Submitted:
26 February 2024
Posted:
26 February 2024
You are already at the latest version
Abstract
Keywords:
1.Introduction
2. Results
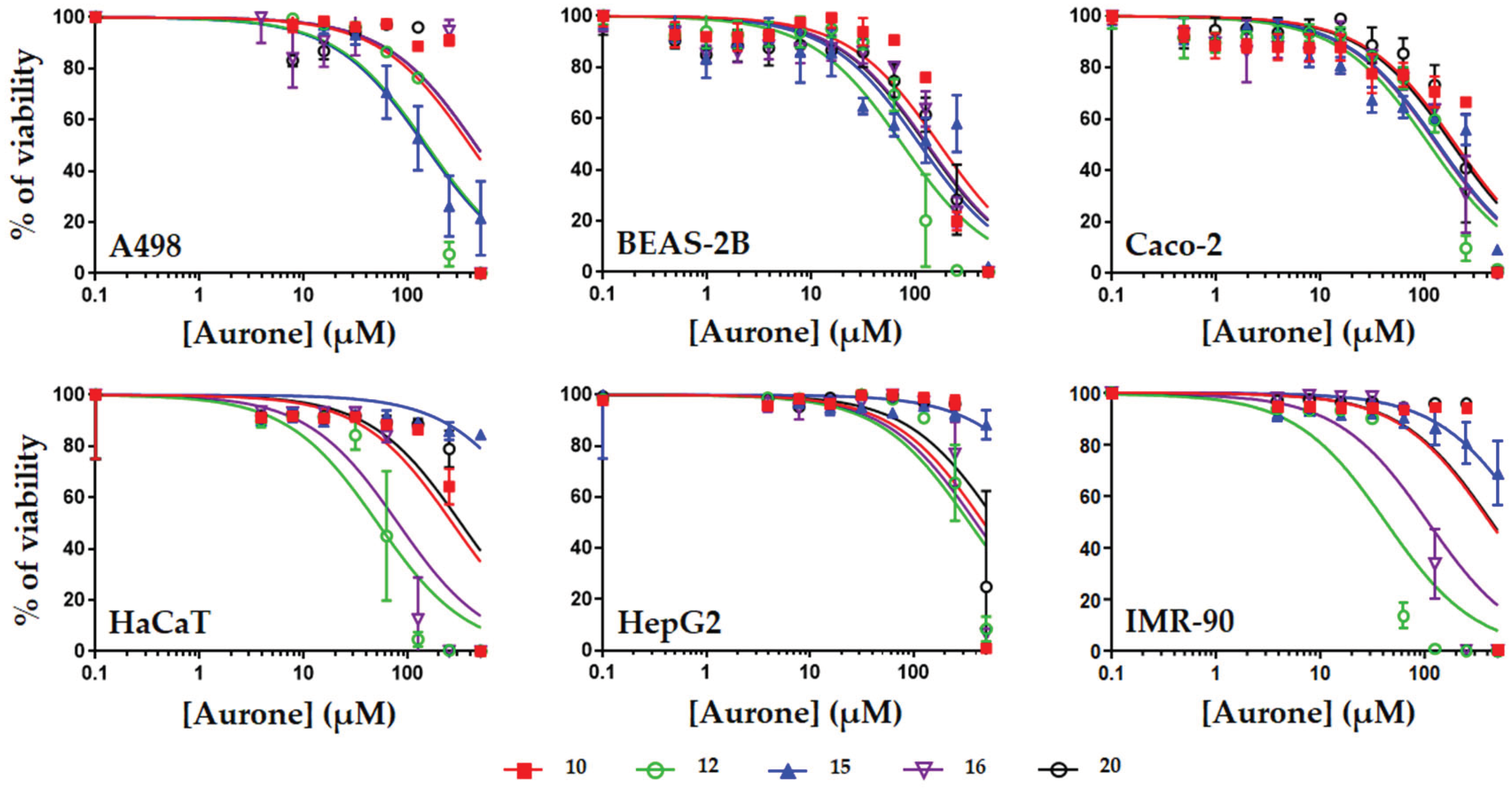
3. Discussion
4. Materials and Methods
4.1. Biology
4.1.1. Microorganism Strains and Growth Conditions
4.1.2. Antimicrobial Activity Assay
4.1.3. Cytotoxic Assays
4.2. Chemistry
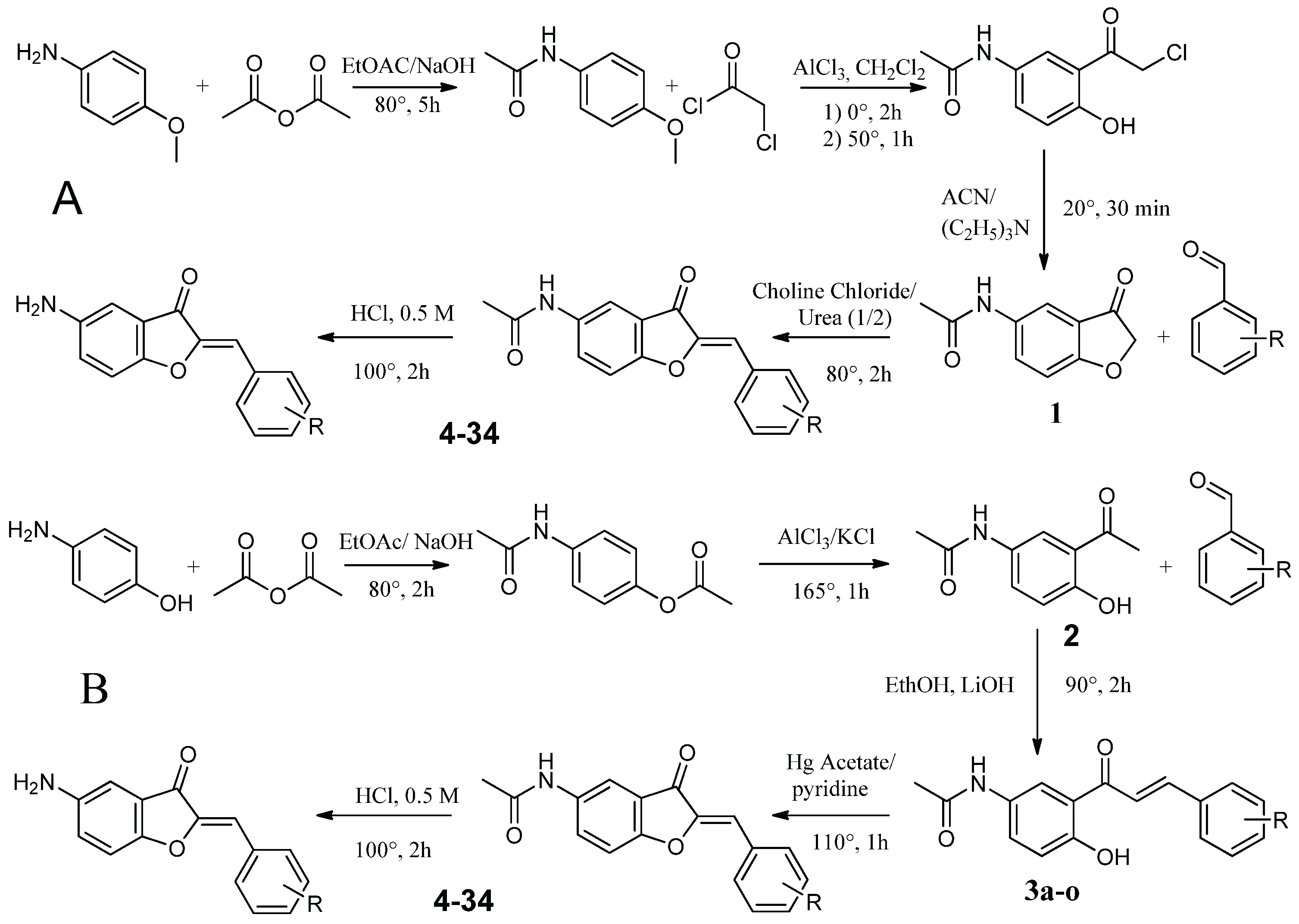
4.2.1. Synthesis route A
4.2.1.1. Synthesis of N-(4-methoxyphenyl)acetamide (1a)
4.2.1.2. Synthesis of 2-chloro-1-{2-hydroxy-5-[(1-hydroxyethyl)amino]cyclohexyl}ethan-1-one (2a)
4.2.1.3. Synthesis of N-(3-oxo-2,3-dihydro-1-benzofuran-5-yl)acetamide (3a)
4.2.1.4. Synthesis of substituted 5-acetamidoaurones
4.2.1.5. Synthesis of substituted 5-aminoaurone
4.2.2. Synthesis Route B
4.2.2.1. Synthesis of 4-acetamidophenyl acetate (1b)
4.2.2.2. Synthesis of N-(3-acetyl-4-hydroxyphenyl)acetamide (2b)
4.2.2.3. Synthesis of substituted of 5-acetamidochalcones (3b)
4.2.2.4. Synthesis of substituted 5-acetamidoaurones
4.2.2.5. Synthesis of substituted 5-aminoaurone
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interests
References
- Abdeen, S.; Kunkle, T.; Salim, N.; Ray, A.-M.; Mammadova, N.; Summers, C.; Stevens, M.; Ambrose, A.J.; Park, Y.; Schultz, P.G.; et al. Sulfonamido-2-Arylbenzoxazole GroEL/ES Inhibitors as Potent Antibacterials against Methicillin-Resistant Staphylococcus Aureus (MRSA). J Med Chem 2018, 61, 7345–7357. [Google Scholar] [CrossRef]
- Cui, X.; Wang, L.; Lü, Y.; Yue, C. Development and Research Progress of Anti-Drug Resistant Fungal Drugs. J Infect Public Health 2022, 15, 986–1000. [Google Scholar] [CrossRef]
- Bastos, R.W.; Rossato, L.; Goldman, G.H.; Santos, D.A. Fungicide Effects on Human Fungal Pathogens: Cross-Resistance to Medical Drugs and Beyond. PLOS Pathogens 2021, 17, e1010073. [Google Scholar] [CrossRef]
- Sati, H.; Alastruey-Izquierdo, A.; Perfect, J.; Govender, N.P.; Harrison, T.S.; Chiller, T.; Sorrell, T.C.; Bongomin, F.; Oladele, R.; Chakrabarti, A.; et al. HIV and Fungal Priority Pathogens. Lancet HIV 2023, 10, e750–e754. [Google Scholar] [CrossRef] [PubMed]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases—Estimate Precision. Journal of Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
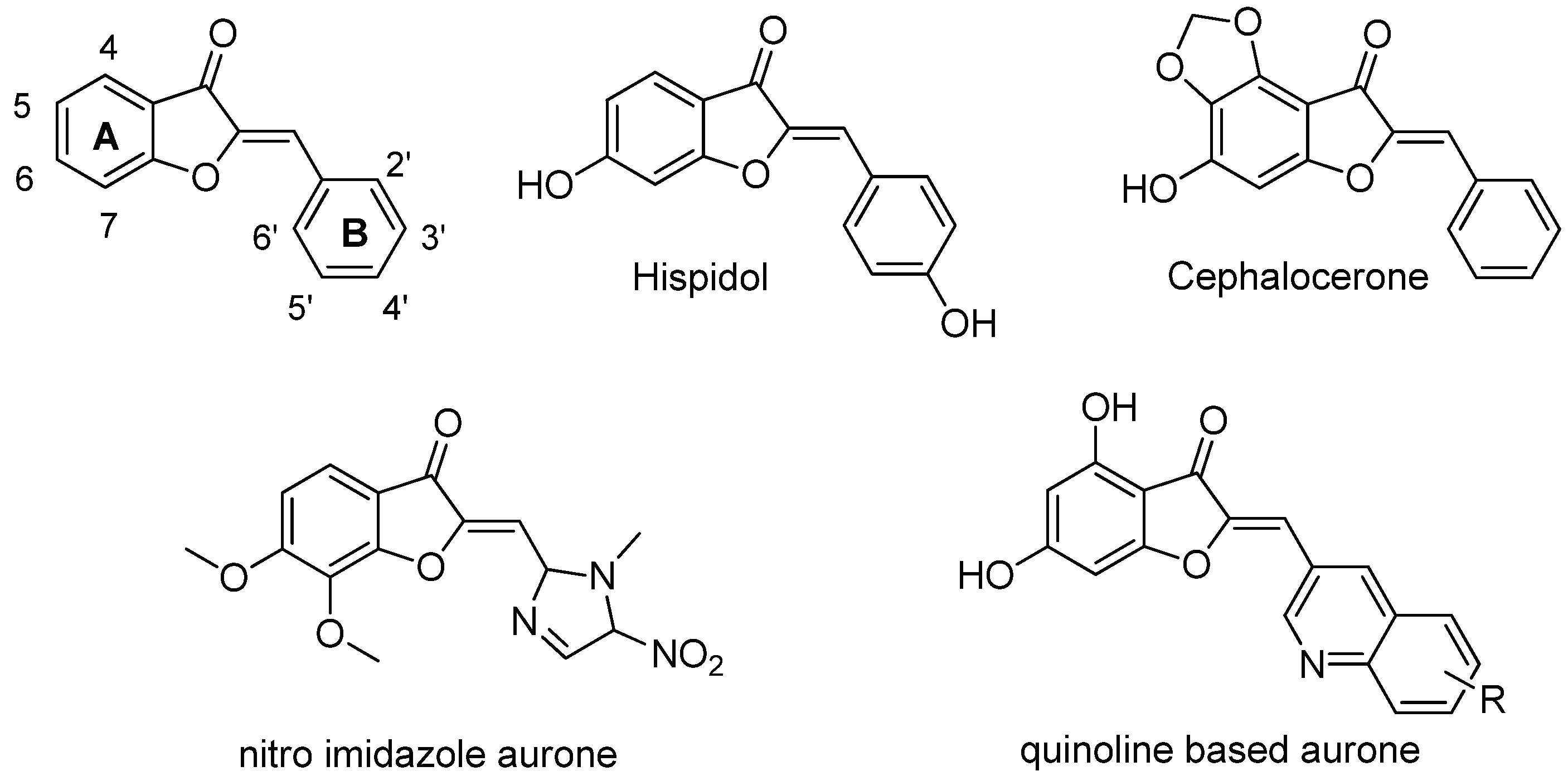
- Haudecoeur, R.; Boumendjel, A. Recent Advances in the Medicinal Chemistry of Aurones. Curr Med Chem 2012, 19, 2861–2875. [Google Scholar] [CrossRef] [PubMed]
- Boumendjel, A. Aurones: A Subclass of Flavones with Promising Biological Potential. Curr Med Chem 2003, 10, 2621–2630. [Google Scholar] [CrossRef] [PubMed]
- Lazinski, L.M.; Royal, G.; Robin, M.; Maresca, M.; Haudecoeur, R. Bioactive Aurones, Indanones, and Other Hemiindigoid Scaffolds: Medicinal Chemistry and Photopharmacology Perspectives. J. Med. Chem. 2022, 65, 12594–12625. [Google Scholar] [CrossRef]
- Boucherle, B.; Peuchmaur, M.; Boumendjel, A.; Haudecoeur, R. Occurrences, Biosynthesis and Properties of Aurones as High-End Evolutionary Products. Phytochemistry 2017, 142, 92–111. [Google Scholar] [CrossRef]
- Pare, P.W.; Dmitrieva, N.; Mabry, T.J. Phytoalexin Aurone Induced in Cephalocereus Senilis Liquid Suspension Culture. Phytochemistry 1991, 30, 1133–1135. [Google Scholar] [CrossRef]
- Pare´, P.W.; Mischke, C.F.; Edwards, R.; Dixon, R.A.; Norman, H.A.; Mabry, T.J. Induction of Phenylpropanoid Pathway Enzymes in Elicitor-Treated Cultures ofCephalocereus Senilis. Phytochemistry 1992, 31, 149–153. [Google Scholar] [CrossRef]
- Farag, M.A.; Deavours, B.E.; de Fátima, A.; Naoumkina, M.; Dixon, R.A.; Sumner, L.W. Integrated Metabolite and Transcript Profiling Identify a Biosynthetic Mechanism for Hispidol in Medicago Truncatula Cell Cultures. Plant Physiol 2009, 151, 1096–1113. [Google Scholar] [CrossRef] [PubMed]
- Olleik, H.; Yahiaoui, S.; Roulier, B.; Courvoisier-Dezord, E.; Perrier, J.; Pérès, B.; Hijazi, A.; Baydoun, E.; Raymond, J.; Boumendjel, A.; et al. Aurone Derivatives as Promising Antibacterial Agents against Resistant Gram-Positive Pathogens. Eur J Med Chem 2019, 165, 133–141. [Google Scholar] [CrossRef]
- Tiwari, K.N.; Monserrat, J.-P.; Hequet, A.; Ganem-Elbaz, C.; Cresteil, T.; Jaouen, G.; Vessières, A.; Hillard, E.A.; Jolivalt, C. In Vitro Inhibitory Properties of Ferrocene-Substituted Chalcones and Aurones on Bacterial and Human Cell Cultures. Dalton Trans. 2012, 41, 6451–6457. [Google Scholar] [CrossRef] [PubMed]
- Hadj-esfandiari, N.; Navidpour, L.; Shadnia, H.; Amini, M.; Samadi, N.; Faramarzi, M.A.; Shafiee, A. Synthesis, Antibacterial Activity, and Quantitative Structure–Activity Relationships of New (Z)-2-(Nitroimidazolylmethylene)-3(2H)-Benzofuranone Derivatives. Bioorganic & Medicinal Chemistry Letters 2007, 17, 6354–6363. [Google Scholar] [CrossRef]
- Kumar, G.; Lathwal, E.; Saroha, B.; Kumar, S.; Kumar, S.; Chauhan, N.S.; Kumar, T. Synthesis and Biological Evaluation of Quinoline-Based Novel Aurones. ChemistrySelect 2020, 5, 3539–3543. [Google Scholar] [CrossRef]
- Olleik, H.; Nicoletti, C.; Lafond, M.; Courvoisier-Dezord, E.; Xue, P.; Hijazi, A.; Baydoun, E.; Perrier, J.; Maresca, M. Comparative Structure-Activity Analysis of the Antimicrobial Activity, Cytotoxicity, and Mechanism of Action of the Fungal Cyclohexadepsipeptides Enniatins and Beauvericin. Toxins (Basel) 2019, 11, 514. [Google Scholar] [CrossRef]
- Berthier, M.; Fauchère, J.-L.; Perrin, J.; Grignon, B.; Oriot, D. Fulminant Meningitis Due to Bacillus Anthracis in 11-Year-Old Girl during Ramadan. The Lancet 1996, 347, 828. [Google Scholar] [CrossRef] [PubMed]
- Doll, J.M.; Zeitz, P.S.; Ettestad, P.; Bucholtz, A.L.; Davis, T.; Gage, K. Cat-Transmitted Fatal Pneumonic Plague in a Person Who Traveled from Colorado to Arizona. The American Journal of Tropical Medicine and Hygiene 1994, 51, 109–114. [Google Scholar] [CrossRef]
- Olleik, H.; Yacoub, T.; Hoffer, L.; Gnansounou, S.M.; Benhaiem-Henry, K.; Nicoletti, C.; Mekhalfi, M.; Pique, V.; Perrier, J.; Hijazi, A.; et al. Synthesis and Evaluation of the Antibacterial Activities of 13-Substituted Berberine Derivatives. Antibiotics 2020, 9, 381. [Google Scholar] [CrossRef]
- Casanova, M.; Olleik, H.; Hdiouech, S.; Roblin, C.; Cavalier, J.-F.; Point, V.; Jeannot, K.; Caron, B.; Perrier, J.; Charriau, S.; et al. Evaluation of the Efficiency of Random and Diblock Methacrylate-Based Amphiphilic Cationic Polymers against Major Bacterial Pathogens Associated with Cystic Fibrosis. Antibiotics (Basel) 2023, 12, 120. [Google Scholar] [CrossRef]
- Loupias, P.; Laumaillé, P.; Morandat, S.; Mondange, L.; Guillier, S.; El Kirat, K.; Da Nascimento, S.; Biot, F.; Taudon, N.; Dassonville-Klimpt, A.; et al. Synthesis and Study of New Siderophore Analog-Ciprofloxacin Conjugates with Antibiotic Activities against Pseudomonas Aeruginosa and Burkholderia Spp. European Journal of Medicinal Chemistry 2023, 245, 114921. [Google Scholar] [CrossRef]
- Broncano-Lavado, A.; Senhaji-Kacha, A.; Santamaría-Corral, G.; Esteban, J.; García-Quintanilla, M. Alternatives to Antibiotics against Mycobacterium Abscessus. Antibiotics 2022, 11, 1322. [Google Scholar] [CrossRef] [PubMed]
- Madani, A.; Ridenour, J.N.; Martin, B.P.; Paudel, R.R.; Abdul Basir, A.; Le Moigne, V.; Herrmann, J.-L.; Audebert, S.; Camoin, L.; Kremer, L.; et al. Cyclipostins and Cyclophostin Analogues as Multitarget Inhibitors That Impair Growth of Mycobacterium Abscessus. ACS Infect. Dis. 2019, 5, 1597–1608. [Google Scholar] [CrossRef] [PubMed]
- Benkhaled, B.T.; Hadiouch, S.; Olleik, H.; Perrier, J.; Ysacco, C.; Guillaneuf, Y.; Gigmes, D.; Maresca, M.; Lefay, C. Elaboration of Antimicrobial Polymeric Materials by Dispersion of Well-Defined Amphiphilic Methacrylic SG1-Based Copolymers. Polym. Chem. 2018, 9, 3127–3141. [Google Scholar] [CrossRef]
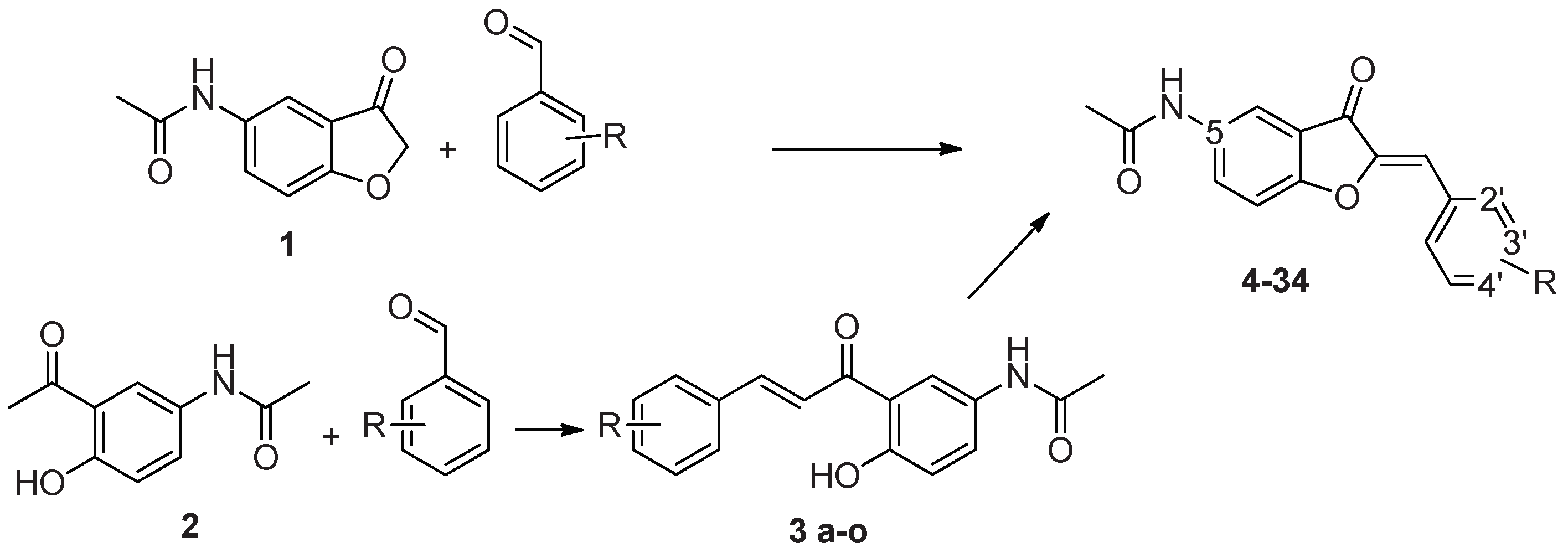
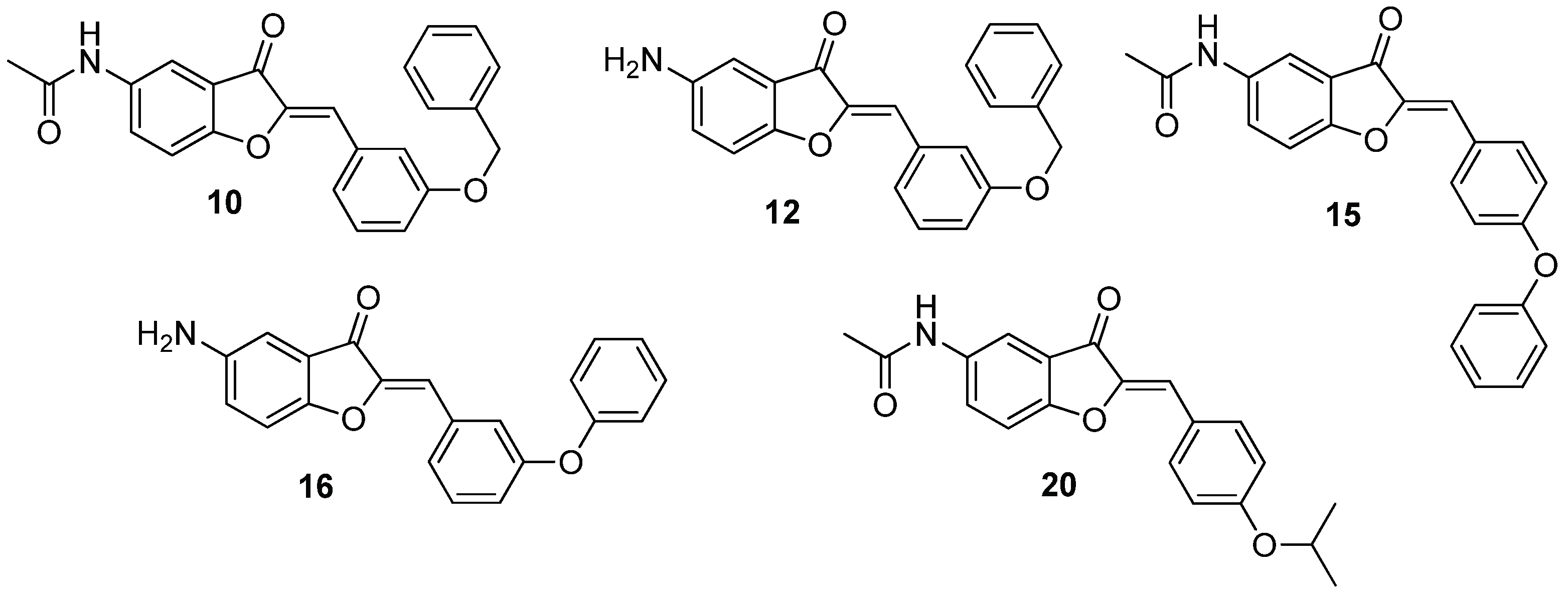
| Compound | 5 | 2’ | 3’ | 4’ |
| 4 | NHCOCH3 | OCH3 | H | H H OCH3 H |
| 5 | NHCOCH3 | H | OCH3 | |
| 6 | NHCOCH3 | H | H | |
| 7 | NH2 | OCH3 | H | |
| 8 | NH2 | H | OCH3 | H |
| 9 | NH2 | H | H | OCH3 |
| 10 | NHCOCH3 | H | Obenzyl | H |
| 11 | NHCOCH3 | H | H | Obenzyl |
| 12 | NH2 | H | Obenzyl | H |
| 13 | NH2 | H | H | Obenzyl |
| 14 | NHCOCH3 | H | Ophenyl | H |
| 15 | NHCOCH3 | H | H | Ophenyl |
| 16 | NH2 | H | Ophenyl | H |
| 17 | NH2 | H | H | Ophenyl |
| 18 | NHCOCH3 | Oisopropyl | H | H |
| 19 | NHCOCH3 | H | Oisopropyl | H |
| 20 | NHCOCH3 | H | H | Oisopropyl |
| 21 | NH2 | Oisopropyl | H | H |
| 22 | NH2 | H | Oisopropyl | H |
| 23 | NH2 | H | H | Oisopropyl |
| 24 | NHCOCH3 | F | H | H |
| 25 | NHCOCH3 | H | F | H |
| 26 | NH2 | F | H | H |
| 27 | NH2 | H | F | H |
| 28 | NH2 | H | H | F |
| 29 | NHCOCH3 | H | CF3 | H |
| 30 | NH2 | H | CF3 | H |
| 31 | NHCOCH3 | H | H | COOH |
| 32 | NHCOCH3 | H | OH | H |
| 33 | NH2 | H | OH | H |
| 34 | NHCOCH3 | H | H | H |
| Gram positive | Gram negative | Mycobacteria | Fungi | |||
|
B. subtilis ATCC6633 |
S. aureus ATCC6538P |
E. coli ATCC8739 |
P. aeruginosa ATCC9027 |
M. smegmatis ATCC700084 |
C. albicans DSM10697 |
|
| Gemifloxacin | 0.03 | 0.06 | 0.06 | 0.25 | 2 | - |
| Amphotericin B | - | - | - | - | - | 0.62 |
| 4 | 50 | 100 | >100 | >100 | >100 | >100 |
| 5 | >100 | 100 | >100 | >100 | 100 | >100 |
| 6 | >100 | >100 | >100 | >100 | >100 | >100 |
| 7 | >100 | >100 | >100 | >100 | >100 | >100 |
| 8 | >100 | >100 | >100 | >100 | >100 | >100 |
| 9 | >100 | >100 | >100 | >100 | >100 | >100 |
| 10 | 3.12 | 12.5 | 12.5 | 25 | 50 | 50 |
| 11 | >100 | >100 | >100 | >100 | >100 | >100 |
| 12 | 25 | 100 | >100 | >100 | 50 | >100 |
| 13 | 100 | >100 | >100 | >100 | >100 | >100 |
| 14 | >100 | >100 | >100 | >100 | >100 | >100 |
| 15 | 50 | 100 | >100 | >100 | 50 | >100 |
| 16 | 25 | 25 | 25 | >100 | 50 | >100 |
| 17 | >100 | >100 | >100 | >100 | >100 | >100 |
| 18 | >100 | >100 | >100 | >100 | >100 | >100 |
| 19 | >100 | >100 | >100 | 25 | >100 | >100 |
| 20 | 25 | 12.5 | 25 | >100 | 50 | 50 |
| 21 | >100 | >100 | >100 | >100 | >100 | >100 |
| 22 | >100 | >100 | >100 | >100 | >100 | >100 |
| 23 | >100 | >100 | >100 | >100 | >100 | >100 |
| 24 | >100 | >100 | >100 | >100 | >100 | >100 |
| 25 | 50 | >100 | >100 | >100 | 100 | >100 |
| 26 | 50 | >100 | >100 | >100 | >100 | >100 |
| 27 | >100 | >100 | >100 | >100 | >100 | >100 |
| 28 | >100 | >100 | >100 | >100 | >100 | >100 |
| 29 | >100 | >100 | >100 | >100 | >100 | >100 |
| 30 | >100 | >100 | >100 | >100 | >100 | >100 |
| 31 | 50 | 100 | >100 | >100 | 100 | >100 |
| 32 | >100 | >100 | >100 | >100 | >100 | >100 |
| 33 | >100 | >100 | >100 | >100 | >100 | >100 |
| 34 | 50 | >100 | >100 | >100 | >100 | >100 |
| Compound | 10 | 12 | 15 | 16 | 20 |
|---|---|---|---|---|---|
| A498 | 398.2+/-164.6 | 152.4+/-31.4 | 145.7+/-17.3 | 452.3+/-147.9 | 453.0+/-46.4 |
| BEAS-2B | 169.0+/-28.3 | 74.6+/-12.1 | 109.5+/-17.4 | 129.6+/-18.3 | 125.9+/-17.2 |
| Caco-2 | 199.6+/-33.5 | 111.7+/-15.0 | 136.8+/-7.2 | 131.7+/-18.0 | 186.5+/-27.8 |
| HaCaT | 268.5+/-51.6 | 51.3+/-9.5 | >500 | 80.4+/-17.8 | 322.9+/-73.2 |
| HepG2 | 472.4+/-145.9 | 343.4+/-68.0 | >500 | 397.8+/-94.1 | >500 |
| IMR-90 | 421.4+/-119.3 | 42.4+/-9.6 | >500 | 116.5+/-29.4 | 437.6+/-128 |
| Mean CC50 | 321.5 | 129.3 | 130.6 | 218.0 | 305.1 |
| Compound | 10 | 12 | 15 | 16 | 20 |
|---|---|---|---|---|---|
| Lowest MIC | 3.12 | 25 | 50 | 25 | 12.5 |
| Lowest CC50 | 169.0 | 42.4 | 109.5 | 80.4 | 125.9 |
| Highest CC50 | 472.4 | 343.4 | >500 | 452.3 | >500 |
| Lowest TI | 54.1 | 1.6 | 2.19 | 3.2 | 10.0 |
| Highest TI | 151.4 | 13.7 | >10 | 18.0 | >40 |
| 10 | 20 | |
|---|---|---|
| Gram positive | ||
| Bacillus anthracis (CNR-charbon_04022) | 12.5 | 6.25 |
| Bacillus cereus (DSM31) | 12.5 | 25 |
| Bacillus subtilis (ATCC6633) | 3.12 | 25 |
| Clostridium botulinum (DSM1985) | 0.78 | 3.12 |
| Clostridium difficile (DSM1296) | 12.5 | 3.12 |
| Clostridium perfringens (ATCC13124) | >100 | >100 |
| Enterococcus faecalis (DSM2570) | 50 | 100 |
| Enterococcus faecium (DSM20477) | 100 | 25 |
| Listeria monocytogenes (DSM20600) | 3.12 | 6.25 |
| Propionibacterium acnes (ATCC6919) | 100 | >100 |
| Staphylococcus aureus (ATCC6538P) | 12.5 | 12.5 |
| MRSA (ATCCBAA-1717) | 12.5 | 25 |
| Streptococcus pyogenes (DSM20565) | 50 | 50 |
| Gram negative | ||
| Acinetobacter baumannii (DSM30007) | 12.5 | 25 |
| Brucella melitensis (NR-256) | 25 | 12.5 |
| Enterobacter cloacae (DSM30054) | >100 | >100 |
| Escherichia coli (ATCC8739) | 12.5 | 25 |
| Francisella tularensis (NR-643) | 50 | 12.5 |
| Helicobacter pylori (ATCC43504) | 12.5 | 12.5 |
| Klebsiella pneumonia (DSM26371) | >100 | >100 |
| Pseudomonas aeruginosa (ATCC9027) | 25 | >100 |
| Salmonella enterica (CIP80.39) | 25 | 50 |
| Shigella flexneri (ATCC12022) | 25 | 50 |
| Vibrio alginolyticus (DSM2171) | 25 | 50 |
| Yersinia pestis (NR-641) | 12.5 | 12.5 |
| Mycobacteria | ||
| Mycobacterium abscessus S (CIP 104536T) | >100 | >100 |
| Mycobacterium abscessus R(CIP 104536T) | >100 | >100 |
| Mycobacterium smegmatis (ATCC700084) | 50 | 50 |
| Mycobacterium tuberculosis H37Rv (mc26230) | >100 | >100 |
| Filamentous fungi | ||
| Aspergillus fumigatus (DSM819) | >100 | >100 |
| Fusarium oxysporum (DSM62316) | 25 | 25 |
| Yeasts | ||
| Candida albicans (DSM10697) | 50 | 50 |
| Candida auris (DSM21092) | 50 | 12.5 |
| Candida glabrata (DSM11226) | 50 | 50 |
| Candida tropicalis (DSM9419) | 100 | 100 |
| Cryptococcus neoformans (DSM11959) | 25 | 25 |
| A498 | BEAS-2B | Caco-2 | HaCaT | HepG-2 | IMR-90 | |
|---|---|---|---|---|---|---|
| CC50 (µM) | 398.2 | 169.0 | 199.6 | 268.5 | 472.4 | 421.4 |
| TI with MIC of 0.78 µM | 510.5 | 216.6 | 255.8 | 344.2 | 605.6 | 540.2 |
| A498 | BEAS-2B | Caco-2 | HaCaT | HepG-2 | IMR-90 | |
|---|---|---|---|---|---|---|
| IC50 (µM) | 453.0 | 125.9 | 186.5 | 322.9 | >500 | 437.6 |
| TI with MIC of 3.12 µM | 145.1 | 40.3 | 59.7 | 103.4 | >160.2 | 140.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





