Submitted:
10 February 2024
Posted:
14 February 2024
You are already at the latest version
Abstract
Keywords:
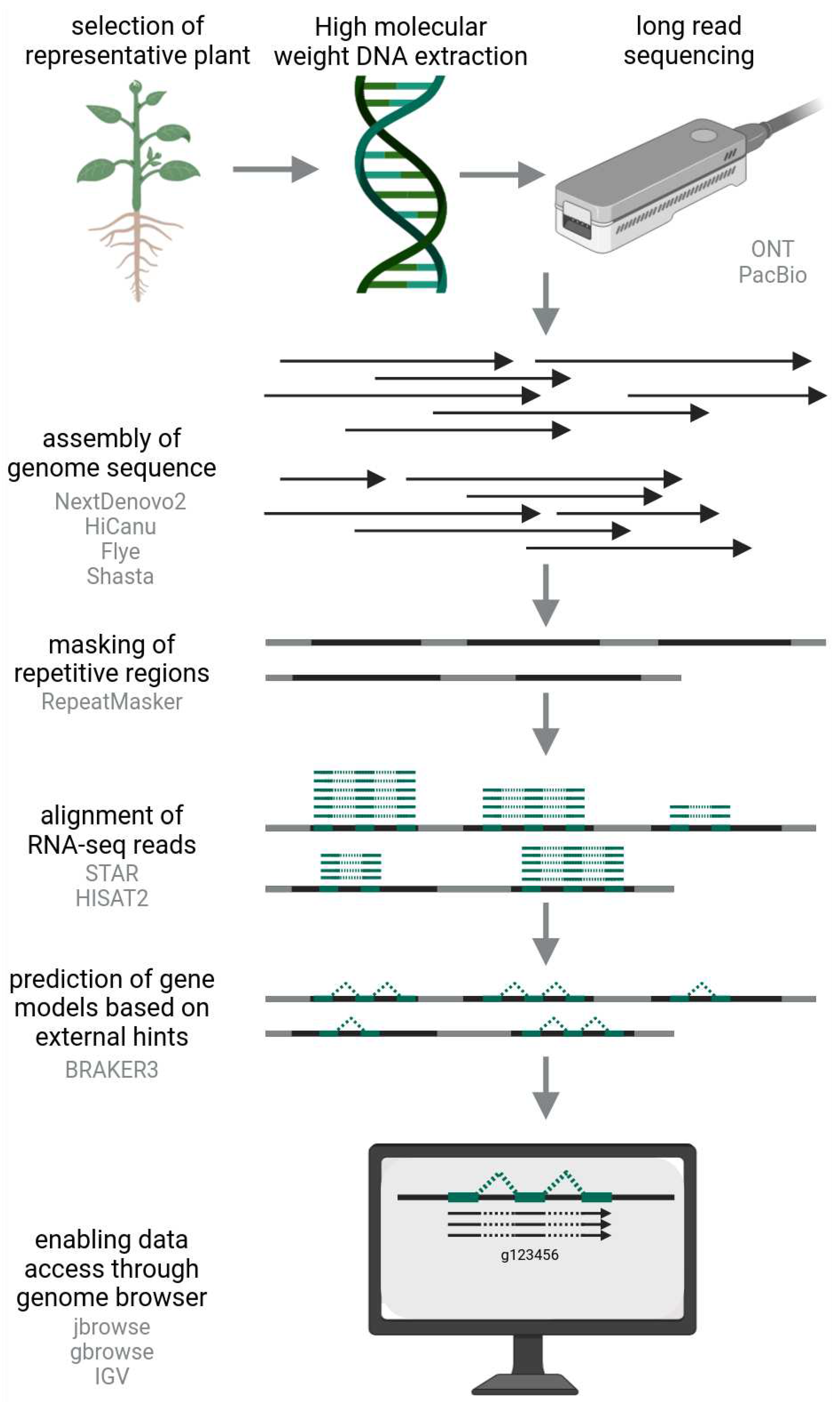
How to Obtain a Gene Sequence?
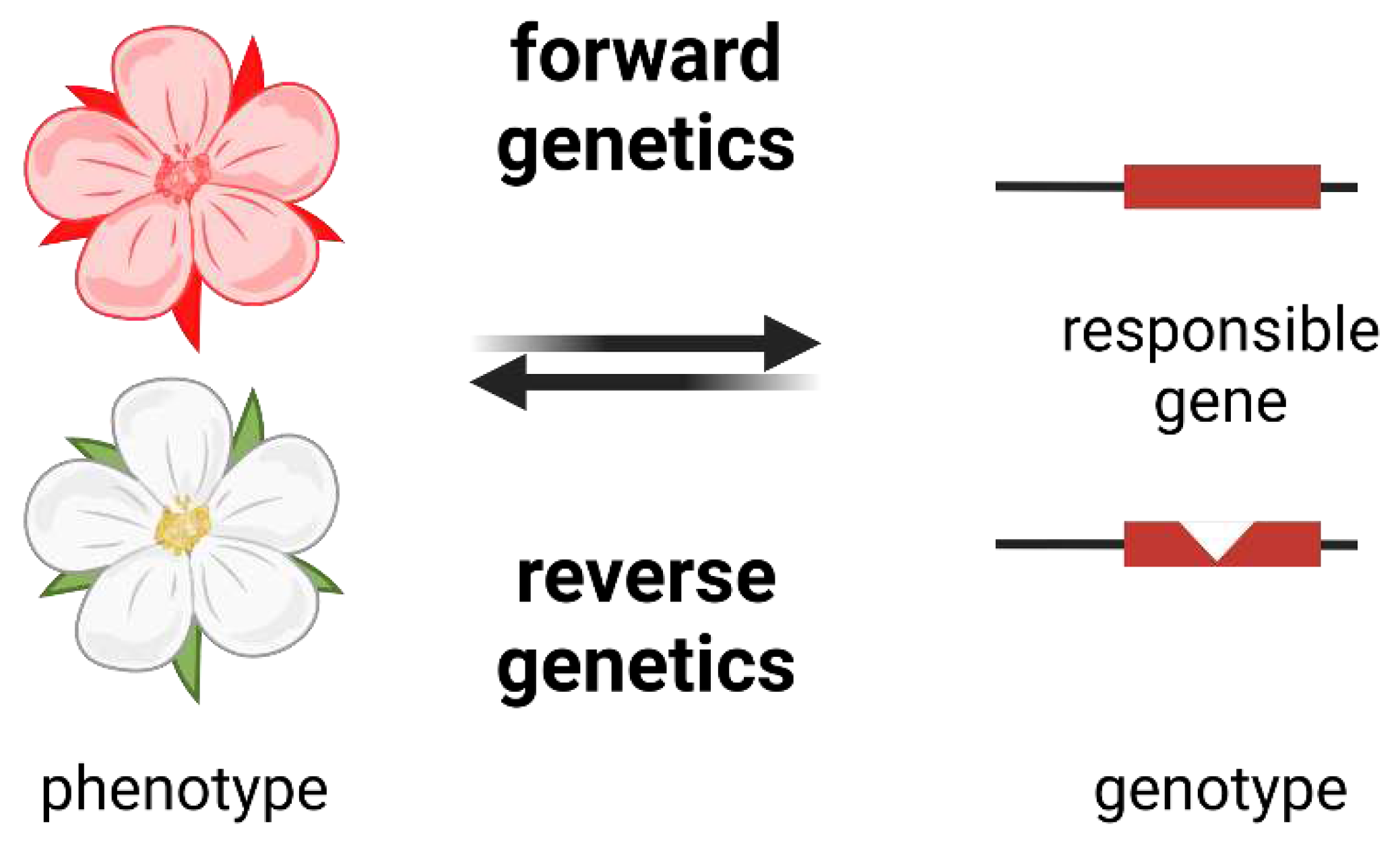
How to Understand the Function of a Gene?
How to Understand the Function of All Genes in a Genome?
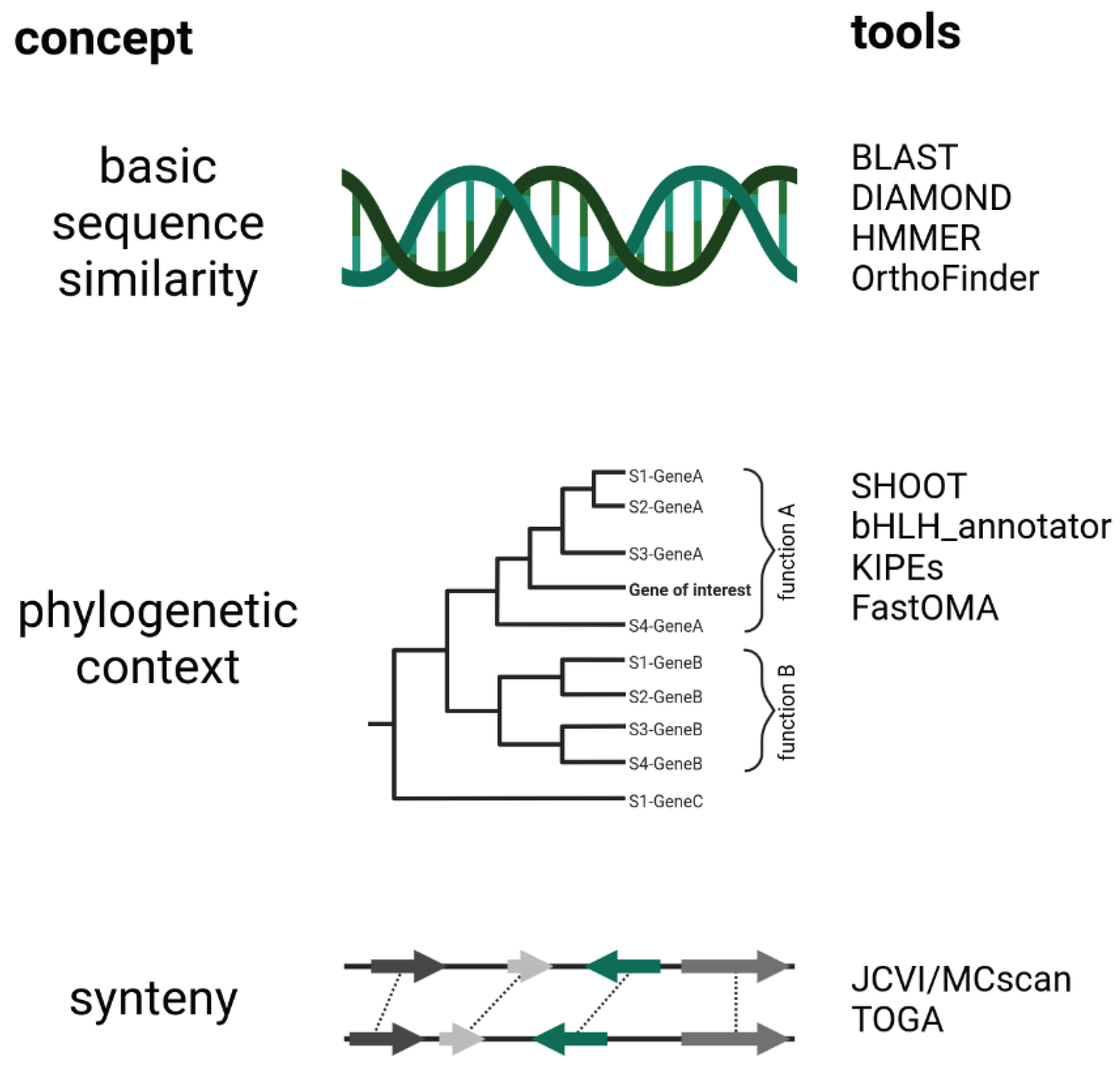
Gene Sequence Similarity Analyses
Phylogenetic Analyses
Synteny Analyses
How Can Artificial Intelligence Improve the Functional Annotation Process?
Summary
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al.2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science (New York, N.Y.) 301: 653–657. [CrossRef]
- Altenhoff AM, Warwick Vesztrocy A, Bernard C, Train C-M, Nicheperovich A, Prieto Baños S, Julca I, Moi D, Nevers Y, Majidian S, et al.2024. OMA orthology in 2024: improved prokaryote coverage, ancestral and extant GO enrichment, a revamped synteny viewer and more in the OMA Ecosystem. Nucleic Acids Research 52: D513–D521. [CrossRef]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. [CrossRef]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [CrossRef]
- Amarasinghe SL, Su S, Dong X, Zappia L, Ritchie ME, Gouil Q. 2020. Opportunities and challenges in long-read sequencing data analysis. Genome Biology 21: 30. [CrossRef]
- Andorf CM, Sen S, Hayford RK, Portwood JL, Cannon EK, Harper LC, Gardiner JM, Sen TZ, Woodhouse MR. 2022. FASSO: An AlphaFold based method to assign functional annotations by combining sequence and structure orthology. : 2022.11.10.516002. [CrossRef]
- Arita M, Karsch-Mizrachi I, Cochrane G, on behalf of the International Nucleotide Sequence Database Collaboration. 2021. The international nucleotide sequence database collaboration. Nucleic Acids Research 49: D121–D124. [CrossRef]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al.2000. Gene Ontology: tool for the unification of biology. Nature Genetics 25: 25–29. [CrossRef]
- Aslam M, She Z, Jakada BH, Fakher B, Greaves JG, Yan M, Chen Y, Zheng P, Cheng Y, Qin Y. 2022. Interspecific complementation-restoration of phenotype in Arabidopsis cuc2cuc3 mutant by sugarcane CUC2 gene. BMC Plant Biology 22: 47. [CrossRef]
- Baasner J-S, Rempel A, Howard D, Pucker B. 2024. NAVIP: Unraveling the Influence of Neighboring Small Sequence Variants on Functional Impact Prediction. : 596718. [CrossRef]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al.2013. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Research 41: D991–D995. [CrossRef]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E. 2015. The arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. genesis 53: 474–485. [CrossRef]
- de Boissier P, Habermann BH. 2020. A Practical Guide to Orthology Resources. In: Pontarotti P, ed. Evolutionary Biology—A Transdisciplinary Approach. Cham: Springer International Publishing, 41–77. [CrossRef]
- Brown D, Sjölander K. 2006. Functional Classification Using Phylogenomic Inference. PLOS Computational Biology 2: e77. [CrossRef]
- Brunet MA, Lucier J-F, Levesque M, Leblanc S, Jacques J-F, Al-Saedi HRH, Guilloy N, Grenier F, Avino M, Fournier I, et al.2021. OpenProt 2021: deeper functional annotation of the coding potential of eukaryotic genomes. Nucleic Acids Research 49: D380–D388. [CrossRef]
- Buchfink B, Reuter K, Drost H-G. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nature Methods 18: 366–368. [CrossRef]
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nature Methods12: 59–60. [CrossRef]
- Buels R, Yao E, Diesh CM, Hayes RD, Munoz-Torres M, Helt G, Goodstein DM, Elsik CG, Lewis SE, Stein L, et al.2016. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biology 17: 66. [CrossRef]
- Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Molecular Biology and Evolution 38: 5825–5829. [CrossRef]
- Caspi R, Billington R, Keseler IM, Kothari A, Krummenacker M, Midford PE, Ong WK, Paley S, Subhraveti P, Karp PD. 2020. The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Research 48: D445–D453. [CrossRef]
- Chen C, Wu Y, Li J, Wang X, Zeng Z, Xu J, Liu Y, Feng J, Chen H, He Y, et al.2023. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Molecular Plant 16: 1733–1742. [CrossRef]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 6: 80–92. [CrossRef]
- Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2016. GenBank. Nucleic Acids Research 44: D67–D72. [CrossRef]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal: For Cell and Molecular Biology 16: 735–743. [CrossRef]
- Cock PJA, Fields CJ, Goto N, Heuer ML, Rice PM. 2010. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Research 38: 1767–1771. [CrossRef]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [CrossRef]
- Debladis E, Lee T-F, Huang Y-J, Lu J-H, Mathioni SM, Carpentier M-C, Llauro C, Pierron D, Mieulet D, Guiderdoni E, et al.2020. Construction and characterization of a knock-down RNA interference line of OsNRPD1 in rice (Oryza sativa ssp japonica cv Nipponbare). Philosophical Transactions of the Royal Society B: Biological Sciences 375: 20190338. [CrossRef]
- Demirer GS, Landry MP. 2021. Efficient Transient Gene Knock-down in Tobacco Plants Using Carbon Nanocarriers. Bio-protocol 11: e3897. [CrossRef]
- Dereeper A, Bocs S, Rouard M, Guignon V, Ravel S, Tranchant-Dubreuil C, Poncet V, Garsmeur O, Lashermes P, Droc G. 2015. The coffee genome hub: a resource for coffee genomes. Nucleic Acids Research 43 (D1). [CrossRef]
- Diesh C, Stevens GJ, Xie P, De Jesus Martinez T, Hershberg EA, Leung A, Guo E, Dider S, Zhang J, Bridge C, et al.2023. JBrowse 2: a modular genome browser with views of synteny and structural variation. Genome Biology 24: 74. [CrossRef]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [CrossRef]
- Dobin A, Gingeras TR. 2015. Mapping RNA-seq Reads with STAR. Current Protocols in Bioinformatics 51: 11.14.1-11.14.19. [CrossRef]
- Dommes AB, Gross T, Herbert DB, Kivivirta KI, Becker A. 2019. Virus-induced gene silencing: empowering genetics in non-model organisms. Journal of Experimental Botany 70: 757–770. [CrossRef]
- Droc G, Martin G, Guignon V, Summo M, Sempéré G, Durant E, Soriano A, Baurens F-C, Cenci A, Breton C, et al.2022. The banana genome hub: a community database for genomics in the Musaceae. Horticulture Research 9: uhac221. [CrossRef]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis. Trends in Plant Science 15: 573–581. [CrossRef]
- Eisen JA. 1998. Phylogenomics: Improving Functional Predictions for Uncharacterized Genes by Evolutionary Analysis. Genome Research 8: 163–167. [CrossRef]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology 20: 238. [CrossRef]
- Emms DM, Kelly S. 2022. SHOOT: phylogenetic gene search and ortholog inference. Genome Biology 23: 85. [CrossRef]
- Fernandez-Pozo N, Menda N, Edwards JD, Saha S, Tecle IY, Strickler SR, Bombarely A, Fisher-York T, Pujar A, Foerster H, et al.2015. The Sol Genomics Network (SGN)--from genotype to phenotype to breeding. Nucleic Acids Research 43: D1036-1041. [CrossRef]
- Fiddes IT, Armstrong J, Diekhans M, Nachtweide S, Kronenberg ZN, Underwood JG, Gordon D, Earl D, Keane T, Eichler EE, et al.2018. Comparative Annotation Toolkit (CAT)—simultaneous clade and personal genome annotation. Genome Research 28: 1029–1038. [CrossRef]
- Fo K, Chuah YS, Foo H, Davey EE, Fullwood M, Thibault G, Mutwil M. 2023. PlantConnectome: knowledge networks encompassing >100,000 plant article abstracts. : 2023.07.11.548541. [CrossRef]
- Fuentes D, Molina M, Chorostecki U, Capella-Gutiérrez S, Marcet-Houben M, Gabaldón T. 2022. PhylomeDB V5: an expanding repository for genome-wide catalogues of annotated gene phylogenies. Nucleic Acids Research 50: D1062–D1068. [CrossRef]
- Gabriel L, Brůna T, Hoff KJ, Ebel M, Lomsadze A, Borodovsky M, Stanke M. 2023. BRAKER3: Fully Automated Genome Annotation Using RNA-Seq and Protein Evidence with GeneMark-ETP, AUGUSTUS and TSEBRA. : 2023.06.10.544449. [CrossRef]
- Gallaher SD, Craig RJ, Ganesan I, Purvine SO, McCorkle SR, Grimwood J, Strenkert D, Davidi L, Roth MS, Jeffers TL, et al.2021. Widespread polycistronic gene expression in green algae. Proceedings of the National Academy of Sciences 118: e2017714118. [CrossRef]
- García-Ríos M, Fujita T, LaRosa PC, Locy RD, Clithero JM, Bressan RA, Csonka LN. 1997. Cloning of a polycistronic cDNA from tomato encoding gamma-glutamyl kinase and gamma-glutamyl phosphate reductase. Proceedings of the National Academy of Sciences of the United States of America 94: 8249–8254. [CrossRef]
- Gene Ontology Consortium. 2021. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Research 49: D325–D334. [CrossRef]
- Gloss AD, Vergnol A, Morton TC, Laurin PJ, Roux F, Bergelson J. 2022. Genome-wide association mapping within a local Arabidopsis thaliana population more fully reveals the genetic architecture for defensive metabolite diversity. Philosophical Transactions of the Royal Society B: Biological Sciences 377: 20200512. [CrossRef]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al.2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40: D1178–D1186. [CrossRef]
- GrandOmics. 2023. NextDenovo.
- Grimplet J, Adam-Blondon A-F, Bert P-F, Bitz O, Cantu D, Davies C, Delrot S, Pezzotti M, Rombauts S, Cramer GR. 2014. The grapevine gene nomenclature system. BMC Genomics 15: 1077. [CrossRef]
- Grützner R, Martin P, Horn C, Mortensen S, Cram EJ, Lee-Parsons CWT, Stuttmann J, Marillonnet S. 2021. High-efficiency genome editing in plants mediated by a Cas9 gene containing multiple introns. Plant Communications 2: 100135. [CrossRef]
- Guignon V, Toure A, Droc G, Dufayard J-F, Conte M, Rouard M. 2021. GreenPhylDB v5: a comparative pangenomic database for plant genomes. Nucleic Acids Research 49: D1464–D1471. [CrossRef]
- Guizard S, Miedzinska K, Smith J, Smith J, Kuo RI, Davey M, Archibald A, Watson M. 2023. nf-core/isoseq: simple gene and isoform annotation with PacBio Iso-Seq long-read sequencing. Bioinformatics 39: btad150. [CrossRef]
- Haak M, Vinke S, Keller W, Droste J, Rückert C, Kalinowski J, Pucker B. 2018. High Quality de Novo Transcriptome Assembly of Croton tiglium. Frontiers in Molecular Biosciences 5. [CrossRef]
- Hart AJ, Ginzburg S, Xu M (Sam), Fisher CR, Rahmatpour N, Mitton JB, Paul R, Wegrzyn JL. 2020. EnTAP: Bringing faster and smarter functional annotation to non-model eukaryotic transcriptomes. Molecular Ecology Resources 20: 591–604. [CrossRef]
- Hu X, Friedberg I. 2019. SwiftOrtho: A fast, memory-efficient, multiple genome orthology classifier. GigaScience 8: giz118. [CrossRef]
- Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, et al.2001. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Research 29: 102–105. [CrossRef]
- Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, et al.2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Research 47: D309–D314. [CrossRef]
- Huynh-Thu VA, Geurts P. 2018. dynGENIE3: dynamical GENIE3 for the inference of gene networks from time series expression data. Scientific Reports 8: 3384. [CrossRef]
- Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P. 2010. Inferring Regulatory Networks from Expression Data Using Tree-Based Methods. PLOS ONE 5: e12776. [CrossRef]
- Irmisch S, Jo S, Roach CR, Jancsik S, Man Saint Yuen M, Madilao LL, O’Neil-Johnson M, Williams R, Withers SG, Bohlmann J. 2018. Discovery of UDP-Glycosyltransferases and BAHD-Acyltransferases Involved in the Biosynthesis of the Antidiabetic Plant Metabolite Montbretin A. The Plant Cell 30: 1864–1886. [CrossRef]
- Irmisch S, Ruebsam H, Jancsik S, Man Saint Yuen M, Madilao LL, Bohlmann J. 2019. Flavonol Biosynthesis Genes and Their Use in Engineering the Plant Antidiabetic Metabolite Montbretin A. Plant Physiology 180: 1277–1290. [CrossRef]
- James GV, Patel V, Nordström KJV, Klasen JR, Salomé PA, Weigel D, Schneeberger K. 2013. User guide for mapping-by-sequencing in Arabidopsis. Genome Biology 14: R61. [CrossRef]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al.2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. [CrossRef]
- Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. 2023. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Research 51: D587–D592. [CrossRef]
- Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 28: 27–30. [CrossRef]
- Keilwagen J, Hartung F, Grau J. 2019. GeMoMa: Homology-Based Gene Prediction Utilizing Intron Position Conservation and RNA-seq Data. Methods in Molecular Biology (Clifton, N.J.) 1962: 161–177. [CrossRef]
- Keilwagen J, Wenk M, Erickson JL, Schattat MH, Grau J, Hartung F. 2016. Using intron position conservation for homology-based gene prediction. Nucleic Acids Research 44: e89. [CrossRef]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology 37: 907–915. [CrossRef]
- Kirilenko BM, Munegowda C, Osipova E, Jebb D, Sharma V, Blumer M, Morales AE, Ahmed A-W, Kontopoulos D-G, Hilgers L, et al.2023. Integrating gene annotation with orthology inference at scale. Science (New York, N.Y.) 380: eabn3107. [CrossRef]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B. 2012. GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Research 40: D1211–D1215. [CrossRef]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nature Biotechnology 37: 540–546. [CrossRef]
- Krishnamurthy N, Brown DP, Kirshner D, Sjölander K. 2006. PhyloFacts: an online structural phylogenomic encyclopedia for protein functional and structural classification. Genome Biology 7: R83. [CrossRef]
- Kuznetsov D, Tegenfeldt F, Manni M, Seppey M, Berkeley M, Kriventseva EV, Zdobnov EM. 2023. OrthoDB v11: annotation of orthologs in the widest sampling of organismal diversity. Nucleic Acids Research 51: D445–D451. [CrossRef]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al.2012. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Research 40: D1202–D1210. [CrossRef]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [CrossRef]
- Lau W, Sattely ES. 2015. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science (New York, N.Y.) 349: 1224–1228. [CrossRef]
- Lee T, Lee I. 2021. Genome-Wide Association Studies in Arabidopsis thaliana: Statistical Analysis and Network-Based Augmentation of Signals. Methods in Molecular Biology (Clifton, N.J.) 2200: 187–210. [CrossRef]
- Lee HY, Seo J-S, Cho JH, Jung H, Kim J-K, Lee JS, Rhee S, Choi YD. 2013. Oryza sativa COI Homologues Restore Jasmonate Signal Transduction in Arabidopsis coi1-1 Mutants. PLOS ONE 8: e52802. [CrossRef]
- Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Research 49: W293–W296. [CrossRef]
- Lipman DJ, Pearson WR. 1985. Rapid and Sensitive Protein Similarity Searches. Science 227: 1435–1441. [CrossRef]
- Lohse M, Nagel A, Herter T, May P, Schroda M, Zrenner R, Tohge T, Fernie AR, Stitt M, Usadel B. 2014. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant, Cell & Environment 37: 1250–1258. [CrossRef]
- Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC. 2003. Virus-induced gene silencing in plants. Methods (San Diego, Calif.) 30: 296–303. [CrossRef]
- Lyons E, Pedersen B, Kane J, Alam M, Ming R, Tang H, Wang X, Bowers J, Paterson A, Lisch D, et al.2008. Finding and Comparing Syntenic Regions among Arabidopsis and the Outgroups Papaya, Poplar, and Grape: CoGe with Rosids. Plant Physiology 148: 1772–1781. [CrossRef]
- Majidian S, Nevers Y, Kharrazi AY, Vesztrocy AW, Pascarelli S, Moi D, Glover N, Altenhoff AM, Dessimoz C. 2024. Orthology inference at scale with FastOMA. : 2024.01.29.577392. [CrossRef]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Molecular Biology and Evolution 38: 4647–4654. [CrossRef]
- Marks RA, Hotaling S, Frandsen PB, VanBuren R. 2021. Representation and participation across 20 years of plant genome sequencing. Nature Plants 7: 1571–1578. [CrossRef]
- Mascher M, Jost M, Kuon J-E, Himmelbach A, Aßfalg A, Beier S, Scholz U, Graner A, Stein N. 2014. Mapping-by-sequencing accelerates forward genetics in barley. Genome Biology 15: R78. [CrossRef]
- Miller JB, Pickett BD, Ridge PG. 2019. JustOrthologs: a fast, accurate and user-friendly ortholog identification algorithm. Bioinformatics 35: 546–552. [CrossRef]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al.2021. Pfam: The protein families database in 2021. Nucleic Acids Research 49: D412–D419. [CrossRef]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Research 35: W182–W185. [CrossRef]
- Muñoz C, Di Genova A, Maass A, Orellana A, Hinrichsen P, Aravena A. 2014. VITIS VINIFERA GENOME ANNOTATION IMPROVEMENT USING NEXT-GENERATION SEQUENCING TECHNOLOGIES AND NCBI PUBLIC DATA. Acta Horticulturae: 349–356. [CrossRef]
- Naake T, Zhu F, Alseekh S, Scossa F, Perez de Souza L, Borghi M, Brotman Y, Mori T, Nakabayashi R, Tohge T, et al.2023. Genome-wide association studies identify loci controlling specialized seed metabolites in Arabidopsis. Plant Physiology: kiad511. [CrossRef]
- Naish M, Alonge M, Wlodzimierz P, Tock AJ, Abramson BW, Schmücker A, Mandáková T, Jamge B, Lambing C, Kuo P, et al.2021. The genetic and epigenetic landscape of the Arabidopsis centromeres. Science 374: eabi7489. [CrossRef]
- Naithani S, Gupta P, Preece J, D’Eustachio P, Elser JL, Garg P, Dikeman DA, Kiff J, Cook J, Olson A, et al.2020. Plant Reactome: a knowledgebase and resource for comparative pathway analysis. Nucleic Acids Research 48: D1093–D1103. [CrossRef]
- Napoli C, Lemieux C, Jorgensen R. 1990. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. The Plant Cell 2: 279–289. [CrossRef]
- Nehrt NL, Clark WT, Radivojac P, Hahn MW. 2011. Testing the ortholog conjecture with comparative functional genomic data from mammals. PLoS computational biology 7: e1002073. [CrossRef]
- Nurk S, Walenz BP, Rhie A, Vollger MR, Logsdon GA, Grothe R, Miga KH, Eichler EE, Phillippy AM, Koren S. 2020. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Research: gr.263566.120. [CrossRef]
- O’Malley RC, Barragan CC, Ecker JR. 2015. A User’s Guide to the Arabidopsis T-DNA Insertional Mutant Collections. Methods in molecular biology (Clifton, N.J.) 1284: 323–342. [CrossRef]
- Ou S, Collins T, Qiu Y, Seetharam AS, Menard CC, Manchanda N, Gent JI, Schatz MC, Anderson SN, Hufford MB, et al.2022. Differences in activity and stability drive transposable element variation in tropical and temperate maize. : 2022.10.09.511471. [CrossRef]
- Ou S, Su W, Liao Y, Chougule K, Agda JRA, Hellinga AJ, Lugo CSB, Elliott TA, Ware D, Peterson T, et al.2019. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biology 20: 275. [CrossRef]
- Palmer J. 2019. funannotate v1.5.3. [CrossRef]
- Pucker B. 2022. Automatic identification and annotation of MYB gene family members in plants. BMC Genomics 23: 220. [CrossRef]
- Pucker B, Holtgräwe D, Sörensen TR, Stracke R, Viehöver P, Weisshaar B. 2016. A De Novo Genome Sequence Assembly of the Arabidopsis thaliana Accession Niederzenz-1 Displays Presence/Absence Variation and Strong Synteny. PLOS ONE 11: e0164321. [CrossRef]
- Pucker B, Irisarri I, Vries J de, Xu B. 2022. Plant genome sequence assembly in the era of long reads: Progress, challenges and future directions. Quantitative Plant Biology 3: e5. [CrossRef]
- Pucker B, Kleinbölting N, Weisshaar B. 2021. Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis. BMC Genomics 22: 599. [CrossRef]
- Pucker B, Pandey A, Weisshaar B, Stracke R. 2020a. The R2R3-MYB gene family in banana (Musa acuminata): Genome-wide identification, classification and expression patterns. PLOS ONE 15: e0239275. [CrossRef]
- Pucker B, Reiher F, Schilbert HM. 2020b. Automatic Identification of Players in the Flavonoid Biosynthesis with Application on the Biomedicinal Plant Croton tiglium. Plants 9: 1103. [CrossRef]
- Ramos-González M, Ramos-González V, Arvanitidou C, Hernández-García J, García-González M, Romero-Campero FJ. 2023. PharaohFUN: PHylogenomic Analysis foR plAnt prOtein History and FUNction elucidation. : 2023.08.01.551440. [CrossRef]
- Rempel A, Choudhary N, Pucker B. 2023. KIPEs3: Automatic annotation of biosynthesis pathways. PLOS ONE 18: e0294342. [CrossRef]
- Rhie A, Walenz BP, Koren S, Phillippy AM. 2020. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biology 21: 245. [CrossRef]
- Riehl K, Riccio C, Miska EA, Hemberg M. 2022. TransposonUltimate: software for transposon classification, annotation and detection. Nucleic Acids Research 50: e64. [CrossRef]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. 2003. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Molecular Biology 53: 247–259. [CrossRef]
- Ruiz MT, Voinnet O, Baulcombe DC. 1998. Initiation and Maintenance of Virus-Induced Gene Silencing. The Plant Cell 10: 937–946. [CrossRef]
- Samuilov S, Rademacher N, Brilhaus D, Flachbart S, Arab L, Kopriva S, Weber APM, Mettler-Altmann T, Rennenberg H. 2018. Knock-Down of the Phosphoserine Phosphatase Gene Effects Rather N- Than S-Metabolism in Arabidopsis thaliana. Frontiers in Plant Science 9.
- Sasaki E, Köcher T, Filiault DL, Nordborg M. 2021. Revisiting a GWAS peak in Arabidopsis thaliana reveals possible confounding by genetic heterogeneity. Heredity 127: 245–252. [CrossRef]
- Sayers EW, Cavanaugh M, Clark K, Ostell J, Pruitt KD, Karsch-Mizrachi I. 2020. GenBank. Nucleic Acids Research 48: D84–D86. [CrossRef]
- Schilbert HM, Pucker B, Ries D, Viehöver P, Micic Z, Dreyer F, Beckmann K, Wittkop B, Weisshaar B, Holtgräwe D. 2022. Mapping-by-Sequencing Reveals Genomic Regions Associated with Seed Quality Parameters in Brassica napus. Genes 13: 1131. [CrossRef]
- Schilbert HM, Schöne M, Baier T, Busche M, Viehöver P, Weisshaar B, Holtgräwe D. 2021. Characterization of the Brassica napus Flavonol Synthase Gene Family Reveals Bifunctional Flavonol Synthases. Frontiers in Plant Science 12.
- Schneeberger K, Weigel D. 2011. Fast-forward genetics enabled by new sequencing technologies. Trends in Plant Science 16: 282–288. [CrossRef]
- Shafin K, Pesout T, Lorig-Roach R, Haukness M, Olsen HE, Bosworth C, Armstrong J, Tigyi K, Maurer N, Koren S, et al.2020. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nature Biotechnology 38: 1044–1053. [CrossRef]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13: 2498–2504. [CrossRef]
- Shi X, Cao S, Wang X, Huang S, Wang Y, Liu Z, Liu W, Leng X, Peng Y, Wang N, et al.2023. The complete reference genome for grapevine (Vitis vinifera L.) genetics and breeding. Horticulture Research 10: uhad061. [CrossRef]
- Sielemann K, Hafner A, Pucker B. 2020. The Reuse of Public Datasets in the Life Sciences: Potential Risks and Rewards.
- Sielemann K, Pucker B, Orsini E, Elashry A, Schulte L, Viehöver P, Müller AE, Schechert A, Weisshaar B, Holtgräwe D. 2023. Genomic characterization of a nematode tolerance locus in sugar beet. BMC Genomics 24: 748. [CrossRef]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. [CrossRef]
- Singh KS, van der Hooft JJJ, van Wees SCM, Medema MH. 2022. Integrative omics approaches for biosynthetic pathway discovery in plants. Natural Product Reports 39: 1876–1896. [CrossRef]
- Sjölander K. 2004. Phylogenomic inference of protein molecular function: advances and challenges. Bioinformatics (Oxford, England) 20: 170–179. [CrossRef]
- Smit A, Hubley R, Green P. 2015. RepeatMasker.
- Sreedasyam A, Plott C, Hossain MS, Lovell JT, Grimwood J, Jenkins JW, Daum C, Barry K, Carlson J, Shu S, et al.2023. JGI Plant Gene Atlas: an updateable transcriptome resource to improve functional gene descriptions across the plant kingdom. Nucleic Acids Research 51: 8383–8401. [CrossRef]
- Stamboulian M, Guerrero RF, Hahn MW, Radivojac P. 2020. The ortholog conjecture revisited: the value of orthologs and paralogs in function prediction. Bioinformatics 36: i219–i226. [CrossRef]
- Stein LD. 2013. Using GBrowse 2.0 to visualize and share next-generation sequence data. Briefings in Bioinformatics 14: 162–171. [CrossRef]
- Stracke R, Werber M, Weisshaar B. 2001. The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology 4: 447–456. [CrossRef]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al.2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research 43: D447-452. [CrossRef]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. 2008. Synteny and Collinearity in Plant Genomes. Science 320: 486–488. [CrossRef]
- Thoben C, Pucker B. 2023. Automatic annotation of the bHLH gene family in plants. : 2023.05.02.539087. [CrossRef]
- Thomas PD, Ebert D, Muruganujan A, Mushayahama T, Albou L-P, Mi H. 2022. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Science 31: 8–22. [CrossRef]
- de la Torre-López J, Ramírez A, Romero JR. 2023. Artificial intelligence to automate the systematic review of scientific literature. Computing 105: 2171–2194. [CrossRef]
- Tran N-V, Greshake Tzovaras B, Ebersberger I. 2018. PhyloProfile: dynamic visualization and exploration of multi-layered phylogenetic profiles. Bioinformatics 34: 3041–3043. [CrossRef]
- Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, FitzGerald LM, Vezzulli S, Reid J, et al.2007. A High Quality Draft Consensus Sequence of the Genome of a Heterozygous Grapevine Variety. PLoS ONE 2: e1326. [CrossRef]
- Velt A, Frommer B, Blanc S, Holtgräwe D, Duchêne É, Dumas V, Grimplet J, Hugueney P, Kim C, Lahaye M, et al.2023. An improved reference of the grapevine genome reasserts the origin of the PN40024 highly homozygous genotype. G3 GenesGenomesGenetics 13: jkad067. [CrossRef]
- Vuruputoor VS, Monyak D, Fetter KC, Webster C, Bhattarai A, Shrestha B, Zaman S, Bennett J, McEvoy SL, Caballero M, et al.2023. Welcome to the big leaves: Best practices for improving genome annotation in non-model plant genomes. Applications in Plant Sciences 11: e11533. [CrossRef]
- Wang K, Wang D, Zheng X, Qin A, Zhou J, Guo B, Chen Y, Wen X, Ye W, Zhou Y, et al.2019. Multi-strategic RNA-seq analysis reveals a high-resolution transcriptional landscape in cotton. Nature Communications 10: 4714. [CrossRef]
- Wlodzimierz P, Rabanal FA, Burns R, Naish M, Primetis E, Scott A, Mandáková T, Gorringe N, Tock AJ, Holland D, et al.2023. Cycles of satellite and transposon evolution in Arabidopsis centromeres. Nature 618: 557–565. [CrossRef]
- Xu L, Dong Z, Fang L, Luo Y, Wei Z, Guo H, Zhang G, Gu YQ, Coleman-Derr D, Xia Q, et al.2019. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Research 47: W52–W58. [CrossRef]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. 2004. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. The Plant Journal: For Cell and Molecular Biology 40: 22–34. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




