Submitted:
19 February 2024
Posted:
19 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. MuSCs isolation and identification
2.2. Animals and samples collection
2.3. Performance measurement of Nanjiang Yellow goat
2.4. Plasmid construction
2.5. Cell culture and transfection
2.6. RNA stability assays
2.7. Total RNA isolation and qPCR
2.8. Luciferase reporter assays
2.9. Extraction of genomic DNA and detection of DNA quality
2.10. PCR amplification and sequencing
2.11. Mass-array genotyping
2.12. Data Analysis
2.13. Bioinformatics Analysis
3. Results
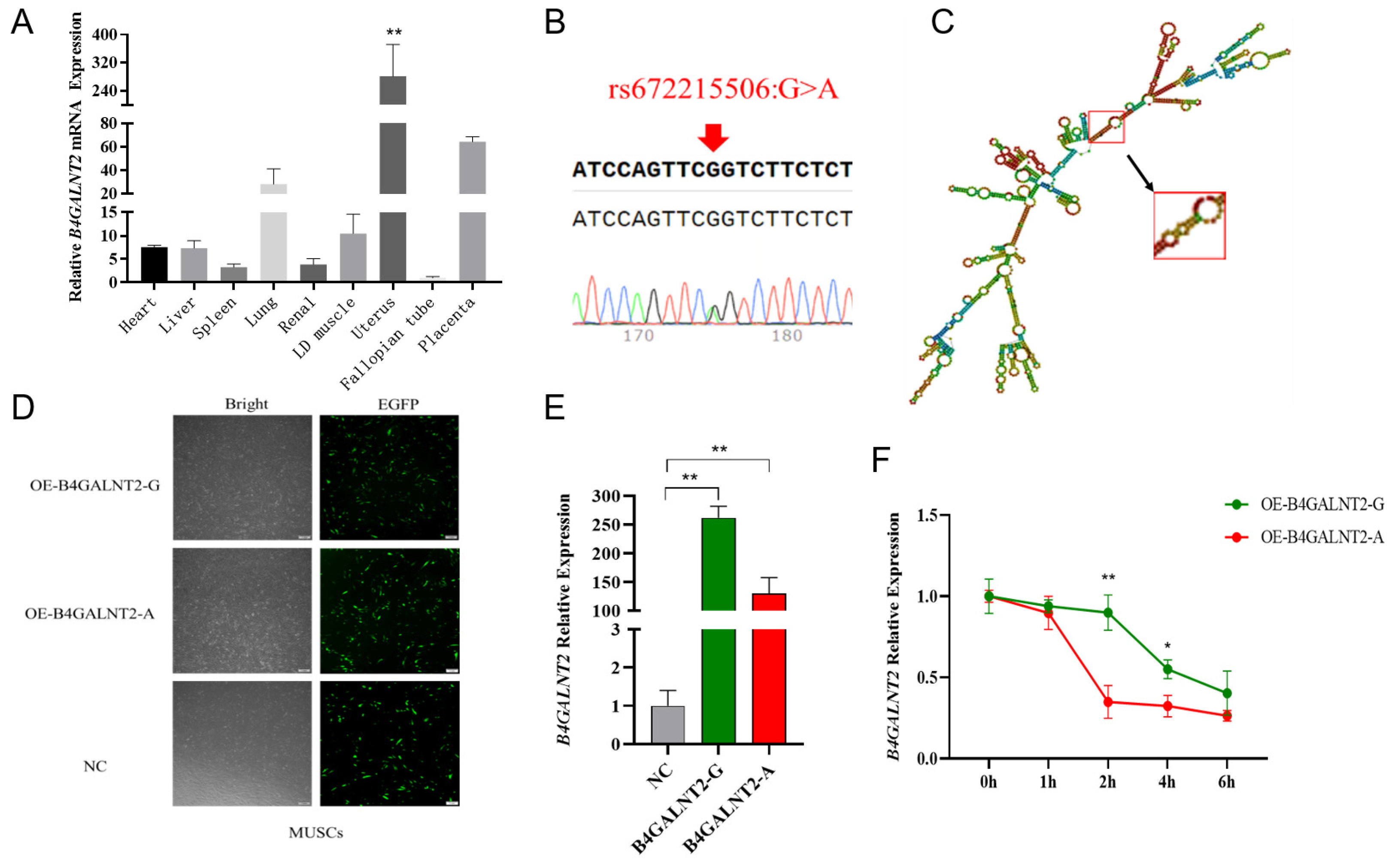
3.1. The synonymous mutation rs672215506 affected the mRNA stability of B4GALNT2 gene
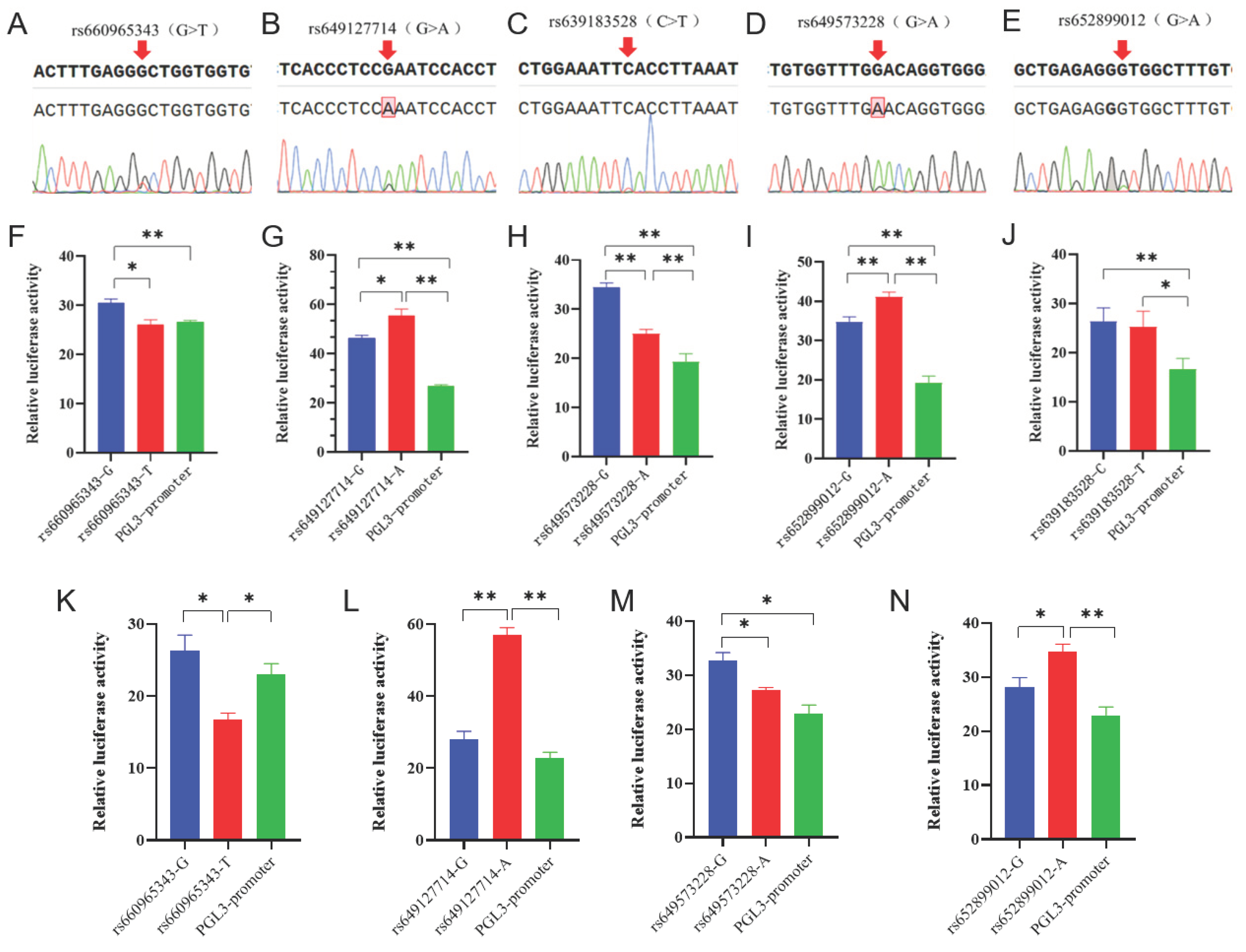
3.2. The detection and functional verification of non-coding SNPs in B4GALNT2.
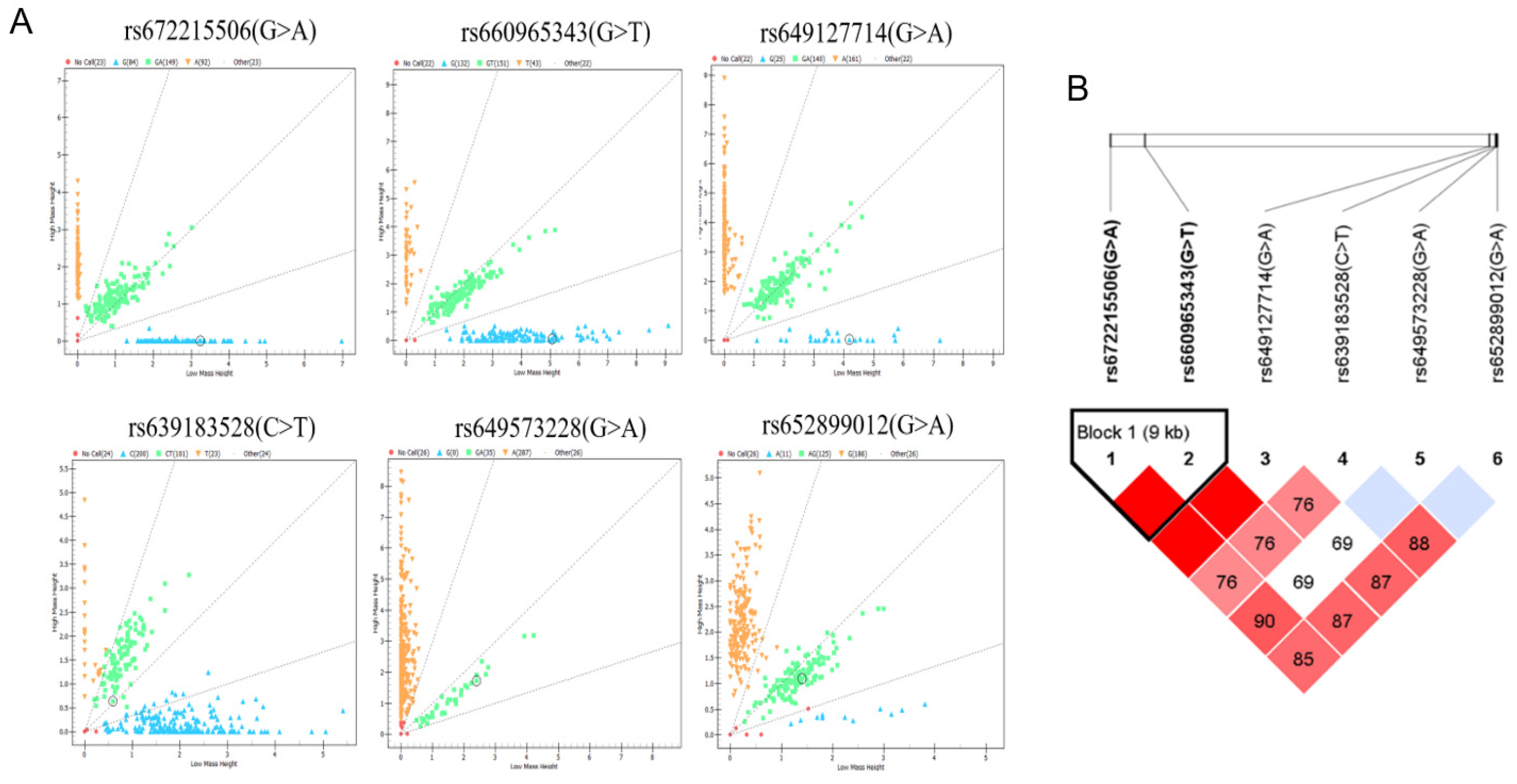
3.3. Population genetic diversity statistics of 6 SNPs in Nanjiang Yellow goat population
3.4. Analysis of linkage disequilibrium and construction of haplotypes
3.5. Association of SNPs and haplotype combinations with growth traits
3.6. Association of SNPs and haplotype combinations with lambing number
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Byrne, G.; Ahmad-Villiers, S.; Du, Z.; McGregor, C. B4GALNT2 and xenotransplantation: A newly appreciated xenogeneic antigen. Xenotransplantation 2018, 25, e12394. [CrossRef]
- Stwora-Wojczyk, M.M.; Kissinger, J.C.; Spitalnik, S.L.; Wojczyk, B.S. O-glycosylation in Toxoplasma gondii: identification and analysis of a family of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. International journal for parasitology 2004, 34, 309-322. [CrossRef]
- Zhao, C.; Cooper, D.K.C.; Dai, Y.; Hara, H.; Cai, Z.; Mou, L. The Sda and Cad glycan antigens and their glycosyltransferase, β1,4GalNAcT-II, in xenotransplantation. Xenotransplantation 2018, 25. [CrossRef]
- Duca, M.; Malagolini, N.; Dall’Olio, F. The story of the Sd(a) antigen and of its cognate enzyme B4GALNT2: What is new? Glycoconjugate journal 2023, 40, 123-133. [CrossRef]
- Groux-Degroote, S.; Vicogne, D.; Cogez, V.; Schulz, C.; Harduin-Lepers, A. B4GALNT2 Controls Sd(a) and SLe(x) Antigen Biosynthesis in Healthy and Cancer Human Colon. Chembiochem : a European journal of chemical biology 2021, 22, 3381-3390. [CrossRef]
- Galeev, A.; Suwandi, A.; Cepic, A.; Basu, M.; Baines, J.F.; Grassl, G.A. The role of the blood group-related glycosyltransferases FUT2 and B4GALNT2 in susceptibility to infectious disease. International journal of medical microbiology : IJMM 2021, 311, 151487. [CrossRef]
- Groux-Degroote, S.; Wavelet, C.; Krzewinski-Recchi, M.A.; Portier, L.; Mortuaire, M.; Mihalache, A.; Trinchera, M.; Delannoy, P.; Malagolini, N.; Chiricolo, M.; et al. B4GALNT2 gene expression controls the biosynthesis of Sda and sialyl Lewis X antigens in healthy and cancer human gastrointestinal tract. The international journal of biochemistry & cell biology 2014, 53, 442-449. [CrossRef]
- Rausch, P.; Steck, N.; Suwandi, A.; Seidel, J.A.; Künzel, S.; Bhullar, K.; Basic, M.; Bleich, A.; Johnsen, J.M.; Vallance, B.A.; et al. Expression of the Blood-Group-Related Gene B4galnt2 Alters Susceptibility to Salmonella Infection. PLoS pathogens 2015, 11, e1005008. [CrossRef]
- Pucci, M.; Gomes Ferreira, I.; Malagolini, N.; Ferracin, M.; Dall’Olio, F. The Sd(a) Synthase B4GALNT2 Reduces Malignancy and Stemness in Colon Cancer Cell Lines Independently of Sialyl Lewis X Inhibition. International journal of molecular sciences 2020, 21. [CrossRef]
- Drouilhet, L.; Mansanet, C.; Sarry, J.; Tabet, K.; Bardou, P.; Woloszyn, F.; Lluch, J.; Harichaux, G.; Viguié, C.; Monniaux, D.; et al. The highly prolific phenotype of Lacaune sheep is associated with an ectopic expression of the B4GALNT2 gene within the ovary. PLoS genetics 2013, 9, e1003809. [CrossRef]
- Drouilhet, L.; Lecerf, F.; Bodin, L.; Fabre, S.; Mulsant, P. Fine mapping of the FecL locus influencing prolificacy in Lacaune sheep. Animal genetics 2009, 40, 804-812. [CrossRef]
- Thomas, P.J.; Xu, R.; Martin, P.T. B4GALNT2 (GALGT2) Gene Therapy Reduces Skeletal Muscle Pathology in the FKRP P448L Mouse Model of Limb Girdle Muscular Dystrophy 2I. The American journal of pathology 2016, 186, 2429-2448. [CrossRef]
- Xu, R.; Singhal, N.; Serinagaoglu, Y.; Chandrasekharan, K.; Joshi, M.; Bauer, J.A.; Janssen, P.M.; Martin, P.T. Deletion of Galgt2 (B4Galnt2) reduces muscle growth in response to acute injury and increases muscle inflammation and pathology in dystrophin-deficient mice. The American journal of pathology 2015, 185, 2668-2684. [CrossRef]
- Cramer, M.L.; Xu, R.; Martin, P.T. Soluble Heparin Binding Epidermal Growth Factor-Like Growth Factor Is a Regulator of GALGT2 Expression and GALGT2-Dependent Muscle and Neuromuscular Phenotypes. Molecular and cellular biology 2019, 39. [CrossRef]
- Gao, Y.; Wang, J.; Chao, L.; Kuan, M.; Yu, H.; Wang, J.; Bao, S.; Liu, Y.; Zhang, W.; Ma, Q.; et al. Association analysis of B4GALNT2 gene polymorphism with lambing number in Mongolian and Uzhumqin sheep. Journal of Agricultural Biotechnology 2022, 30, 1510-1523.
- Rong, X.; Shao, S.; Liang, P.; Zhang, T.; Zou, S.; Meng, K.; Qiang, H.; Feng, D. Analysis of polymorphisms in B4GALNT2 and ESR1 genes and their association with lambing number in sheep. China Animal Husbandry & Veterinary Medicine 2021, 48, 3332-3342. [CrossRef]
- Guo, X.; Wang, X.; Liang, B.; Di, R.; Liu, Q.; Hu, W.; He, X.; Zhang, J.; Zhang, X.; Chu, M. Molecular Cloning of the B4GALNT2 Gene and Its Single Nucleotide Polymorphisms Association with Litter Size in Small Tail Han Sheep. Animals : an open access journal from MDPI 2018, 8. [CrossRef]
- Sa, C.; Wu, T.; Ma, Y.; He, Y.; Zhu, L.; He, T.; Wu, Y.; Liu, B. Analysis of polymorphisms in four candidate genes for multiparous traits and their association with lambing number in cashmere goats. China Animal Husbandry & Veterinary Medicine 2023, 50, 1037-1047. [CrossRef]
- Yang, B.G.; Yuan, Y.; Zhou, D.K.; Ma, Y.H.; Mahrous, K.F.; Wang, S.Z.; He, Y.M.; Duan, X.H.; Zhang, W.Y.; E, G. Genome-wide selection signal analysis of Australian Boer goat reveals artificial selection imprinting on candidate genes related to muscle development. Animal genetics 2021, 52, 550-555. [CrossRef]
- Zhao, W.; Chen, L.; Zhong, T.; Wang, L.; Guo, J.; Dong, Y.; Feng, J.; Song, T.; Li, L.; Zhang, H. The differential proliferation and differentiation ability of skeletal muscle satellite cell in Boer and Nanjiang brown goats. Small Ruminant Research 2018, 169, 99-107. [CrossRef]
- Chen, L.; Zhang, C.; Ma, W.; Huang, J.; Zhao, Y.; Liu, H. METTL3-mediated m6A modification stabilizes TERRA and maintains telomere stability. Nucleic Acids Research 2022, 50, 11619-11634. [CrossRef]
- Chaney, J.L.; Clark, P.L. Roles for Synonymous Codon Usage in Protein Biogenesis. Annual review of biophysics 2015, 44, 143-166. [CrossRef]
- Otsuka, H.; Sasai, H.; Nakama, M.; Aoyama, Y.; Abdelkreem, E.; Ohnishi, H.; Konstantopoulou, V.; Sass, J.O.; Fukao, T. Exon 10 skipping in ACAT1 caused by a novel c.949G>A mutation located at an exonic splice enhancer site. Molecular medicine reports 2016, 14, 4906-4910. [CrossRef]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: the causes and consequences of codon bias. Nature reviews. Genetics 2011, 12, 32-42. [CrossRef]
- Szewczuk, M.; Zych, S.; Wojcik, J.; Czerniawska-Piatkowska, E. Association of two SNPs in the coding region of the insulin-like growth factor 1 receptor (IGF1R) gene with growth-related traits in Angus cattle. J Appl Genet 2013, 54, 305-308. [CrossRef]
- Cui, C.; Jiang, H.; Liang, Y.; Xiao, C.; Liu, Y.; Jin, H.; Cao, Y. Analysis of HLF gene polymorphism and its association with muscle fatty acid and amino acid contents in sheep. Heilongjiang Animal Husbandry and Veterinary Medicine 2022.
- Cheng, Y.; Liu, S.; Wang, G.; Wei, W.; Huang, S.; Yang, R.; Geng, H.; Li, H.; Song, J.; Sun, L.; et al. Porcine IGF1 synonymous mutation alter gene expression and protein binding affinity with IGF1R. Int J Biol Macromol 2018, 116, 23-30. [CrossRef]
- Jolma, A.; Yan, J.; Whitington, T.; Toivonen, J.; Nitta, K.R.; Rastas, P.; Morgunova, E.; Enge, M.; Taipale, M.; Wei, G.; et al. DNA-binding specificities of human transcription factors. Cell 2013, 152, 327-339. [CrossRef]
- Hoogendoorn, B.; Coleman, S.L.; Guy, C.A.; Smith, S.K.; O’Donovan, M.C.; Buckland, P.R. Functional analysis of polymorphisms in the promoter regions of genes on 22q11. Hum Mutat 2004, 24, 35-42. [CrossRef]
- Zhang, Z.; Liu, C.; Hao, W.; Yin, W.; Ai, S.; Zhao, Y.; Duan, Z. Novel Single Nucleotide Polymorphisms and Haplotype of MYF5 Gene Are Associated with Body Measurements and Ultrasound Traits in Grassland Short-Tailed Sheep. Genes (Basel) 2022, 13. [CrossRef]
- Chen, W.; Xu, H.; Chen, X.; Liu, Z.; Zhang, W.; Xia, D. Functional and Activity Analysis of Cattle UCP3 Promoter with MRFs-Related Factors. Int J Mol Sci 2016, 17. [CrossRef]
- Wang, D. Analysis of transcriptional regulation of non-coding region of FSHR gene in lake sheep. Nanjing Agricultural University.
- Kamarudin, N.J.; Wang, V.C.; Tan, X.T.; Ramesh, A.; Ling, M.H. A Simulation Study on the Effects of Founding Population Size and Number of Alleles Per Locus on the Observed Population Genetic Profile: Implications to Broodstock Management. 2020.
- Barrandeguy, M.E.; García, M. The Sensitiveness of Expected Heterozygosity and Allelic Richness Estimates for Analyzing Population Genetic Diversity; Genetic Diversity [Working Title]: 2021.
- Silió, L.; Rodríguez, M.C.; Fernández, A.; Barragán, C.; Benítez, R.; Óvilo, C.; Fernández, A.I. Measuring inbreeding and inbreeding depression on pig growth from pedigree or SNP-derived metrics. Journal of Animal Breeding and Genetics 2013, 130, 349-360. [CrossRef]
- Penedo, M.; Weisenberger, M.E.; Boyce, W.M.; Johnson, C.K.J.A.o.O.; Environmental Medicine, 1. Wildlife translocation: the conservation implications of pathogen exposure and genetic heterozygosity. 2011, 11, 1-7.
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W.J.A.j.o.h.g. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. 1980, 32, 314.
- Jiang, Y.; Wang, S.; Zhu, L.; Yang, H.; Hong, Q. Analysis of muscle growth inhibitor gene polymorphism in black goats on Yunnan. Chinese Herbivore Science 2020, 5-7.
- Ma, X.; Du, L.; Zhang, L.; Xuan, J.; Wang, H.; Yuan, Z.; Wu, M.; Zhu, C.; Liu, R.; Wei, C. Association of RIPK2 gene polymorphisms with growth traits in Uzhumqin sheep. Chinese Agricultural Science 2016, 49, 17.
- Peng, Y.; Liu, J.; Zhao, S.; Xu, Z.; Zuo, B. Detection of SNPs in porcine RXRB gene and their association analysis with growth and fattening and reproductive traits. Journal of Animal Husbandry and Veterinary Science 2021, 52, 03.
- Meng, K.; Zhang, T.; Liang, P.; Shao, S.; Zou, S.; Rong, X.; Qiang, H.; Feng, D. Analysis of polymorphisms in sheep MYF5 gene and their association with growth traits. Journal of Agricultural Biotechnology 2022, 030.
- Wu, S.-B.; Franks, T.K.; Hunt, P.; Wirthensohn, M.G.; Gibson, J.P.; Sedgley, M. Discrimination of SNP genotypes associated with complex haplotypes by high resolution melting analysis in almond: implications for improved marker efficiencies. Molecular Breeding 2009, 25, 351-357. [CrossRef]
- Li, L.; Wu, D.; et al. Correlation analysis between LHβ gene polymorphism and reproductive performance of Nanjiang yellow sheep. Livestock and Veterinary Medicine 2006, 3-5.
| Locus | Genotype | Genotype frequency | Allele frequency | Ho | He | Ne | P | PIC |
|---|---|---|---|---|---|---|---|---|
| rs672215506(G>A) | AA | 0.28 | 0.51(A) | 0.5 | 0.5 | 2 | 0.16 | 0.37 |
| GA | 0.46 | 0.49(G) | ||||||
| GG | 0.26 | |||||||
| rs660965343(G>T) | GG | 0.4 | 0.64(G) | 0.54 | 0.46 | 1.85 | 1 | 0.35 |
| GT | 0.46 | 0.36(T) | ||||||
| TT | 0.14 | |||||||
| rs649127714(G>A) | GG | 0.08 | 0.29(G) | 0.59 | 0.41 | 1.7 | 0.58 | 0.33 |
| GA | 0.43 | 0.71(A) | ||||||
| AA | 0.49 | |||||||
| rs639183528(C>T) | CC | 0.62 | 0.77(C) | 0.65 | 0.35 | 1.54 | 0.07 | 0.29 |
| CT | 0.31 | 0.23(T) | ||||||
| TT | 0.07 | |||||||
| rs649573228(G>A) | AA | 0.89 | 0.95(A) | 0.9 | 0.1 | 1.11 | 0.75 | 0.1 |
| GA | 0.11 | 0.05(G) | ||||||
| rs652899012(G>A) | GG | 0.58 | 0.65 | 0.35 | 1.54 | 0.09 | 0.29 | |
| AG | 0.39 | 0.77(G) | ||||||
| AA | 0.03 | 0.23(A) |
| Haplotype and Haplotype combination | Type | Genotype | Frequency |
|---|---|---|---|
| Haplotype | H1 | GG | 0.487 |
| H2 | AG | 0.141 | |
| H3 | AT | 0.372 | |
| H1H3 | GGAT | 0.379 | |
| Haplotype combination | H2H2 | GGGG | 0.241 |
| H3H3 | ATAT | 0.124 | |
| H2H3 | AGAT | 0.118 | |
| H1H2 | GGAG | 0.112 | |
| H1H1 | AGAG | 0.026 |
| Locus | Genotype | Number | Birth weight |
|---|---|---|---|
| rs672215506(G>A) | AA | 92 | 2.29±0.28a |
| GA | 149 | 2.26±0.29ab | |
| GG | 84 | 2.20±0.31b | |
| rs660965343(G>T) | TT | 43 | 2.29±0.26a |
| GT | 151 | 2.27±0.33ab | |
| GG | 132 | 2.22±0.27b | |
| rs649127714(G>A) | AA | 161 | 2.27±0.34 |
| GA | 140 | 2.24±0.24 | |
| GG | 25 | 2.22±0.30 | |
| rs639183528(C>T) | CC | 200 | 2.26±0.32 |
| CT | 101 | 2.24±0.26 | |
| TT | 23 | 2.23±0.25 | |
| rs649573228(G>A) | AA | 287 | 2.25±0.31 |
| GA | 35 | 2.24±0.24 | |
| AA | 11 | 2.26±0.26 | |
| rs652899012(G>A) | AG | 125 | 2.22±0.27 |
| GG | 186 | 2.27±0.32 |
| Locus | rs672215506(G>A) | rs660965343(G>T) | rs649127714(G>A) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | AA | GA | GG | TT | GT | GG | AA | GA | GG | |
| Number | 60 | 88 | 49 | 26 | 89 | 82 | 100 | 82 | 15 | |
| BW-6 | 25.99±3.87 | 25.83±4.45 | 26.28±4.58 | 25.42±3.67b | 25.99±4.16ab | 26.17±4.64a | 26.66±4.42A | 25.26±3.91B | 25.53±5.03B | |
| BL-6 | 58.85±5.21 | 59.06±5.42 | 59.73±5.53 | 58.00±4.43B | 59.16±5.23A | 59.54±5.79A | 59.79±5.59A | 58.37±4.98B | 59.33±5.77AB | |
| BH-6 | 55.80±4.44B | 56.11±4.96A | 56.11±4.96A | 55.23±3.92B | 56.21±4.79A | 56.43±4.80A | 56.71±4.99A | 55.50±4.24B | 56.27±4.67AB | |
| CC-6 | 65.19±4.02 | 65.06±4.56 | 65.71±4.83 | 64.37±3.65B | 65.17±4.23AB | 65.65±4.92A | 65.89±4.69Aa | 64.54±4.09Bb | 65.07±4.48ABb | |
| BW-12 | 34.08±4.54 | 34.22±5.04 | 34.49±5.31 | 33.37±4.18Bb | 34.37±4.80ABa | 34.39±5.33Aa | 35.02±5.23A | 33.38±4.33B | 33.83±5.55B | |
| BL-12 | 65.70±4.76Bb | 66.11±5.14ABb | 66.80±5.65Aa | 64.73±4.19Bc | 66.13±5.01Ab | 66.63±5.54Aa | 66.72±5.46A | 65.44±4.68B | 66.33±5.39AB | |
| BH-12 | 62.75±4.04b | 63.13±4.55b | 63.73±4.71a | 62.15±3.56Bb | 63.08±4.37ABa | 63.57±4.74Aa | 63.61±4.69a | 62.55±4.03b | 63.53±4.70a | |
| CC-12 | 74.86±4.93 | 75.26±5.28 | 75.87±5.20 | 74.12±4.73Bb | 75.23±4.92ABa | 75.73±5.49Aa | 75.76±5.51 | 74.76±4.74 | 75.10±4.73 | |
| BW-18 | 47.23±6.43 | 48.39±7.62 | 48.20±7.45 | 46.33±6.08B | 48.20±7.37A | 48.29±7.39A | 48.67±7.79a | 47.10±6.08b | 48.30±8.80ab | |
| BL-18 | 72.78±4.80 | 73.56±5.42 | 73.53±5.65 | 71.35±4.23B | 73.48±5.11A | 73.76±5.68A | 73.77±5.58a | 72.82±4.84b | 73.00±5.66b | |
| BH-18 | 69.17±3.95 | 69.30±4.31 | 69.41±4.38 | 68.38±3.42B | 69.25±4.22AB | 69.61±4.40A | 69.69±4.43A | 68.70±3.69AB | 69.80±5.10B | |
| CC-18 | 85.52±4.66 | 85.85±5.10 | 85.71±4.88 | 84.46±4.34B | 85.93±4.62A | 85.88±5.33A | 86.32±5.13a | 85.04±4.37b | 85.37±5.80b | |
| Locus | rs639183528(C>T) | rs649573228(G>A) | rs652899012(G>A) | |||||||
| Genotype | CC | CT | TT | AA | GA | AA | AG | GG | ||
| Number | 119 | 63 | 15 | 174 | 21 | 7 | 71 | 117 | ||
| BW-6 | 26.38±4.45 | 25.47±3.85 | 25.10±4.69 | 26.14±4.28 | 24.86±4.56 | 25.71±5.19 | 25.19±4.00 | 26.46±4.41 | ||
| BL-6 | 59.45±5.54a | 58.89±5.03ab | 58.00±5.57b | 59.30±5.41A | 58.05±5.31B | 60.14±6.15A | 58.49±5.09B | 59.56±5.52AB | ||
| BH-6 | 56.45±4.97 | 55.76±4.08 | 55.67±4.82 | 56.32±4.70A | 55.14±4.67B | 57.29±5.22A | 55.48±4.16B | 56.53±4.97AB | ||
| CC-6 | 65.55±4.72 | 64.92±4.07 | 64.40±3.96 | 65.44±4.47A | 64.00±4.42B | 65.71±4.03a | 64.65±4.23b | 65.62±4.64ab | ||
| BW-12 | 34.60±5.17 | 33.79±4.47 | 33.30±5.02 | 34.43±4.95A | 32.67±4.91B | 34.07±5.76 | 33.48±4.48 | 34.71±5.18 | ||
| BL-12 | 66.39±5.34 | 65.87±4.76 | 65.47±5.44 | 66.29±5.22A | 65.24±4.82B | 66.57±6.16a | 65.58±4.72b | 66.51±5.39ab | ||
| BH-12 | 63.34±4.66 | 62.86±3.95 | 63.00±4.77 | 63.29±4.46A | 62.24±4.41B | 63.86±5.24a | 62.65±4.01b | 63.43±4.67ab | ||
| CC-12 | 75.45±5.34 | 75.30±5.02 | 73.97±3.95 | 75.41±5.18 | 74.52±5.06 | 75.36±5.02 | 74.80±4.80 | 75.58±5.40 | ||
| BW-18 | 48.41±7.58 | 47.40±6.42 | 47.10±7.66 | 48.14±7.34 | 47.05±6.45 | 47.79±8.32 | 47.20±6.63 | 48.53±7.55 | ||
| BL-18 | 73.61±5.52 | 72.98±4.91 | 72.40±5.03 | 73.40±5.32a | 73.00±5.2b | 73.29±5.88 | 72.77±4.96 | 73.7±5.48 | ||
| BH-18 | 69.47±4.34 | 68.94±3.82 | 69.27±4.76 | 69.4±4.24A | 68.57±4.01B | 68.71±4.82 | 68.83±3.81 | 69.55±4.34 | ||
| CC-18 | 86.02±5.10 | 85.33±4.53 | 84.87±4.81 | 85.84±4.92A | 84.60±4.91B | 85.00±4.93 | 85.08±4.72 | 86.13±5.03 | ||
| Combined Haplotypes | H2H3 | H3H3 | H1H2 | H1H3 | H2H2 |
|---|---|---|---|---|---|
| Number | 27 | 26 | 26 | 75 | 49 |
| BW-6 | 26.30±3.63a | 25.42±3.67b | 25.77±4.67ab | 25.66±4.42ab | 26.28±4.58ab |
| BL-6 | 59.52±5.4Aa | 58±4.43Bb | 59.19±6.06Aa | 58.8±5.29ABab | 59.73±5.53Aa |
| BH-6 | 56.11±4.46Aa | 55.23±3.92Bb | 55.77±5.04ABa | 56.07±5.02ABa | 56.73±4.51Aa |
| CC-6 | 65.70±3.71Aa | 64.37±3.65Bc | 65.35±4.91Aab | 64.74±4.40ABbc | 65.71±4.83Aab |
| BW-12 | 34.33±4.46a | 33.37±4.18b | 33.83±5.30ab | 34.14±4.95ab | 34.49±5.31a |
| BL-12 | 66.07±4.82Aab | 64.73±4.19Bc | 66.00±5.29Aab | 66.02±5.09Ab | 66.80±5.65Aa |
| BH-12 | 62.85±4.03ABa | 62.15±3.56Bb | 63.00±4.67ABa | 63.01±4.47ABb | 63.73±4.71Aa |
| CC-12 | 75.35±4.90Aa | 74.12±4.73Bb | 75.46±6.05Aa | 74.88±4.94Bb | 75.87±5.20Aa |
| BW-18 | 47.76±6.53Aa | 46.33±6.08Bb | 48.38±7.45Aa | 47.93±7.45Ba | 48.20±7.45Aa |
| BL-18 | 73.56±4.55Aab | 71.35±4.23Bc | 73.81±5.64Aa | 73.2±5.22Ab | 73.53±5.65Aab |
| BH-18 | 69.52±4.05Aab | 68.38±3.42Bc | 69.69±4.34Aa | 69.03±4.16Ab | 69.41±4.38Aab |
| CC-18 | 86.33±4.07Aa | 84.46±4.34Bb | 86.08±5.73Aa | 85.61±4.88ABab | 85.71±4.88ABa |
| Combined Haplotypes | Number | Primiparity | Multiparity |
|---|---|---|---|
| H2H3 | 21 | 1.52±0.51 | 1.82±0.26Cc |
| H3H3 | 23 | 1.74±0.45 | 1.95±0.15BCb |
| H1H2 | 18 | 1.67±0.49 | 1.94±0.24BCb |
| H1H3 | 47 | 1.57±0.54 | 1.98±0.21Bb |
| H2H2 | 34 | 1.65±0.54 | 2.13±0.17Aa |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





