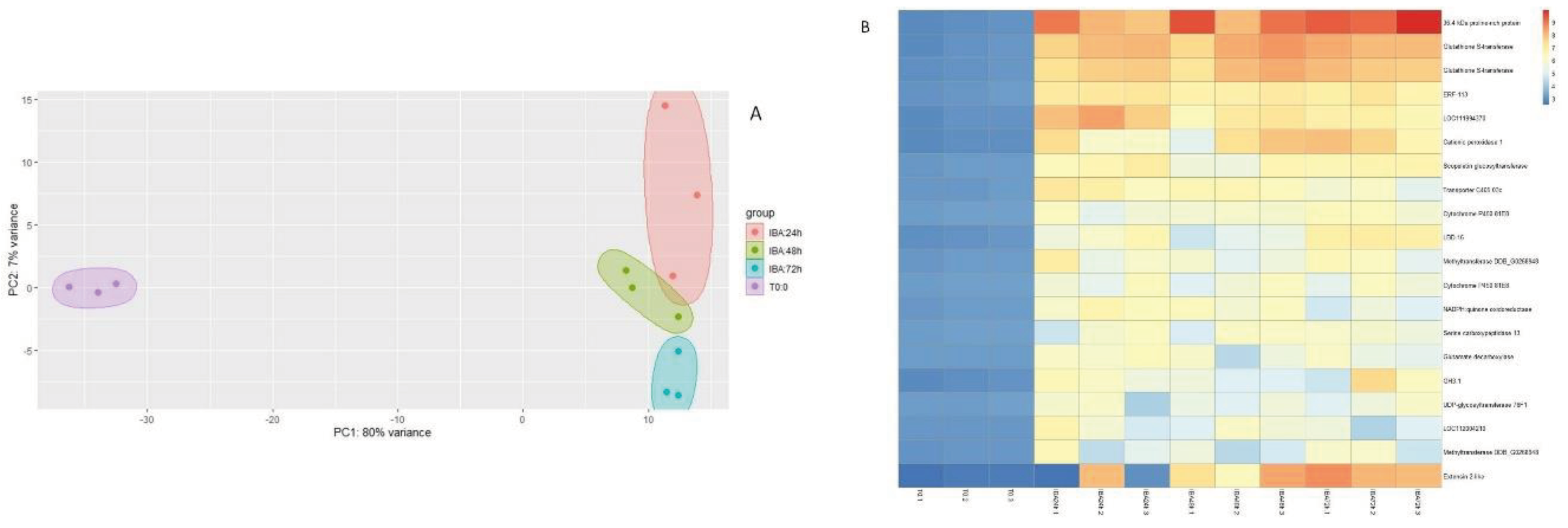
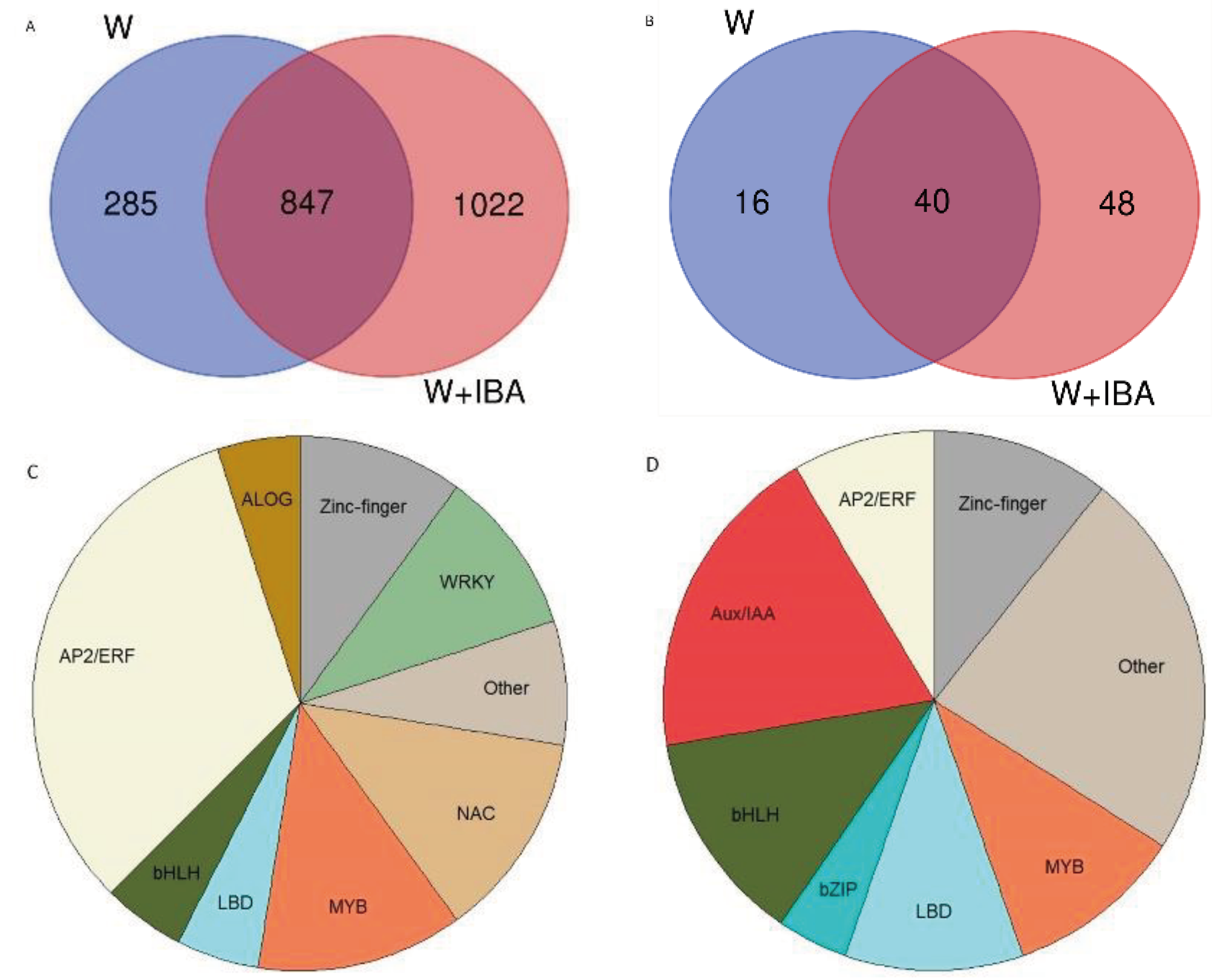
3.2. Transcriptomic Analysis
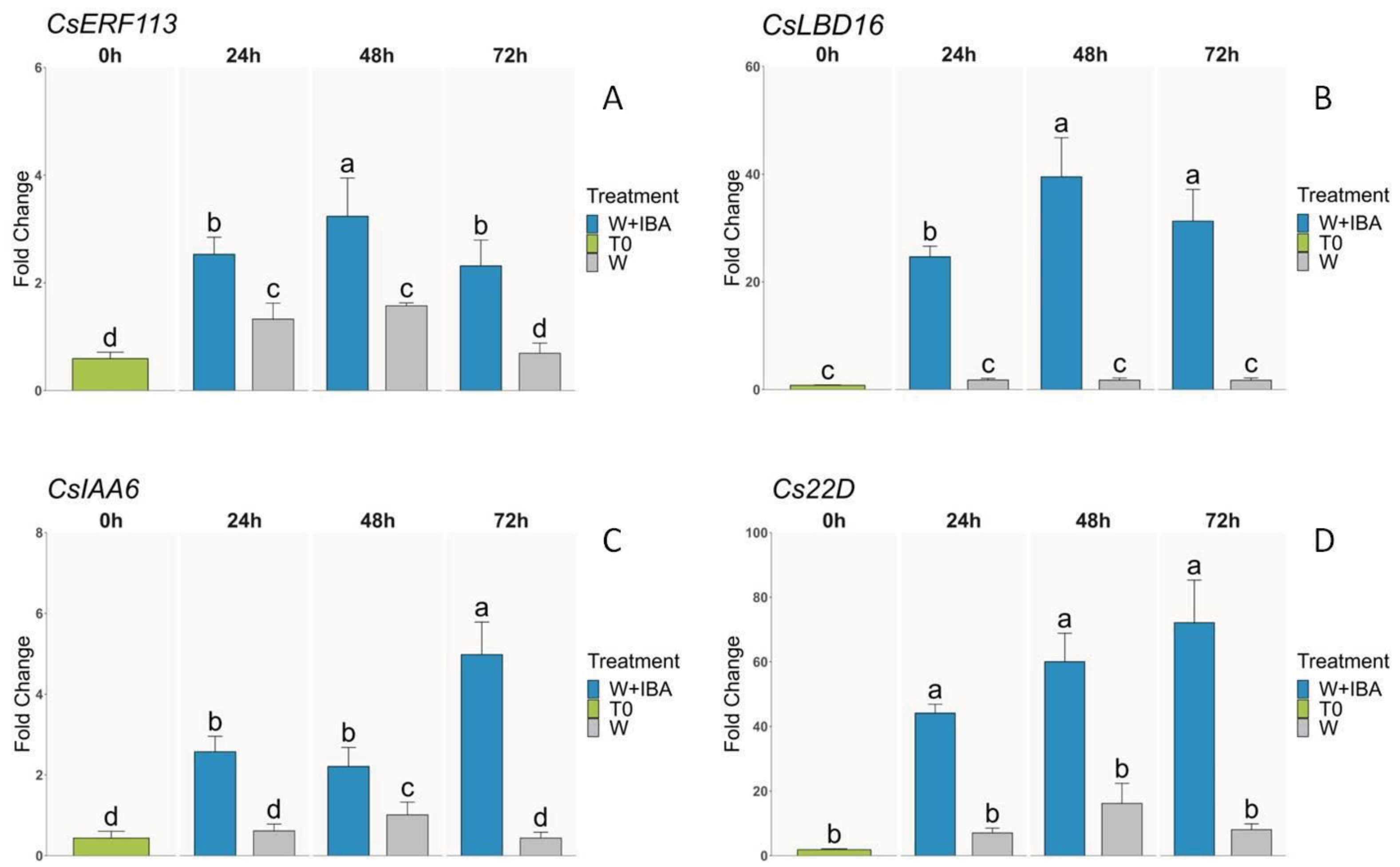
3.3. qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guan, L.; Murphy, A.S.; Peer, W.A.; Gan, L.; Li, Y.; Cheng, Z.-M. (Max) Physiological and Molecular Regulation of Adventitious Root Formation. CRC. Crit. Rev. Plant Sci. 2015, 34, 506–521. [Google Scholar] [CrossRef]
- Druege, U.; Franken, P.; Hajirezaei, M.R. Plant Hormone Homeostasis, Signaling, and Function during Adventitious Root Formation in Cuttings. Front. Plant Sci. 2016, 7, 381. [Google Scholar] [CrossRef]
- Gonin; Bergougnoux; Nguyen; Gantet; Champion What Makes Adventitious Roots? Plants 2019, 8, 240. [CrossRef]
- Vielba; Vidal, N.; San-José, M.C.; Rico, S.; Sánchez, C.; José, M.C.S.; Rico, S.; Sánchez, C. Recent Advances in Adventitious Root Formation in Chestnut. Plants 2020, 9, 1543. [Google Scholar] [CrossRef]
- Bannoud, F.; Bellini, C. Adventitious Rooting in Populus Species: Update and Perspectives. Front. Plant Sci. 2021, 12, 7. [Google Scholar] [CrossRef]
- Brunoni, F.; Vielba, J.M.; Sánchez, C. Plant Growth Regulators in Tree Rooting. Plants 2022, 11, 805. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Favero, D.S.; Sakamoto, Y.; Iwase, A.; Coleman, D.; Rymen, B.; Sugimoto, K. Molecular Mechanisms of Plant Regeneration. Annu. Rev. Plant Biol. 2019, 70, 377–406. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Heyman, J.; Lu, R.; De Veylder, L. Evolution of wound-activated regeneration pathways in the plant kingdom. Eur. J. Cell Biol. 2023, 102, 151291. [Google Scholar] [CrossRef] [PubMed]
- Vielba; Vidal, N.; Ricci, A.; Castro, R.; Covelo, P.; San-Jose, M.C.; Sanchez, C. Effects of auxin and urea derivatives on adventitious rooting in chestnut and oak microshoots. Isr. J. Plant Sci. 2020, 67, 52–68. [Google Scholar] [CrossRef]
- Ballester, A.; San-José, M.C.; Vidal, N.; Fernández-Lorenzo, J.L.; Vieitez, A.M. Anatomical and Biochemical Events during in vitro Rooting of Microcuttings from Juvenile and Mature Phases of Chestnut. Ann. Bot. 1999, 83, 619–629. [Google Scholar] [CrossRef]
- Vielba, J.M.; Rico, S.; Sevgin, N.; Castro-Camba, R.; Covelo, P.; Vidal, N.; Sanchez, C.; Sánchez, C. Transcriptomics Analysis Reveals a Putative Role for Hormone Signaling and MADS-Box Genes in Mature Chestnut Shoots Rooting Recalcitrance. Plants (Basel) 2022, 11, 3486. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.C.; Vieitez, A.M. In vitro morphogenetic competence of basal sprouts and crown branches of mature chestnut. Tree Physiol. 1991, 8, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Gresshoff, P.M.; Doy, C.H. Development and differentiation of haploid Lycopersicon esculentum (tomato). Planta 1972, 107, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Cribari-Neto, F.; Zeileis, A. Beta Regression in R. J. Stat. Softw. 2010, 34. [Google Scholar] [CrossRef]
- Lenth, R. _emmeans: Estimated Marginal Means, aka Least-Squares Means_. 2023.
- R Core Team R: A language and environment for statistical computing. 2021.
- De Coster, W.; Rademakers, R. NanoPack2: population-scale evaluation of long-read sequencing data. Bioinformatics 2023, 39. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.M.; Usié, A.; Barbosa, P.; Barros, P.M.; Capote, T.; Chaves, I.; Simões, F.; Abreu, I.; Carrasquinho, I.; Faro, C.; et al. The draft genome sequence of cork oak. Sci. Data 2018, 5, 180069. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- da Costa, C.T.; de Almeida, M.R.; Ruedell, C.M.; Schwambach, J.; Maraschin, F.S.; Fett-Neto, A.G. When stress and development go hand in hand: main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 2013, 4, 133. [Google Scholar] [CrossRef]
- Druege, U.; Hilo, A.; Pérez-Pérez, J.M.; Klopotek, Y.; Acosta, M.; Shahinnia, F.; Zerche, S.; Franken, P.; Hajirezaei, M.R. Molecular and physiological control of adventitious rooting in cuttings: phytohormone action meets resource allocation. Ann. Bot. 2019, 123, 929–949. [Google Scholar] [CrossRef]
- Castro-Camba, R.; Neves, M.; Correia, S.; Canhoto, J.; Vielba, J.M. Ethylene Action-Inhibition Improves Adventitious Root Induction in Adult Chestnut Tissues. Preprint 2023. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Kawamura, A.; Suzuki, T.; Segami, S.; Maeshima, M.; Polyn, S.; De Veylder, L.; Sugimoto, K. Transcriptional activation of auxin biosynthesis drives developmental reprogramming of differentiated cells. Plant Cell 2022, 34, 4348–4365. [Google Scholar] [CrossRef]
- Bairu, M.W.; Novák, O.; Doležal, K.; Van Staden, J. Changes in endogenous cytokinin profiles in micropropagated Harpagophytum procumbens in relation to shoot-tip necrosis and cytokinin treatments. Plant Growth Regul. 2011, 63, 105–114. [Google Scholar] [CrossRef]
- Lup, S.D.; Tian, X.; Xu, J.; Pérez-Pérez, J.M. Wound signaling of regenerative cell reprogramming. Plant Sci. 2016, 250, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Gordon, S.P.; Meyerowitz, E.M. Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 2011, 21, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Fehér, A. Callus, Dedifferentiation, Totipotency, Somatic Embryogenesis: What These Terms Mean in the Era of Molecular Plant Biology? Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Heyman, J.; Canher, B.; Bisht, A.; Christiaens, F.; De Veylder, L. Emerging role of the plant ERF transcription factors in coordinating wound defense responses and repair. J. Cell Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cai, G.; Zhang, K.; Sun, H.; Huang, L.; Ren, W.; Ding, Y.; Wang, N. PdeERF114 recruits PdeWRKY75 to regulate callus formation in poplar by modulating the accumulation of H 2 O 2 and the relaxation of cell walls. New Phytol. 2024, 241, 732–746. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Xu, C.; Xu, K.; Hu, Y. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012, 22, 1169–1180. [Google Scholar] [CrossRef]
- Jing, T.; Ardiansyah, R.; Xu, Q.; Xing, Q.; Müller-Xing, R. Reprogramming of Cell Fate During Root Regeneration by Transcriptional and Epigenetic Networks. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Wang, R.; Bai, T.; Gao, H.; Cui, Y.; Zhou, R.; Wang, Z.; Song, S.; Jiao, J.; Wang, M.; Wan, R.; et al. Genome-wide identification of LBD transcription factors in apple and the function of MdLBD16a in adventitious rooting and callus development. Sci. Hortic. (Amsterdam). 2023, 317, 112048. [Google Scholar] [CrossRef]
- Xu, L. De novo root regeneration from leaf explants: wounding, auxin, and cell fate transition. Curr. Opin. Plant Biol. 2018, 41, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Korasick, D.A.; Westfall, C.S.; Lee, S.G.; Nanao, M.H.; Dumas, R.; Hagen, G.; Guilfoyle, T.J.; Jez, J.M.; Strader, L.C. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. 2014, 111, 5427–5432. [Google Scholar] [CrossRef] [PubMed]
- Karannagoda, N.; Spokevicius, A.; Hussey, S.; Cassan-Wang, H.; Grima-Pettenati, J.; Bossinger, G. Eucalyptus grandis AUX/INDOLE-3-ACETIC ACID 13 (EgrIAA13) is a novel transcriptional regulator of xylogenesis. Plant Mol. Biol. 2022, 109, 51–65. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.-J.; Zhang, J.-Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef]
- Ma, G.; Li, M.; Wu, Y.; Jiang, C.; Chen, Y.; Xing, D.; Zhao, Y.; Liu, Y.; Jiang, X.; Xia, T.; et al. Camellia sinensis CsMYB4a participates in regulation of stamen growth by interaction with auxin signaling transduction repressor CsAUX/IAA4. Crop J. 2023. [Google Scholar] [CrossRef]
- Lakehal, A.; Chaabouni, S.; Cavel, E.; Le Hir, R.; Ranjan, A.; Raneshan, Z.; Novák, O.; Păcurar, D.I.; Perrone, I.; Jobert, F.; et al. A Molecular Framework for the Control of Adventitious Rooting by TIR1/AFB2-Aux/IAA-Dependent Auxin Signaling in Arabidopsis. Mol. Plant 2019, 12, 1499–1514. [Google Scholar] [CrossRef]
| Clone | IBA (μM) | Rooting (%) | Root number | Root length (cm) | Callus (%) | Apical necrosis (%) | T50 | Active growth (%) |
|---|---|---|---|---|---|---|---|---|
| P2BS | 0 | 0 b | - | - | 100 ± 0 a | 83.3 ± 4.1 a | - | - |
| 2.5 | 97.2 ± 4.8 a | 3.0 ± 1.4 a | 4.4 ± 1.8 a | 2.8 ± 1.4 c | 2.8 ± 4.8 e | 8 a | 100 ± 0 a | |
| 5 | 97.2 ± 4.8 a | 3.7 ± 1.0 a | 4.1 ± 0.8 a | 2.8 ± 1.4 c | 27.7 ± 4.1 d | 7 a | 69.9 ± 5.8 b | |
| 10 | 94.4 ± 4.8 a | 4.3 ± 1.9 a | 4.2 ± 1.5 a | 5.3 ± 2.1 c | 27.7 ± 4.1 d | 8 a | 30.3 ± 4.6 c | |
| 12.5 | 94.4 ± 9.3 a | 4.2 ± 1.9 a | 4.1 ± 1.2 a | 27.7 ± 4.6 b | 55.3 ± 9.2 c | 8 a | 20.4 ± 11.7 d | |
| 25 | 93.2 ± 4.6 a | 4.0 ± 1.8 a | 3.5 ± 1.4 a | 100 ± 0 a | 72.0 ± 5.1 b | 7 a | 14.0 ± 5.2 d |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





