Submitted:
03 February 2024
Posted:
05 February 2024
You are already at the latest version
Abstract
Keywords:
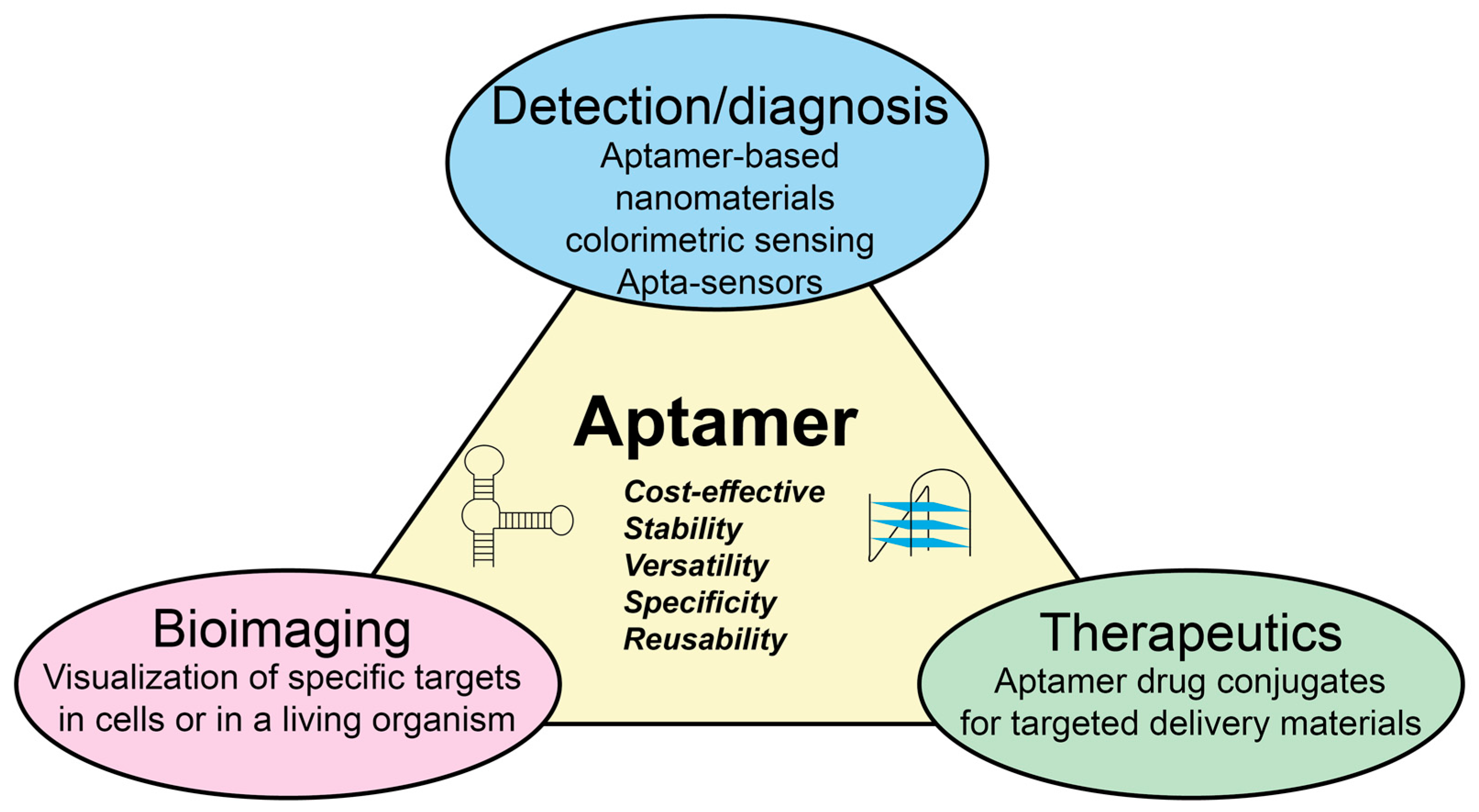
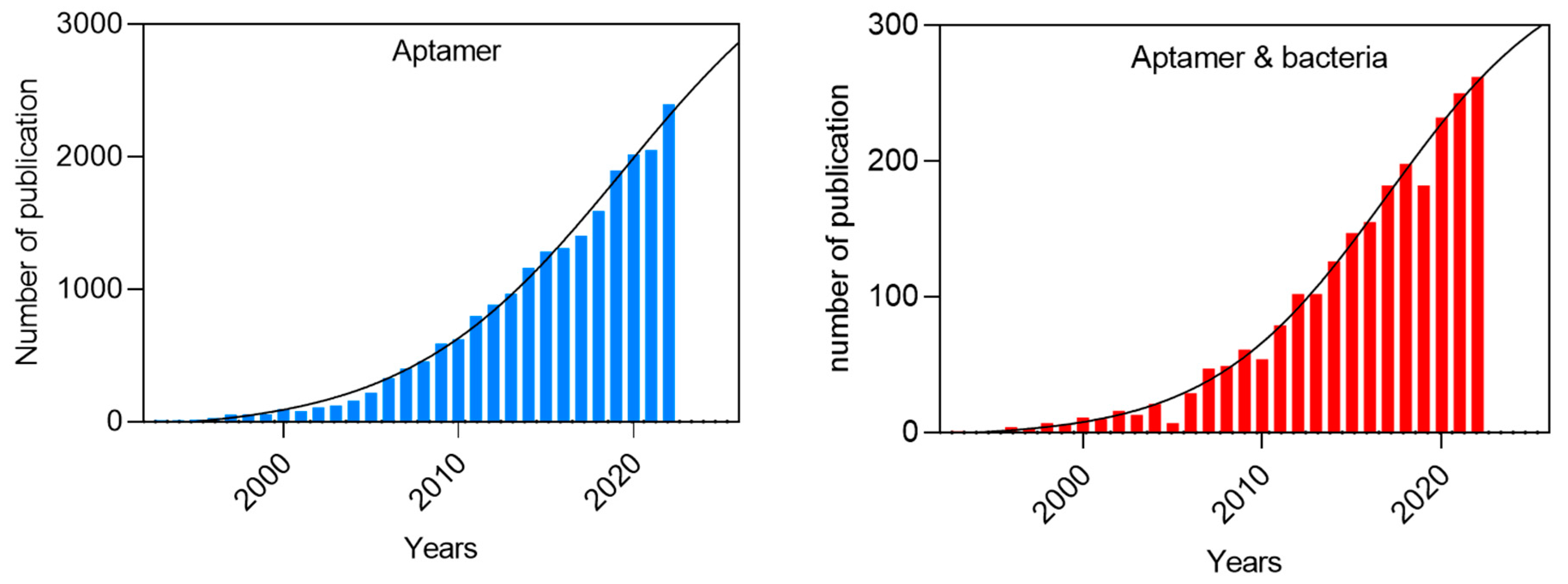
1. Introduction
2. Aptamer Selection Strategies for Bacterial Pathogen Detection
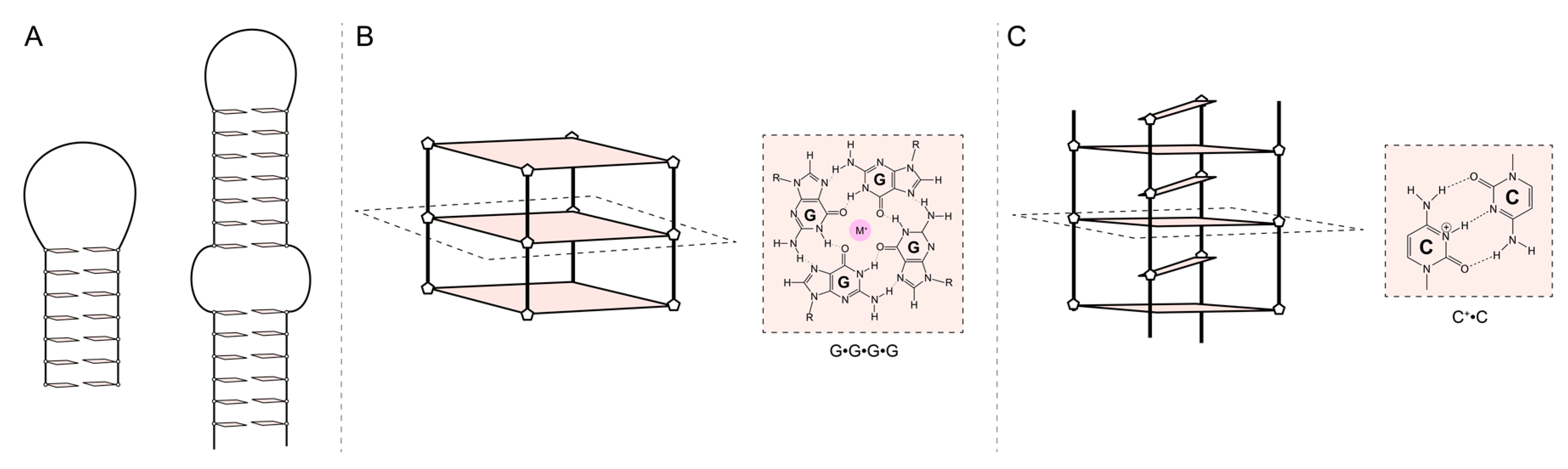
3. Aptamer characterization: affinity, stability, and structure
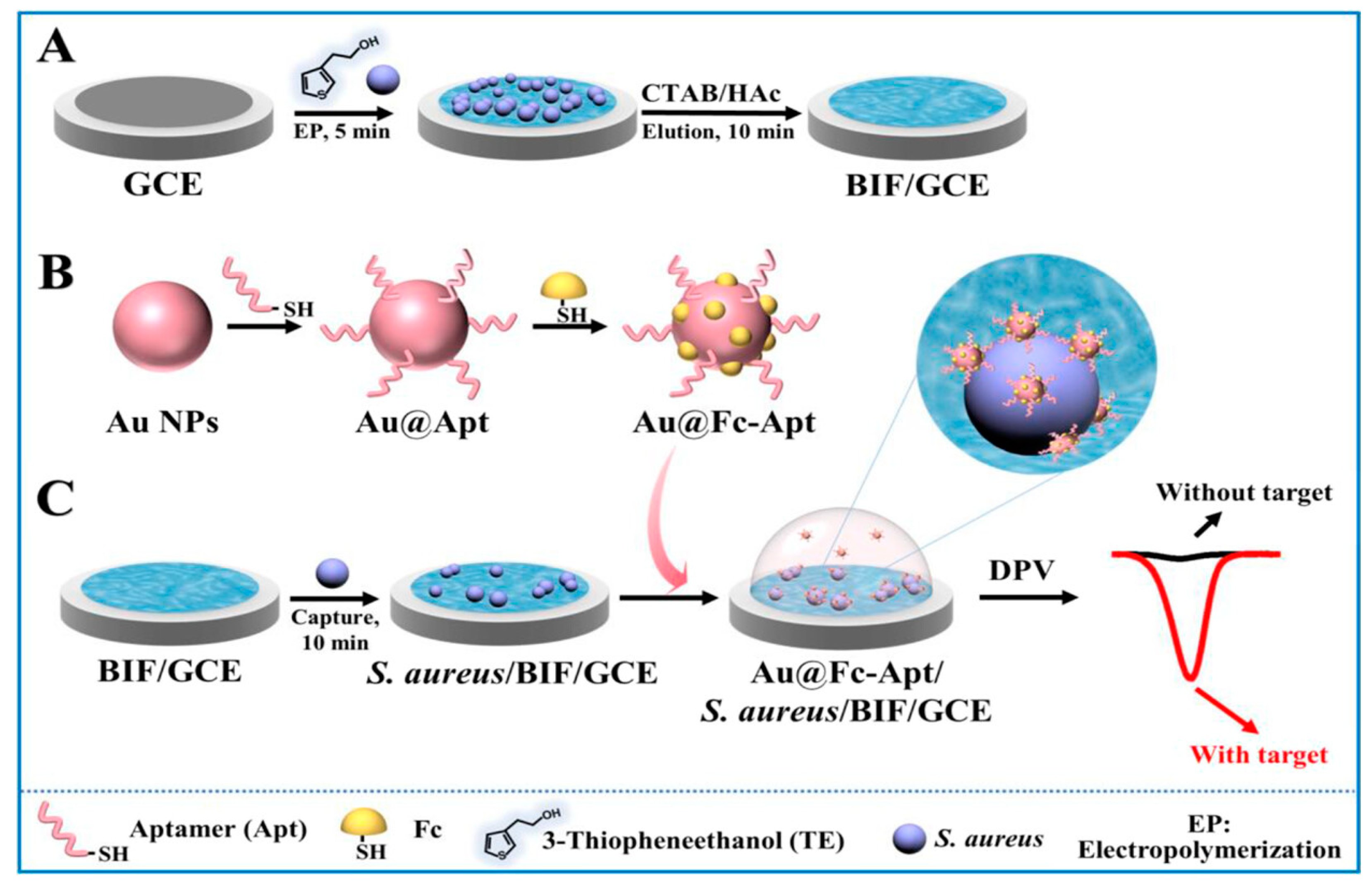
4. Bacterial aptasensors
4.1. Detection of bacterial cell
4.2. Bacterial toxin detection
4.3. Bacterial biomarker detection
5. Conclusions
6. Future directions
Author Contributions
Funding
Conflicts of Interest
References
- J. C. de M Campos, L. C. Antunes, and R. B. Ferreira, “Global priority pathogens: virulence, antimicrobial resistance and prospective treatment options,” Future Microbiology, vol. 15, no. 8, pp. 649–677, May 2020. [CrossRef]
- “WHO. 2022.” [Online]. Available: https://cdn.who.int/media/docs/default-source/gho-documents/world-health-statistic-reports/worldhealthstatistics_2022.pdf.
- P. S. Mead et al., “Food-Related Illness and Death in the United States,” Emerging Infectious Diseases, vol. 5, no. 5, pp. 607–625, 1999. [CrossRef]
- M. Marin, M. V. Nikolic, and J. Vidic, “Rapid point-of-need detection of bacteria and their toxins in food using gold nanoparticules,” Comp Rev Food Sci Food Saf., vol. 20, no. 6, pp. 5880–5900, Oct. 2021. [CrossRef]
- J. Vidic, C. Chaix, M. Manzano, and M. Heyndrickx, “Food Sensing: Detection of Bacillus cereus Spores in Dairy Products,” Biosensors, vol. 10, no. 3, p. 15, Feb. 2020. [CrossRef]
- N. Ramarao, S.-L. Tran, M. Marin, and J. Vidic, “Advanced Methods for Detection of Bacillus cereus and Its Pathogenic Factors,” Sensors, vol. 20, no. 9, p. 2667, May 2020. [CrossRef]
- P. Setlow and E. A. Johnson, “Spores and Their Significance,” in Food Microbiology, M. P. Doyle, F. Diez-Gonzalez, and C. Hill, Eds., Washington, DC, USA: ASM Press, 2019, pp. 23–63. [CrossRef]
- K. S. Ikuta et al., “Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019,” The Lancet, vol. 400, no. 10369, pp. 2221–2248, Dec. 2022. [CrossRef]
- E. J. Scallan Walter, H. Q. McLean, and P. M. Griffin, “Hospital Discharge Data Underascertain Enteric Bacterial Infections Among Children,” Foodborne Pathogens and Disease, vol. 17, no. 9, pp. 530–532, Sep. 2020. [CrossRef]
- R. Chowdary Akkina, V. Payala, and S. Sushma Maganti, “Tools for Rapid Detection and Control of Foodborne Microbial Pathogens,” in Foodborne Pathogens - Recent Advances in Control and Detection, A. Lamas, C. Manuel Franco, and P. Regal, Eds., IntechOpen, 2023. [CrossRef]
- K. Mullis, F. Faloona, S. Scharf, R. Saiki, G. Horn, and H. Erlich, “Specific Enzymatic Amplification of DNA In Vitro: The Polymerase Chain Reaction,” Cold Spring Harbor Symposia on Quantitative Biology, vol. 51, no. 0, pp. 263–273, Jan. 1986. [CrossRef]
- B. Malorny, P. T. Tassios, P. Rådström, N. Cook, M. Wagner, and J. Hoorfar, “Standardization of diagnostic PCR for the detection of foodborne pathogens,” International Journal of Food Microbiology, vol. 83, no. 1, pp. 39–48, May 2003. [CrossRef]
- S. Toze, “PCR and the detection of microbial pathogens in water and wastewater,” Water Research, vol. 33, no. 17, pp. 3545–3556, Dec. 1999. [CrossRef]
- J. E. Butler, “Enzyme-Linked Immunosorbent Assay,” Journal of Immunoassay, vol. 21, no. 2–3, pp. 165–209, May 2000. [CrossRef]
- A. Klancnik, M. Kovac, N. Toplak, S. Piskernik, and B. Jersek, “PCR in Food Analysis,” in Polymerase Chain Reaction, P. Hernandez-Rodriguez, Ed., InTech, 2012. [CrossRef]
- K. J. Land, D. I. Boeras, X.-S. Chen, A. R. Ramsay, and R. W. Peeling, “REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes,” Nat Microbiol, vol. 4, no. 1, pp. 46–54, Dec. 2018. [CrossRef]
- D. R. Thévenot, K. Toth, R. A. Durst, and G. S. Wilson, “ELECTROCHEMICAL BIOSENSORS: RECOMMENDED DEFINITIONS AND CLASSIFICATION *,” Analytical Letters, vol. 34, no. 5, pp. 635–659, Mar. 2001. [CrossRef]
- K.-Y. Wang, Y.-L. Zeng, X.-Y. Yang, W.-B. Li, and X.-P. Lan, “Utility of aptamer-fluorescence in situ hybridization for rapid detection of Pseudomonas aeruginosa,” Eur J Clin Microbiol Infect Dis, vol. 30, no. 2, pp. 273–278, Feb. 2011. [CrossRef]
- J. Zhou and J. Rossi, “Aptamers as targeted therapeutics: current potential and challenges,” Nat Rev Drug Discov, vol. 16, no. 3, pp. 181–202, Mar. 2017. [CrossRef]
- S. Y. Toh, M. Citartan, S. C. B. Gopinath, and T.-H. Tang, “Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay,” Biosensors and Bioelectronics, vol. 64, pp. 392–403, Feb. 2015. [CrossRef]
- E. Dausse, S. Da Rocha Gomes, and J.-J. Toulmé, “Aptamers: a new class of oligonucleotides in the drug discovery pipeline?,” Current Opinion in Pharmacology, vol. 9, no. 5, pp. 602–607, Oct. 2009. [CrossRef]
- T. Wandtke, E. Wędrowska, M. Szczur, G. Przybylski, M. Libura, and P. Kopiński, “Aptamers—Diagnostic and Therapeutic Solution in SARS-CoV-2,” IJMS, vol. 23, no. 3, p. 1412, Jan. 2022. [CrossRef]
- T. Wandtke, J. Woźniak, and P. Kopiński, “Aptamers in Diagnostics and Treatment of Viral Infections,” Viruses, vol. 7, no. 2, pp. 751–780, Feb. 2015. [CrossRef]
- T. Wang, C. Chen, L. M. Larcher, R. A. Barrero, and R. N. Veedu, “Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development,” Biotechnology Advances, vol. 37, no. 1, pp. 28–50, Jan. 2019. [CrossRef]
- P. Röthlisberger and M. Hollenstein, “Aptamer chemistry,” Advanced Drug Delivery Reviews, vol. 134, pp. 3–21, Sep. 2018. [CrossRef]
- K. Y. Chan, A. B. Kinghorn, M. Hollenstein, and J. A. Tanner, “Chemical Modifications for a Next Generation of Nucleic Acid Aptamers,” ChemBioChem, vol. 23, no. 15, p. e202200006, Aug. 2022. [CrossRef]
- N. E. Trunzo and K. L. Hong, “Recent Progress in the Identification of Aptamers Against Bacterial Origins and Their Diagnostic Applications,” IJMS, vol. 21, no. 14, p. 5074, Jul. 2020. [CrossRef]
- J. Yi et al., “The research of aptamer biosensor technologies for detection of microorganism,” Appl Microbiol Biotechnol, vol. 104, no. 23, pp. 9877–9890, Dec. 2020. [CrossRef]
- F. Rizzotto, M. Marin, C. Péchoux, S. Auger, and J. Vidic, “Colorimetric aptasensor for detection of Bacillus cytotoxicus spores in milk and ready-to-use food,” Heliyon, vol. 9, no. 7, p. e17562, Jul. 2023. [CrossRef]
- I. Bobrinetskiy et al., “Advances in Nanomaterials-Based Electrochemical Biosensors for Foodborne Pathogen Detection,” Nanomaterials, vol. 11, no. 10, p. 2700, Oct. 2021. [CrossRef]
- S. Sharifi et al., “Detection of pathogenic bacteria via nanomaterials-modified aptasensors,” Biosensors and Bioelectronics, vol. 150, p. 111933, Feb. 2020. [CrossRef]
- M. Liu, F. Yue, Q. Kong, Z. Liu, Y. Guo, and X. Sun, “Aptamers against Pathogenic Bacteria: Selection Strategies and Apta-assay/Aptasensor Application for Food Safety,” Agricultural and food chemistry, vol. 70, no. 18, pp. 5477–5498, May 2022. [CrossRef]
- Andrew D. Ellington and Jack W. Szostak, “In vitro selection of RNA molecules that bind specific ligands,” Nature, vol. 346, no. 6287, pp. 818–822, Aug. 1990. [CrossRef]
- C. Tuerk and L. Gold, “Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase,” Science, New Series, vol. 249, no. 4968, pp. 505–510, 1990. [CrossRef]
- T. Sampson, “Aptamers and SELEX: the technology,” World Patent Information, vol. 25, no. 2, pp. 123–129, Jun. 2003. [CrossRef]
- E. Dausse et al., “Aptamer selection by direct microfluidic recovery and surface plasmon resonance evaluation,” Biosensors and Bioelectronics, vol. 80, pp. 418–425, Jun. 2016. [CrossRef]
- N. Komarova and A. Kuznetsov, “Inside the Black Box: What Makes SELEX Better?,” Molecules, vol. 24, no. 19, p. 3598, Oct. 2019. [CrossRef]
- M. Kohlberger and G. Gadermaier, “SELEX: Critical factors and optimization strategies for successful aptamer selection,” Biotech and App Biochem, vol. 69, no. 5, pp. 1771–1792, Oct. 2022. [CrossRef]
- C. Zhu et al., “Recent progress of SELEX methods for screening nucleic acid aptamers,” Talanta, vol. 266, p. 124998, Jan. 2024. [CrossRef]
- K. Wang, M. Wang, T. Ma, W. Li, and H. Zhang, “Review on the Selection of Aptamers and Application in Paper-Based Sensors,” Biosensors, vol. 13, no. 1, p. 39, Dec. 2022. [CrossRef]
- B. Mondal, S. Ramlal, P. S. Lavu, B. N, and J. Kingston, “Highly Sensitive Colorimetric Biosensor for Staphylococcal Enterotoxin B by a Label-Free Aptamer and Gold Nanoparticles,” Front. Microbiol., vol. 9, p. 179, Feb. 2018. [CrossRef]
- Y. Huang et al., “A multicolor time-resolved fluorescence aptasensor for the simultaneous detection of multiplex Staphylococcus aureus enterotoxins in the milk,” Biosensors and Bioelectronics, vol. 74, pp. 170–176, Dec. 2015. [CrossRef]
- E. Frohnmeyer, F. Frisch, S. Falke, C. Betzel, and M. Fischer, “Highly affine and selective aptamers against cholera toxin as capture elements in magnetic bead-based sandwich ELAA,” Journal of Biotechnology, vol. 269, pp. 35–42, Mar. 2018. [CrossRef]
- J. G. Bruno, A. M. Richarte, M. P. Carrillo, and A. Edge, “An aptamer beacon responsive to botulinum toxins,” Biosensors and Bioelectronics, vol. 31, no. 1, pp. 240–243, Jan. 2012. [CrossRef]
- A. Subekin, R. Alieva, V. Kukushkin, I. Oleynikov, and E. Zavyalova, “Rapid SERS Detection of Botulinum Neurotoxin Type A,” Nanomaterials, vol. 13, no. 18, p. 2531, Sep. 2023. [CrossRef]
- N. A. Molejon et al., “Selection of G-rich ssDNA aptamers for the detection of enterotoxins of the cholera toxin family,” Analytical Biochemistry, vol. 669, p. 115118, May 2023. [CrossRef]
- S. R. Han and S.-W. Lee, “In vitro selection of RNA aptamer specific to Staphylococcus aureus,” Ann Microbiol, vol. 64, no. 2, pp. 883–885, Jun. 2014. [CrossRef]
- I. M. Ferreira, C. M. De Souza Lacerda, L. S. De Faria, C. R. Corrêa, and A. S. R. De Andrade, “Selection of Peptidoglycan-Specific Aptamers for Bacterial Cells Identification,” Appl Biochem Biotechnol, vol. 174, no. 7, pp. 2548–2556, Dec. 2014. [CrossRef]
- J. Moon, G. Kim, S. Park, J. Lim, and C. Mo, “Comparison of Whole-Cell SELEX Methods for the Identification of Staphylococcus Aureus-Specific DNA Aptamers,” Sensors, vol. 15, no. 4, pp. 8884–8897, Apr. 2015. [CrossRef]
- K. M. Wijesinghe, G. Sabbih, C. H. Algama, R. Syed, M. K. Danquah, and S. Dhakal, “FRET-Based Single-Molecule Detection of Pathogen Protein IsdA Using Computationally Selected Aptamers,” Anal. Chem., vol. 95, no. 26, pp. 9839–9846, Jul. 2023. [CrossRef]
- S. Ohuchi, “Cell-SELEX Technology,” BioResearch Open Access, vol. 1, no. 6, pp. 265–272, Dec. 2012. [CrossRef]
- K. Sefah, D. Shangguan, X. Xiong, M. B. O’Donoghue, and W. Tan, “Development of DNA aptamers using Cell-SELEX,” Nat Protoc, vol. 5, no. 6, pp. 1169–1185, Jun. 2010. [CrossRef]
- K. N. Morris, K. B. Jensen, C. M. Julin, M. Weil, and L. Gold, “High affinity ligands from in vitro selection: Complex targets,” Proc. Natl. Acad. Sci. U.S.A., vol. 95, no. 6, pp. 2902–2907, Mar. 1998. [CrossRef]
- Y.-W. Zhao, H.-X. Wang, G.-C. Jia, and Z. Li, “Application of Aptamer-Based Biosensor for Rapid Detection of Pathogenic Escherichia coli,” Sensors, vol. 18, no. 8, p. 2518, Aug. 2018. [CrossRef]
- W. Zhao et al., “Ultrasensitive dual-enhanced sandwich strategy for simultaneous detection of Escherichia coli and Staphylococcus aureus based on optimized aptamers-functionalized magnetic capture probes and graphene oxide-Au nanostars SERS tags,” Journal of Colloid and Interface Science, vol. 634, pp. 651–663, Mar. 2023. [CrossRef]
- H. P. Dwivedi, R. D. Smiley, and L.-A. Jaykus, “Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX,” Appl Microbiol Biotechnol, vol. 87, no. 6, pp. 2323–2334, Aug. 2010. [CrossRef]
- J. G. Bruno, T. Phillips, M. P. Carrillo, and R. Crowell, “Plastic-Adherent DNA Aptamer-Magnetic Bead and Quantum Dot Sandwich Assay for Campylobacter Detection,” J Fluoresc, vol. 19, no. 3, pp. 427–435, May 2009. [CrossRef]
- M. Alibolandi et al., “Smart AS1411-aptamer conjugated pegylated PAMAM dendrimer for the superior delivery of camptothecin to colon adenocarcinoma in vitro and in vivo,” International Journal of Pharmaceutics, vol. 519, no. 1–2, pp. 352–364, Mar. 2017. [CrossRef]
- X. Cao et al., “Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus,” Nucleic Acids Research, vol. 37, no. 14, pp. 4621–4628, Aug. 2009. [CrossRef]
- T. T.-Q. Nguyen, E. R. Kim, and M. B. Gu, “A new cognate aptamer pair-based sandwich-type electrochemical biosensor for sensitive detection of Staphylococcus aureus,” Biosensors and Bioelectronics, vol. 198, p. 113835, Feb. 2022. [CrossRef]
- Y.-C. Chang, C.-Y. Yang, R.-L. Sun, Y.-F. Cheng, W.-C. Kao, and P.-C. Yang, “Rapid single cell detection of Staphylococcus aureus by aptamer-conjugated gold nanoparticles,” Sci Rep, vol. 3, no. 1, p. 1863, May 2013. [CrossRef]
- S. H. Lim, Y. C. Ryu, and B. H. Hwang, “Aptamer-immobilized Gold Nanoparticles Enable Facile and On-site Detection of Staphylococcus aureus,” Biotechnol Bioproc E, vol. 26, no. 1, pp. 107–113, Feb. 2021. [CrossRef]
- A. Manfredini, E. Malusà, and L. Canfora, “Aptamer-based technology for detecting Bacillus subtilis in soil,” Appl Microbiol Biotechnol, vol. 107, no. 22, pp. 6963–6972, Nov. 2023. [CrossRef]
- H. P. Dwivedi, R. D. Smiley, and L.-A. Jaykus, “Selection of DNA aptamers for capture and detection of Salmonella Typhimurium using a whole-cell SELEX approach in conjunction with cell sorting,” Appl Microbiol Biotechnol, vol. 97, no. 8, pp. 3677–3686, Apr. 2013. [CrossRef]
- S. H. Suh, H. P. Dwivedi, S. J. Choi, and L.-A. Jaykus, “Selection and characterization of DNA aptamers specific for Listeria species,” Analytical Biochemistry, vol. 459, pp. 39–45, Aug. 2014. [CrossRef]
- P. Setlow, “Resistance of spores of Bacillus species to ultraviolet light,” Environ and Mol Mutagen, vol. 38, no. 2–3, pp. 97–104, Jan. 2001. [CrossRef]
- P. Setlow and G. Christie, “New Thoughts on an Old Topic: Secrets of Bacterial Spore Resistance Slowly Being Revealed,” Microbiol Mol Biol Rev, vol. 87, no. 2, pp. e00080-22, Jun. 2023. [CrossRef]
- V. Mazzaracchio et al., “A label-free impedimetric aptasensor for the detection of Bacillus anthracis spore simulant,” Biosensors and Bioelectronics, vol. 126, pp. 640–646, Feb. 2019. [CrossRef]
- C. Zhou et al., “Aptamer-Conjugated Polydiacetylene Colorimetric Paper Chip for the Detection of Bacillus thuringiensis Spores,” Sensors, vol. 20, no. 11, p. 3124, Jun. 2020. [CrossRef]
- M. Ikanovic et al., “Fluorescence Assay Based on Aptamer-Quantum Dot Binding to Bacillus thuringiensis Spores,” J Fluoresc, vol. 17, no. 2, pp. 193–199, Feb. 2007. [CrossRef]
- A. Asif, H. Mohsin, R. Tanvir, and Y. Rehman, “Revisiting the Mechanisms Involved in Calcium Chloride Induced Bacterial Transformation,” Front. Microbiol., vol. 8, p. 2169, Nov. 2017. [CrossRef]
- K. Raval and T. Ganatra, “Basics, types and applications of molecular docking: A review,” IJCAAP, vol. 7, no. 1, pp. 12–16, Mar. 2022. [CrossRef]
- C. Dominguez, R. Boelens, and A. M. J. J. Bonvin, “HADDOCK: A Protein−Protein Docking Approach Based on Biochemical or Biophysical Information,” J. Am. Chem. Soc., vol. 125, no. 7, pp. 1731–1737, Feb. 2003. [CrossRef]
- G. M. Morris, R. Huey, and A. J. Olson, “Using AutoDock for Ligand-Receptor Docking,” CP in Bioinformatics, vol. 24, no. 1, Dec. 2008. [CrossRef]
- S. Soon and N. Aina Nordin, “In silico predictions and optimization of aptamers against Streptococcus agalactiae surface protein using computational docking,” Materials Today: Proceedings, vol. 16, pp. 2096–2100, 2019. [CrossRef]
- A. Escamilla-Gutiérrez, M. G. Córdova-Espinoza, A. Sánchez-Monciváis, B. Tecuatzi-Cadena, A. G. Regalado-García, and K. Medina-Quero, “In silico selection of aptamers for bacterial toxins detection,” Journal of Biomolecular Structure and Dynamics, vol. 41, no. 20, pp. 10909–10918, Dec. 2023. [CrossRef]
- M. Moradi, H. Mohabatkar, M. Behbahani, and G. Dini, “Application of G-quadruplex aptamer conjugated MSNs to deliver ampicillin for suppressing S. aureus biofilm on mice bone,” Arabian Journal of Chemistry, vol. 15, no. 11, p. 104274, Nov. 2022. [CrossRef]
- R. Selvam, I. H. Y. Lim, J. C. Lewis, C. H. Lim, M. K. K. Yap, and H. S. Tan, “Selecting antibacterial aptamers against the BamA protein in Pseudomonas aeruginosa by incorporating genetic algorithm to optimise computational screening method,” Sci Rep, vol. 13, no. 1, p. 7582, May 2023. [CrossRef]
- K. Yuan et al., “Extracellular Milieu and Membrane Receptor Dual-Driven DNA Nanorobot for Accurate in Vivo Tumor Imaging,” CCS Chem, vol. 4, no. 5, pp. 1597–1609, May 2022. [CrossRef]
- S. B. Ebrahimi, D. Samanta, H. F. Cheng, L. I. Nathan, and C. A. Mirkin, “Forced Intercalation (FIT)-Aptamers,” J. Am. Chem. Soc., vol. 141, no. 35, pp. 13744–13748, Sep. 2019. [CrossRef]
- G. N. Parkinson, M. P. H. Lee, and S. Neidle, “Crystal structure of parallel quadruplexes from human telomeric DNA,” Nature, vol. 417, no. 6891, pp. 876–880, Jun. 2002. [CrossRef]
- R. F. Macaya, P. Schultze, F. W. Smith, J. A. Roe, and J. Feigon, “Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution.,” Proc. Natl. Acad. Sci. U.S.A., vol. 90, no. 8, pp. 3745–3749, Apr. 1993. [CrossRef]
- M. C. Cowperthwaite and A. D. Ellington, “Bioinformatic Analysis of the Contribution of Primer Sequences to Aptamer Structures,” J Mol Evol, vol. 67, no. 1, pp. 95–102, Jul. 2008. [CrossRef]
- M. McKeague et al., “Analysis of In Vitro Aptamer Selection Parameters,” J Mol Evol, vol. 81, no. 5–6, pp. 150–161, Dec. 2015. [CrossRef]
- P. C. Sabeti, P. J. Unrau, and D. P. Bartel, “Accessing rare activities from random RNA sequences: the importance of the length of molecules in the starting pool,” Chemistry & Biology, vol. 4, no. 10, pp. 767–774, Oct. 1997. [CrossRef]
- R. Thevendran and M. Citartan, “Assays to Estimate the Binding Affinity of Aptamers,” Talanta, vol. 238, p. 122971, Feb. 2022. [CrossRef]
- H. Kaur and L.-Y. L. Yung, “Probing High Affinity Sequences of DNA Aptamer against VEGF165,” PLoS ONE, vol. 7, no. 2, p. e31196, Feb. 2012. [CrossRef]
- J. Moon, G. Kim, S. Lee, and S. Park, “Identification of Salmonella Typhimurium-specific DNA aptamers developed using whole-cell SELEX and FACS analysis,” Journal of Microbiological Methods, vol. 95, no. 2, pp. 162–166, Nov. 2013. [CrossRef]
- N. Duan, M. Ye, M. Lu, X. Chen, and S. Wu, “DNA aptamers selection and characterization for development of impedimetric aptasensor for Bacillus cereus at different growing stages,” Advanced Agrochem, vol. 2, no. 3, pp. 284–290, Sep. 2023. [CrossRef]
- D. Yılmaz, T. Muslu, A. Parlar, H. Kurt, and M. Yüce, “SELEX against whole-cell bacteria resulted in lipopolysaccharide binding aptamers,” Journal of Biotechnology, vol. 354, pp. 10–20, Aug. 2022. [CrossRef]
- J.-L. Mergny and L. Lacroix, “Analysis of Thermal Melting Curves,” Oligonucleotides, vol. 13, no. 6, pp. 515–537, Dec. 2003. [CrossRef]
- Y. Luo, A. Granzhan, D. Verga, and J.-L. Mergny, “FRET-MC: A fluorescence melting competition assay for studying G4 structures in vitro,” Biopolymers, vol. 112, no. 4, Dec. 2020. [CrossRef]
- V. Esposito et al., “A straightforward modification in the thrombin binding aptamer improving the stability, affinity to thrombin and nuclease resistance,” Org. Biomol. Chem., vol. 12, no. 44, pp. 8840–8843, 2014. [CrossRef]
- J.-L. Mergny, “Thermal difference spectra: a specific signature for nucleic acid structures,” Nucleic Acids Research, vol. 33, no. 16, pp. e138–e138, Sep. 2005. [CrossRef]
- I. Smirnov and R. H. Shafer, “Effect of Loop Sequence and Size on DNA Aptamer Stability,” Biochemistry, vol. 39, no. 6, pp. 1462–1468, Feb. 2000. [CrossRef]
- S. M. Reilly, R. K. Morgan, T. A. Brooks, and R. M. Wadkins, “Effect of Interior Loop Length on the Thermal Stability and p K a of i-Motif DNA,” Biochemistry, vol. 54, no. 6, pp. 1364–1370, Feb. 2015. [CrossRef]
- S. Ahmed, M. Kaushik, S. Chaudhary, and S. Kukreti, “Structural polymorphism of a cytosine-rich DNA sequence forming i-motif structure: Exploring pH based biosensors,” International Journal of Biological Macromolecules, vol. 111, pp. 455–461, May 2018. [CrossRef]
- B. I. Kankia and L. A. Marky, “Folding of the Thrombin Aptamer into a G-Quadruplex with Sr 2+ : Stability, Heat, and Hydration,” J. Am. Chem. Soc., vol. 123, no. 44, pp. 10799–10804, Nov. 2001. [CrossRef]
- N. Zhang et al., “Structural Biology for the Molecular Insight between Aptamers and Target Proteins,” IJMS, vol. 22, no. 8, p. 4093, Apr. 2021. [CrossRef]
- D. Shangguan, Z. Tang, P. Mallikaratchy, Z. Xiao, and W. Tan, “Optimization and Modifications of Aptamers Selected from Live Cancer Cell Lines,” ChemBioChem, vol. 8, no. 6, pp. 603–606, Apr. 2007. [CrossRef]
- T. Bing, X. Yang, H. Mei, Z. Cao, and D. Shangguan, “Conservative secondary structure motif of streptavidin-binding aptamers generated by different laboratories,” Bioorganic & Medicinal Chemistry, vol. 18, no. 5, pp. 1798–1805, Mar. 2010. [CrossRef]
- G. Xu et al., “Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer,” Nucleic Acids Research, vol. 51, no. 14, pp. 7666–7674, Aug. 2023. [CrossRef]
- M. Gellert, M. N. Lipsett, and D. R. Davies, “HELIX FORMATION BY GUANYLIC ACID,” Proc. Natl. Acad. Sci. U.S.A., vol. 48, no. 12, pp. 2013–2018, Dec. 1962. [CrossRef]
- K. W. Lim and A. T. Phan, “Structural Basis of DNA Quadruplex–Duplex Junction Formation,” Angew Chem Int Ed, vol. 52, no. 33, pp. 8566–8569, Aug. 2013. [CrossRef]
- R. Stoltenburg, P. Krafčiková, V. Víglaský, and B. Strehlitz, “G-quadruplex aptamer targeting Protein A and its capability to detect Staphylococcus aureus demonstrated by ELONA,” Sci Rep, vol. 6, no. 1, p. 33812, Sep. 2016. [CrossRef]
- K. Gehring, J.-L. Leroy, and M. Gueron, “A tetrameric DNA structure with protonated cytosine· cytosine base pairs,” vol. 363, 1993. [CrossRef]
- X.-J. Lu, “3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures,” Nucleic Acids Research, vol. 31, no. 17, pp. 5108–5121, Sep. 2003. [CrossRef]
- A. R. Gruber, R. Lorenz, S. H. Bernhart, R. Neubock, and I. L. Hofacker, “The Vienna RNA Websuite,” Nucleic Acids Research, vol. 36, no. Web Server, pp. W70–W74, May 2008. [CrossRef]
- M. Biesiada, K. J. Purzycka, M. Szachniuk, J. Blazewicz, and R. W. Adamiak, “Automated RNA 3D Structure Prediction with RNAComposer,” in RNA Structure Determination, vol. 1490, D. H. Turner and D. H. Mathews, Eds., in Methods in Molecular Biology, vol. 1490., New York, NY: Springer New York, 2016, pp. 199–215. [CrossRef]
- B. Gao, Y.-T. Zheng, A.-M. Su, B. Sun, X.-G. Xi, and X.-M. Hou, “Remodeling the conformational dynamics of I-motif DNA by helicases in ATP-independent mode at acidic environment,” iScience, vol. 25, no. 1, p. 103575, Jan. 2022. [CrossRef]
- J. Kypr, I. Kejnovska, D. Renciuk, and M. Vorlickova, “Circular dichroism and conformational polymorphism of DNA,” Nucleic Acids Research, vol. 37, no. 6, pp. 1713–1725, Jan. 2009. [CrossRef]
- P. Bielecka, A. Dembska, and B. Juskowiak, “Monitoring of pH Using an i-Motif-Forming Sequence Containing a Fluorescent Cytosine Analogue, tC,” Molecules, vol. 24, no. 5, p. 952, Mar. 2019. [CrossRef]
- C. Chen et al., “Study of pH-Induced Folding and Unfolding Kinetics of the DNA i-Motif by Stopped-Flow Circular Dichroism,” Langmuir, vol. 28, no. 51, pp. 17743–17748, Dec. 2012. [CrossRef]
- T. Santos et al., “Stabilization of a DNA aptamer by ligand binding,” Biochimie, vol. 200, pp. 8–18, Sep. 2022. [CrossRef]
- P.-H. Lin, R.-H. Chen, C.-H. Lee, Y. Chang, C.-S. Chen, and W.-Y. Chen, “Studies of the binding mechanism between aptamers and thrombin by circular dichroism, surface plasmon resonance and isothermal titration calorimetry,” Colloids and Surfaces B: Biointerfaces, vol. 88, no. 2, pp. 552–558, Dec. 2011. [CrossRef]
- R. Troisi, N. Balasco, I. Autiero, L. Vitagliano, and F. Sica, “Structural Insights into Protein–Aptamer Recognitions Emerged from Experimental and Computational Studies,” IJMS, vol. 24, no. 22, p. 16318, Nov. 2023. [CrossRef]
- S. S. Wijmenga and B. N. M. Van Buuren, “The use of NMR methods for conformational studies of nucleic acids,” Progress in Nuclear Magnetic Resonance Spectroscopy, vol. 32, no. 4, pp. 287–387, Jun. 1998. [CrossRef]
- T. Someya, S. Baba, M. Fujimoto, G. Kawai, T. Kumasaka, and K. Nakamura, “Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq,” Nucleic Acids Research, vol. 40, no. 4, pp. 1856–1867, Feb. 2012. [CrossRef]
- E. Menichelli et al., “Discovery of small molecules that target a tertiary-structured RNA,” Proc. Natl. Acad. Sci. U.S.A., vol. 119, no. 48, p. e2213117119, Nov. 2022. [CrossRef]
- R. Troisi et al., “Steric hindrance and structural flexibility shape the functional properties of a guanine-rich oligonucleotide,” Nucleic Acids Research, vol. 51, no. 16, pp. 8880–8890, Sep. 2023. [CrossRef]
- C. Kratschmer and M. Levy, “Effect of Chemical Modifications on Aptamer Stability in Serum,” Nucleic Acid Therapeutics, vol. 27, no. 6, pp. 335–344, Dec. 2017. [CrossRef]
- C. G. Peng and M. J. Damha, “G-quadruplex induced stabilization by 2′-deoxy-2′-fluoro-d-arabinonucleic acids (2′F-ANA),” Nucleic Acids Research, vol. 35, no. 15, pp. 4977–4988, Aug. 2007. [CrossRef]
- J. P. Elskens, J. M. Elskens, and A. Madder, “Chemical Modification of Aptamers for Increased Binding Affinity in Diagnostic Applications: Current Status and Future Prospects,” International Journal of Molecular Sciences, vol. 21, no. 12, p. 4522, Jun. 2020. [CrossRef]
- M. R. Dunn, R. M. Jimenez, and J. C. Chaput, “Analysis of aptamer discovery and technology,” Nat Rev Chem, vol. 1, no. 10, p. 0076, Oct. 2017. [CrossRef]
- Y. Kasahara and M. Kuwahara, “Artificial Specific Binders Directly Recovered from Chemically Modified Nucleic Acid Libraries,” Journal of Nucleic Acids, vol. 2012, pp. 1–13, 2012. [CrossRef]
- J. Byun, “Recent Progress and Opportunities for Nucleic Acid Aptamers,” Life, vol. 11, no. 3, p. 193, Feb. 2021. [CrossRef]
- F. Odeh et al., “Aptamers Chemistry: Chemical Modifications and Conjugation Strategies,” Molecules, vol. 25, no. 1, p. 3, Dec. 2019. [CrossRef]
- A. R. Chandrasekaran, “Nuclease resistance of DNA nanostructures,” Nat Rev Chem, vol. 5, no. 4, pp. 225–239, Feb. 2021. [CrossRef]
- Q. Liu et al., “Enhanced Stability of DNA Nanostructures by Incorporation of Unnatural Base Pairs,” ChemPhysChem, vol. 18, no. 21, pp. 2977–2980, Nov. 2017. [CrossRef]
- Y. Tabuchi, J. Yang, and M. Taki, “Relative Nuclease Resistance of a DNA Aptamer Covalently Conjugated to a Target Protein,” IJMS, vol. 23, no. 14, p. 7778, Jul. 2022. [CrossRef]
- A. Lacroix, T. G. W. Edwardson, M. A. Hancock, M. D. Dore, and H. F. Sleiman, “Development of DNA Nanostructures for High-Affinity Binding to Human Serum Albumin,” J. Am. Chem. Soc., vol. 139, no. 21, pp. 7355–7362, May 2017. [CrossRef]
- N. Ponnuswamy et al., “Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation,” Nat Commun, vol. 8, no. 1, p. 15654, May 2017. [CrossRef]
- J.-W. Keum and H. Bermudez, “Enhanced resistance of DNA nanostructures to enzymatic digestion,” Chem. Commun., no. 45, p. 7036, 2009. [CrossRef]
- T. Gerling, M. Kube, B. Kick, and H. Dietz, “Sequence-programmable covalent bonding of designed DNA assemblies,” Sci. Adv., vol. 4, no. 8, p. eaau1157, Aug. 2018. [CrossRef]
- R. El-Khoury and M. J. Damha, “End-ligation can dramatically stabilize i-motifs at neutral pH,” Chem. Commun., vol. 59, no. 25, pp. 3715–3718, 2023. [CrossRef]
- J. Hahn, S. F. J. Wickham, W. M. Shih, and S. D. Perrault, “Addressing the Instability of DNA Nanostructures in Tissue Culture,” ACS Nano, vol. 8, no. 9, pp. 8765–8775, Sep. 2014. [CrossRef]
- Y. Lian, F. He, H. Wang, and F. Tong, “A new aptamer/graphene interdigitated gold electrode piezoelectric sensor for rapid and specific detection of Staphylococcus aureus,” Biosensors and Bioelectronics, vol. 65, pp. 314–319, Mar. 2015. [CrossRef]
- Q. Kang et al., “A novel Aptamer-induced CHA amplification strategy for ultrasensitive detection of Staphylococcus aureus and NIR-triggered photothermal bactericidal Activity based on aptamer-modified magnetic Fe3O4@AuNRs,” Sensors and Actuators B: Chemical, vol. 382, p. 133554, May 2023. [CrossRef]
- A. Abbaspour, F. Norouz-Sarvestani, A. Noori, and N. Soltani, “Aptamer-conjugated silver nanoparticles for electrochemical dual-aptamer-based sandwich detection of staphylococcus aureus,” Biosensors and Bioelectronics, vol. 68, pp. 149–155, Jun. 2015. [CrossRef]
- M. Marin, F. Rizzotto, V. Léguillier, C. Péchoux, E. Borezee-Durant, and J. Vidic, “Naked-eye detection of Staphylococcus aureus in powdered milk and infant formula using gold nanoparticles,” Journal of Microbiological Methods, vol. 201, p. 106578, Oct. 2022. [CrossRef]
- Y. S. Kim, M. Y. Song, J. Jurng, and B. C. Kim, “Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cell–systematic evolution of ligands by exponential enrichment approach,” Analytical Biochemistry, vol. 436, no. 1, pp. 22–28, May 2013. [CrossRef]
- J. A. DeGrasse, “A Single-Stranded DNA Aptamer That Selectively Binds to Staphylococcus aureus Enterotoxin B,” PLoS ONE, vol. 7, no. 3, p. e33410, Mar. 2012. [CrossRef]
- N. Alizadeh, M. Y. Memar, B. Mehramuz, S. S. Abibiglou, F. Hemmati, and H. Samadi Kafil, “Current advances in aptamer-assisted technologies for detecting bacterial and fungal toxins,” J Appl Microbiol, vol. 124, no. 3, pp. 644–651, Mar. 2018. [CrossRef]
- J. Chen et al., “Multichannel-Structured Three-Dimensional Chip for Highly Sensitive Pathogenic Bacteria Detection Based on Fast DNA-Programmed Signal Polymerization,” Anal. Chem., vol. 90, no. 20, pp. 12019–12026, Oct. 2018. [CrossRef]
- F. Bakhshandeh et al., “A universal bacterial sensor created by integrating a light modulating aptamer complex with photoelectrochemical signal readout,” Biosensors and Bioelectronics, vol. 235, p. 115359, Sep. 2023. [CrossRef]
- Y. Zhang, Y. Liu, Y. Yang, L. Li, X. Tao, and E. Song, “Rapid detection of pathogenic bacteria based on a universal dual-recognition FRET sensing system constructed with aptamer-quantum dots and lectin-gold nanoparticles,” Chinese Chemical Letters, vol. 34, no. 8, p. 108102, Aug. 2023. [CrossRef]
- H.-S. Shin, V. Gedi, J.-K. Kim, and D. Lee, “Detection of Gram-negative bacterial outer membrane vesicles using DNA aptamers,” Sci Rep, vol. 9, no. 1, p. 13167, Sep. 2019. [CrossRef]
- J. C. Niles and M. A. Marletta, “Utilizing RNA Aptamers To Probe a Physiologically Important Heme-Regulated Cellular Network,” ACS Chem. Biol., vol. 1, no. 8, pp. 515–524, Sep. 2006. [CrossRef]
- J. G. Bruno, M. P. Carrillo, T. Phillips, and C. J. Andrews, “A Novel Screening Method for Competitive FRET-Aptamers Applied to E. coli Assay Development,” J Fluoresc, vol. 20, no. 6, pp. 1211–1223, Nov. 2010. [CrossRef]
- R. B. Queirós, N. de-los-Santos-Álvarez, J. P. Noronha, and M. G. F. Sales, “A label-free DNA aptamer-based impedance biosensor for the detection of E. coli outer membrane proteins,” Sensors and Actuators B: Chemical, vol. 181, pp. 766–772, May 2013. [CrossRef]
- D. Jiang et al., “Ultra-sensitive photoelectrochemical aptamer biosensor for detecting E. coli O157:H7 based on nonmetallic plasmonic two-dimensional hydrated defective tungsten oxide nanosheets coupling with nitrogen-doped graphene quantum dots (dWO3•H2O@N-GQDs),” Biosensors and Bioelectronics, vol. 183, p. 113214, Jul. 2021. [CrossRef]
- H. Kaur, M. Shorie, and P. Sabherwal, “Biolayer interferometry-SELEX for Shiga toxin antigenic-peptide aptamers & detection via chitosan-WSe2 aptasensor,” Biosensors and Bioelectronics, vol. 167, p. 112498, Nov. 2020. [CrossRef]
- F. R. W. Schmitz, K. Cesca, A. Valério, D. De Oliveira, and D. Hotza, “Colorimetric detection of Pseudomonas aeruginosa by aptamer-functionalized gold nanoparticles,” Appl Microbiol Biotechnol, vol. 107, no. 1, pp. 71–80, Jan. 2023. [CrossRef]
- H. Zheng, R. Sheng, H. Li, W. Ahmad, and Q. Chen, “Rapid and selective detection of Bacillus cereus in food using cDNA-based up-conversion fluorescence spectrum copy and aptamer modified magnetic separation,” Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, vol. 267, p. 120618, Feb. 2022. [CrossRef]
- J. Yan et al., “Dual recognition strategy for the rapid and precise detection of Bacillus cereus using post-modified nano-MOF and aptamer,” Sensors and Actuators B: Chemical, vol. 386, p. 133745, Jul. 2023. [CrossRef]
- Z. Zhou et al., “Portable dual-aptamer microfluidic chip biosensor for Bacillus cereus based on aptamer tailoring and dumbbell-shaped probes,” Journal of Hazardous Materials, vol. 445, p. 130545, Mar. 2023. [CrossRef]
- C. Y. Effah et al., “A SERS bioassay based on vancomycin-modified PEI-interlayered nanocomposite and aptamer-functionalized SERS tags for synchronous detection of Acinetobacter baumannii and Klebsiella pneumoniae,” Food Chemistry, vol. 423, p. 136242, Oct. 2023. [CrossRef]
- R. Abedi, J. B. Raoof, M. Mohseni, and A. Bagheri Hashkavayi, “Development of a label-free impedimetric aptasensor for the detection of Acinetobacter baumannii bacteria,” Analytical Biochemistry, vol. 679, p. 115288, Oct. 2023. [CrossRef]
- A. Vishwakarma, Y. Meganathan, and M. Ramya, “Aptamer-based assay for rapid detection, surveillance, and screening of pathogenic Leptospira in water samples,” Sci Rep, vol. 13, no. 1, p. 13379, Aug. 2023. [CrossRef]
- M. Sun, N. Ma, H. Shi, L.-Z. Cheong, W. Yang, and Z. Qiao, “A HCR based multivalent aptamer amplifier for ultrasensitive detection of Salmonella,” Sensors and Actuators B: Chemical, vol. 375, p. 132860, Jan. 2023. [CrossRef]
- M. Tavassoli, A. Khezerlou, H. Hamishehkar, A. Ehsani, and B. Khalilzadeh, “An ultrasensitive aptamer-based fluorescent on/off system for trace amount evaluation of Yersinia enterocolitica in food samples,” Microchimica Acta, vol. 190, no. 253, Jun. 2023. [CrossRef]
- P. Luo, Y. Liu, Y. Xia, H. Xu, and G. Xie, “Aptamer biosensor for sensitive detection of toxin A of Clostridium difficile using gold nanoparticles synthesized by Bacillus stearothermophilus,” Biosensors and Bioelectronics, vol. 54, pp. 217–221, Apr. 2014. [CrossRef]
- J. Hu, Z. Shen, L. Tan, J. Yuan, and N. Gan, “Electrochemical aptasensor for simultaneous detection of foodborne pathogens based on a double stirring bars-assisted signal amplification strategy,” Sensors and Actuators B: Chemical, vol. 345, p. 130337, Oct. 2021. [CrossRef]
- J. Vidic et al., “Point-of-Need DNA Testing for Detection of Foodborne Pathogenic Bacteria,” Sensors, vol. 19, no. 5, p. 1100, Mar. 2019. [CrossRef]
- M. Majdinasab, A. Hayat, and J. L. Marty, “Aptamer-based assays and aptasensors for detection of pathogenic bacteria in food samples,” TrAC Trends in Analytical Chemistry, vol. 107, pp. 60–77, Oct. 2018. [CrossRef]
- C. Fischer et al., “Food Sensing: Aptamer-Based Trapping of Bacillus cereus Spores with Specific Detection via Real Time PCR in Milk,” J. Agric. Food Chem., vol. 63, no. 36, pp. 8050–8057, Sep. 2015. [CrossRef]
- Y. S. Kim, J. Chung, M. Y. Song, J. Jurng, and B. C. Kim, “Aptamer cocktails: Enhancement of sensing signals compared to single use of aptamers for detection of bacteria,” Biosensors and Bioelectronics, vol. 54, pp. 195–198, Apr. 2014. [CrossRef]
- X. Lin, P. P. Liu, J. Yan, D. Luan, T. Sun, and X. Bian, “Dual Synthetic Receptor-Based Sandwich Electrochemical Sensor for Highly Selective and Ultrasensitive Detection of Pathogenic Bacteria at the Single-Cell Level,” Anal. Chem., vol. 95, no. 13, pp. 5561–5567, Apr. 2023. [CrossRef]
- Z. Ding, et al., “Rolling Circle Amplification/G-Quadruplex-Based Dual Signal Ratiometric Electrochemical Aptasensor for Ultrasensitive Detection of Pathogenic Bacteria,” ChemElectroChem, vol. 10, no. e202300257, Aug. 2023. [CrossRef]
- V. V. Demidov, “Rolling-circle amplification in DNA diagnostics: the power of simplicity,” Expert Review of Molecular Diagnostics, vol. 2, no. 6, pp. 542–548, Nov. 2002. [CrossRef]
- W. Zhao, M. M. Ali, M. A. Brook, and Y. Li, “Rolling Circle Amplification: Applications in Nanotechnology and Biodetection with Functional Nucleic Acids,” Angew Chem Int Ed, vol. 47, no. 34, pp. 6330–6337, Aug. 2008. [CrossRef]
- M. M. Ali et al., “Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine,” Chem. Soc. Rev., vol. 43, no. 10, p. 3324, 2014. [CrossRef]
- F.-T. Zhang, J. Nie, D.-W. Zhang, J.-T. Chen, Y.-L. Zhou, and X.-X. Zhang, “Methylene Blue as a G-Quadruplex Binding Probe for Label-Free Homogeneous Electrochemical Biosensing,” Anal. Chem., vol. 86, no. 19, pp. 9489–9495, Oct. 2014. [CrossRef]
- J. G. Bruno and M. P. Carrillo, “Development of Aptamer Beacons for Rapid Presumptive Detection of Bacillus Spores,” J Fluoresc, vol. 22, no. 3, pp. 915–924, May 2012. [CrossRef]
- T. Martinović, U. Andjelković, M. Š. Gajdošik, D. Rešetar, and D. Josić, “Foodborne pathogens and their toxins,” Journal of Proteomics, vol. 147, pp. 226–235, Sep. 2016. [CrossRef]
- M. Otto, “Staphylococcus aureus toxins,” Current Opinion in Microbiology, vol. 17, pp. 32–37, Feb. 2014. [CrossRef]
- A.-M. Tătaru, A. Canciu, M. Tertiș, C. Cristea, and A. Cernat, “Staphylococcus aureus – Review on potential targets for sensors development,” Bioelectrochemistry, vol. 153, p. 108492, Oct. 2023. [CrossRef]
- J. Mainil, “Escherichia coli virulence factors,” Veterinary Immunology and Immunopathology, vol. 152, no. 1–2, pp. 2–12, Mar. 2013. [CrossRef]
- W. K. Smits, D. Lyras, D. B. Lacy, M. H. Wilcox, and E. J. Kuijper, “Clostridium difficile infection,” Nat Rev Dis Primers, vol. 2, no. 1, p. 16020, Apr. 2016. [CrossRef]
- C. L. Pickett, E. C. Pesci, D. L. Cottle, G. Russell, A. N. Erdem, and H. Zeytin, “Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB gene,” Infect Immun, vol. 64, no. 6, pp. 2070–2078, Jun. 1996. [CrossRef]
- R. L. T. Churchill, H. Lee, and J. C. Hall, “Detection of Listeria monocytogenes and the toxin listeriolysin O in food,” Journal of Microbiological Methods, vol. 64, no. 2, pp. 141–170, Feb. 2006. [CrossRef]
- A. Platt-Samoraj, “Toxigenic Properties of Yersinia enterocolitica Biotype 1A,” Toxins, vol. 14, no. 2, p. 118, Feb. 2022. [CrossRef]
- G. Sakaguchi, “Clostridium botulinum toxin,” Pharmacology & therapeutics, vol. 19, no. 2, pp. 165–194, 1983. [CrossRef]
- A. Rajkovic, “Microbial toxins and low level of foodborne exposure,” Trends in Food Science & Technology, vol. 38, no. 2, pp. 149–157, Aug. 2014. [CrossRef]
- S. Mousavi Nodoushan, N. Nasirizadeh, J. Amani, R. Halabian, and A. A. Imani Fooladi, “An electrochemical aptasensor for staphylococcal enterotoxin B detection based on reduced graphene oxide and gold nano-urchins,” Biosensors and Bioelectronics, vol. 127, pp. 221–228, Feb. 2019. [CrossRef]
- M. Gholamzad, M. R. Khatami, S. Ghassemi, Z. Vaise Malekshahi, and M. B. Shooshtari, “Detection of Staphylococcus Enterotoxin B (SEB) Using an Immunochromatographic Test Strip,” Jundishapur J Microbiol, vol. 8, no. 9, Sep. 2015. [CrossRef]
- B. Jin et al., “Lateral flow aptamer assay integrated smartphone-based portable device for simultaneous detection of multiple targets using upconversion nanoparticles,” Sensors and Actuators B: Chemical, vol. 276, pp. 48–56, Dec. 2018. [CrossRef]
- C. Roca et al., “Selection of an Aptamer against the Enzyme 1-deoxy-D-xylulose-5-phosphate Reductoisomerase from Plasmodium falciparum,” Pharmaceutics, vol. 14, no. 11, p. 2515, Nov. 2022. [CrossRef]
- K. Wang et al., “Neutralization of Staphylococcal Enterotoxin B by an Aptamer Antagonist,” Antimicrob Agents Chemother, vol. 59, no. 4, pp. 2072–2077, Apr. 2015. [CrossRef]
- L. Rotariu, F. Lagarde, N. Jaffrezic-Renault, and C. Bala, “Electrochemical biosensors for fast detection of food contaminants – trends and perspective,” TrAC Trends in Analytical Chemistry, vol. 79, pp. 80–87, May 2016. [CrossRef]
- L. Xu, D. Li, S. Ramadan, Y. Li, and N. Klein, “Facile biosensors for rapid detection of COVID-19,” Biosensors and Bioelectronics, vol. 170, p. 112673, Dec. 2020. [CrossRef]
- Y. Lu, Z. Shi, and Q. Liu, “Smartphone-based biosensors for portable food evaluation,” Current Opinion in Food Science, vol. 28, pp. 74–81, Aug. 2019. [CrossRef]
- J. R. Choi, “Development of Point-of-Care Biosensors for COVID-19,” Front. Chem., vol. 8, p. 517, May 2020. [CrossRef]
- V. Soheili, S. M. Taghdisi, K. Abnous, and M. Ebrahimi, “Point-of-care detection of Escherichia coli O157:H7 in water using AuNPs-based aptasensor,” Iran J Basic Med Sci, vol. 23, no. 7, 2020. [CrossRef]
- H. Yang, Y. Zhou, and J. Liu, “G-quadruplex DNA for construction of biosensors,” TrAC Trends in Analytical Chemistry, vol. 132, p. 116060, Nov. 2020. [CrossRef]
- J. Dejeu, A. Van Der Heyden, N. Spinelli, E. Defrancq, and L. Coche-Guérente, “Recent progress in the design of G-quadruplex–based electrochemical aptasensors,” Current Opinion in Electrochemistry, vol. 30, p. 100812, Dec. 2021. [CrossRef]
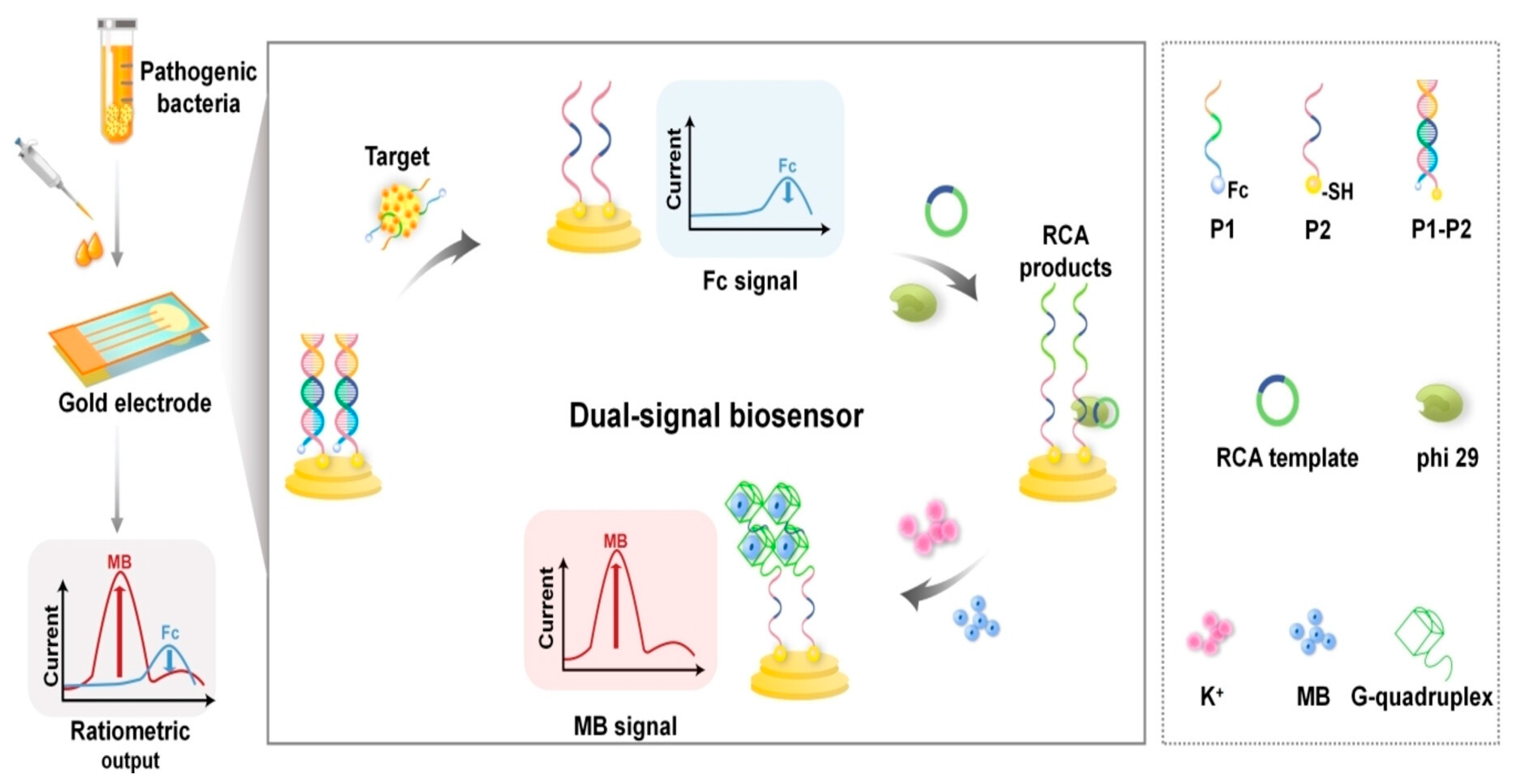
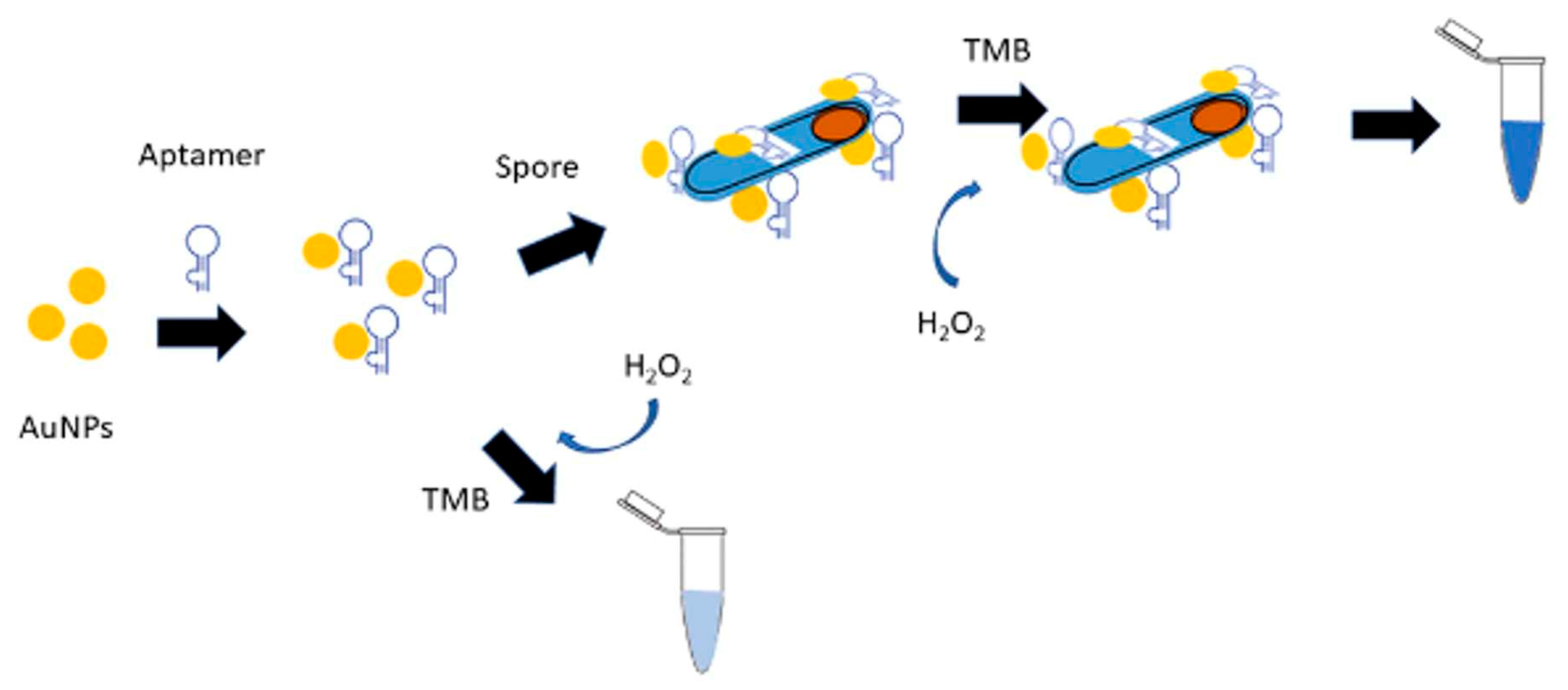
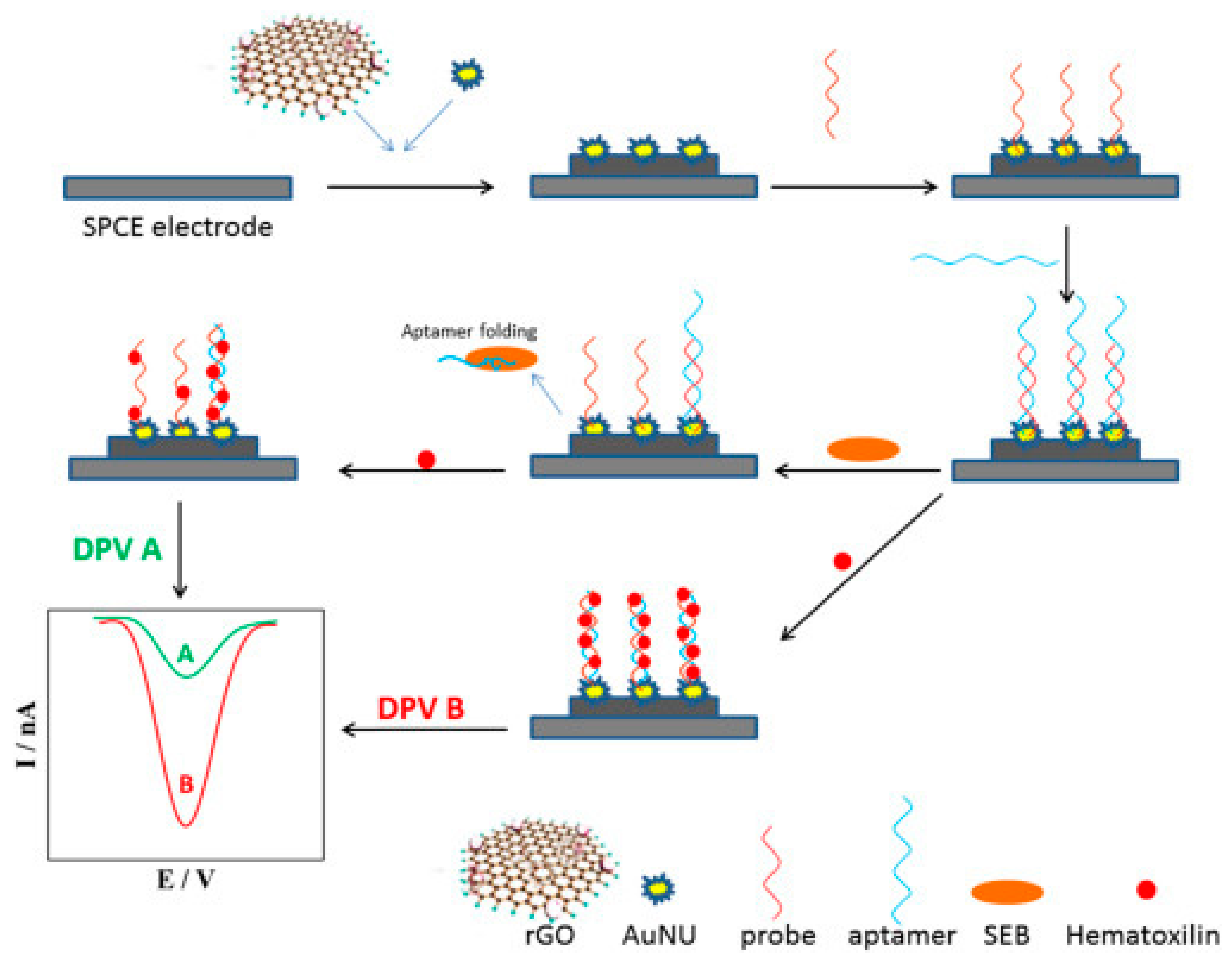
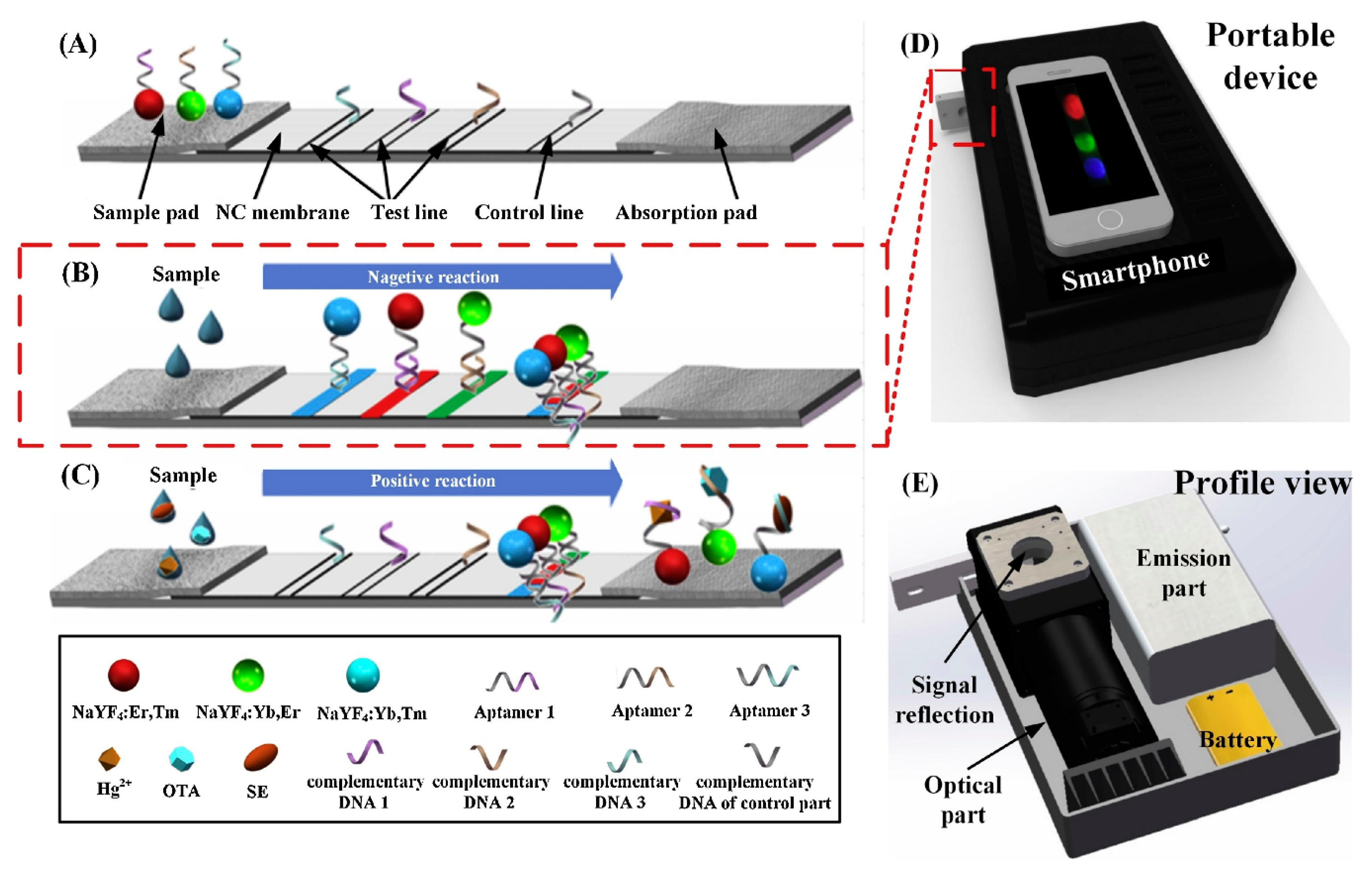
| Bacterium | Aptamer | DNA or RNA | Target | Kd (nM) | Detection method | LOD | Linear range (CFU/mL) | Ref |
|---|---|---|---|---|---|---|---|---|
| S. aureus | SA20 SA23 SA34 SA31 SA43 |
DNA | Whole cell | 70.86±39.22 61.50±22.43 72.42±35.23 82.86±33.20 210.7±135.9 |
Fluorescence |
/ | / | [59] |
| S. aureus | T1 T2 T3 A14 |
RNA & DNA |
IsdA | 2.2 ± 0.5 1.0 ± 0.3 0.7 ± 0.4 4 ± 2 |
FRET-switch | 113 pM 17 pM 11 pM 485 pM |
/ | [50] |
| S. aureus | H1 | DNA | / | / | Piezoelectric sensor | 41 CFU/mL | 4.1 x 101 to 4.1 x 105 | [137] |
| S. aureus | H1 H2 cApt |
DNA | / | / | Fluorescence | 101 CFU/mL | 102-106 | [138] |
| S. aureus | Apt1 Apt2 |
DNA | Whole cell | 35 129 |
Colorimetric | 7,5 – 8,4 * 104 CFU/mL | 104 - 108 | [139,140] |
| S. aureus | SH-Apt2 | DNA | Whole cell | 210.7 | Surface-enhanced Raman scattering (SERS) | / | / | [55,141] |
| S. aureus | APTseb1 | DNA | Staphylococcal Enterotoxin B (SEB) | / | SDS-Page | / | / | [142] |
| S. aureus | G1 #2 #18 |
RNA | Teichoic acid | / | SDS-Page | / | / | [47] |
| S. aureus | AT-27 AT-33 AT-36 AT-49 |
DNA | α-toxin | / | Neutralization | / | / | [143] |
| S. aureus | H1 H2 |
DNA | Whole cell | / | Fluorescence | 4-8 CFU/mL | 45–4.5 × 106 | [144] |
| S. aureus | Antibac1&2 | DNA | Peptidoglycan | 415 + 0.047 1261 + 0.280 |
Photoelectrochemical sensor | 82 pg/mL | / | [48,145] |
| E. coli | SH-Apt1 | DNA | Whole cell | 25,2 | Surface-enhanced Raman scattering (SERS) | / | / | [55,123] |
| E.coli | / | DNA | Whole cell | / | Optical | 45 CFU/mL | 102 -10e8 | [146] |
| E. coli | GN6 GN12 |
DNA | Outer membrane vesicles (OMV) | 29,94 20.36 59.70 24.80 38,98 53.83 |
Enzyme-linked aptamer assay (ELAA) | / | / | [147] |
| E. coli | 6-3 8-1 8-7 8-8 8-12 8-13 8-19 8-35 |
RNA | Heme | 188 309 256 371 445 425 |
Functional assay | / | / | [148] |
| E. coli | ECA I ECA II |
DNA | Outer membrane proteins (OMP) | / | Electrochemistry | / | 1*10-7–2*10-6M | [149,150] |
| E. coli | / | DNA | Whole cell | / | Plasmonic sensor | 0.05 CFU/mL | 0.1-104 | [151] |
| E. coli | Stx1 stx2 |
DNA | Shiga toxin Viz, stx1 & stx2 |
47 pM 29 pM |
/ | 44,5 pg/ml 41,3 pg/mg |
50 pg/ml – 100 ng/mg | [152] |
| P. aeroginosa | F23 | DNA | Whole cell | 17.27 ± 5.00 | colorimetric | 104 CFU/mL | / | [153] |
| B. cereus | / | DNA | Whole cell | / | Fluorescence | 22 CFU/mL | 49-49*106 | [154] |
| B. cereus | / | DNA | Whole Cell | / | Fluorescence | 4 CFU/mL | 20-2*108 | [155] |
| B. cereus | B15 B16 |
DNA | Whole cell | 16,13 20.67 |
Electrochemical sensor | 10 CFU/mL | / | [89] |
| B. cereus | 13-18 13-24 |
DNA | Whole cell | 22.75 36.72 |
Microfluidic chip | 9.27 CFU/mL | / | [156] |
| Acinetobacter baumannii | AB K2 |
DNA | Whole cell | 5.377 6.8 |
SERS | 10 CFU/mL | 10–105 | [157] |
| Acinetobacterer baumanni | / | DNA | Whole cell | / | Electrochemical sensor | 150 CFU/mL | 1*103–1.0*108 | [158] |
| Klebsiella pneumoniae | K2 | DNA | Whole cell | / | SERS | 10 CFU/mL | 10–105 | [157] |
| Leptospira interrogans | LAP3 | DNA | Outer Membrane protein | 133,13 | Colorimetric | 57 CFU/mL | 60–6*105 | [159] |
| Salmonella | Multi-apt | DNA | Multi | 11.72 | Fluorescence | 7 CFU/mL | 10-107 | [160] |
| Yersinia enterocolitica | / | DNA | Whole cell | / | Fluorescence | 3 CFU/mL | 10–109 | [161] |
| Bacillus cytotoxicus | BAS6R | DNA | Spore | / | Colorimetric | 102 – 104 CFU/mL | 103 - 104 | [29] |
| Vibrio cholerae | CT916 | Cholera toxin (CT) | 48,5 | ELAA | 2,1 – 2,4 ng/ml | 0-10 ng/mL | [43] | |
| Clostridium difficile | No name | DNA G-quadruplex |
Toxin A (TOA) | Electrochemistry | 1 nM | 0-200 ng/mL | [162] | |
| S. typhimurium | H2 | DNA | Whole cell | / | Fluorescence | 4-8 CFU/mL | 36–3.6 × 106 | [144] |
| S. Typhimurium | Apt S.T | DNA | Whole cell | 10 | Fluorescence | 30 CFU/mL | 102-106 | [163] |
| Vibrio parahaemolyticus | Apt VP | DNA | Whole cell | 16,88 | Fluorescence | 10 CFU/mL | 102-106 | [163] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





