Submitted:
03 February 2024
Posted:
05 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
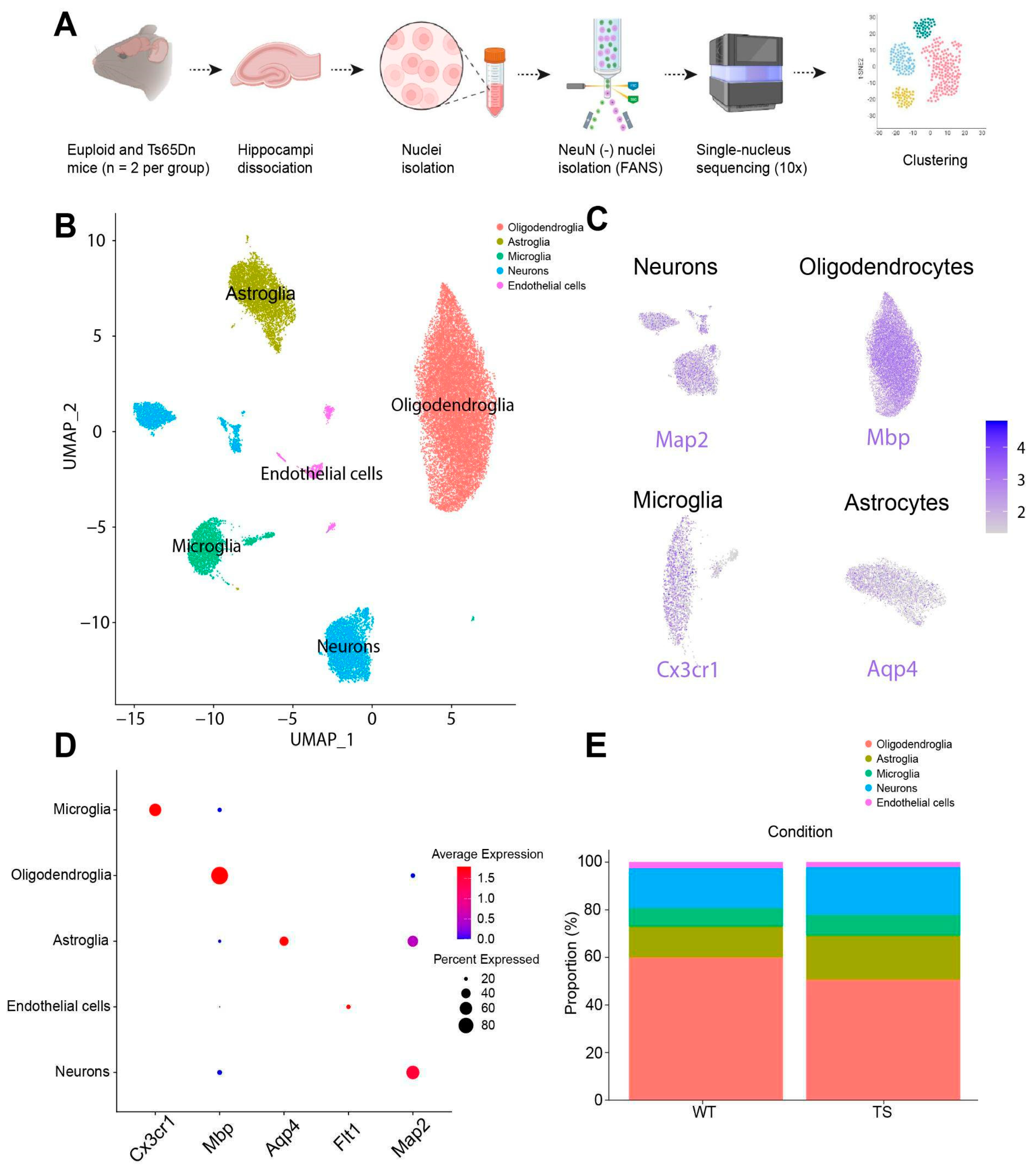
- Unbiased identification of different glial subtypes in WT and Ts65Dn hippocampus
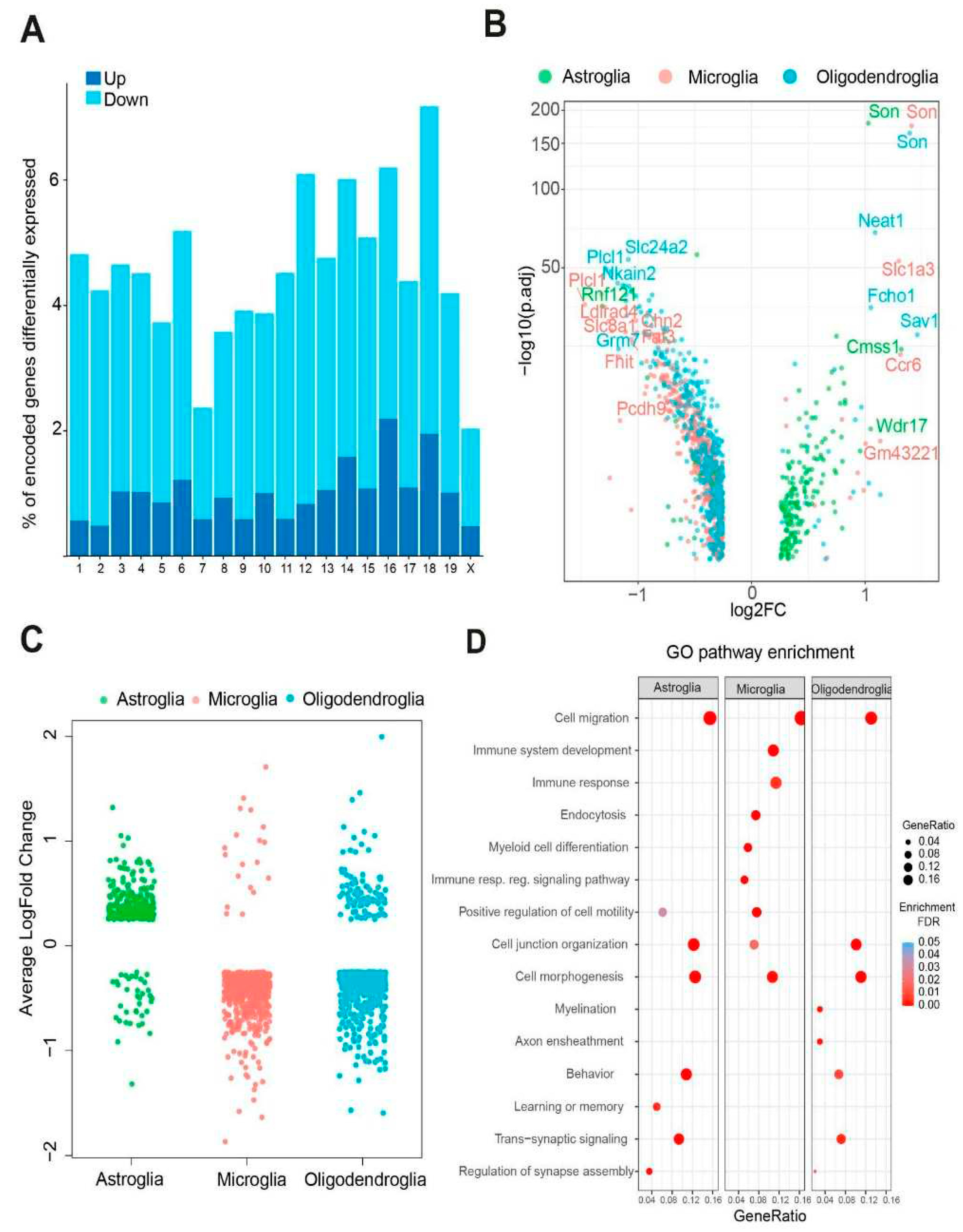
- Trisomic glial cells exhibit cell-type-specific transcriptomic alterations
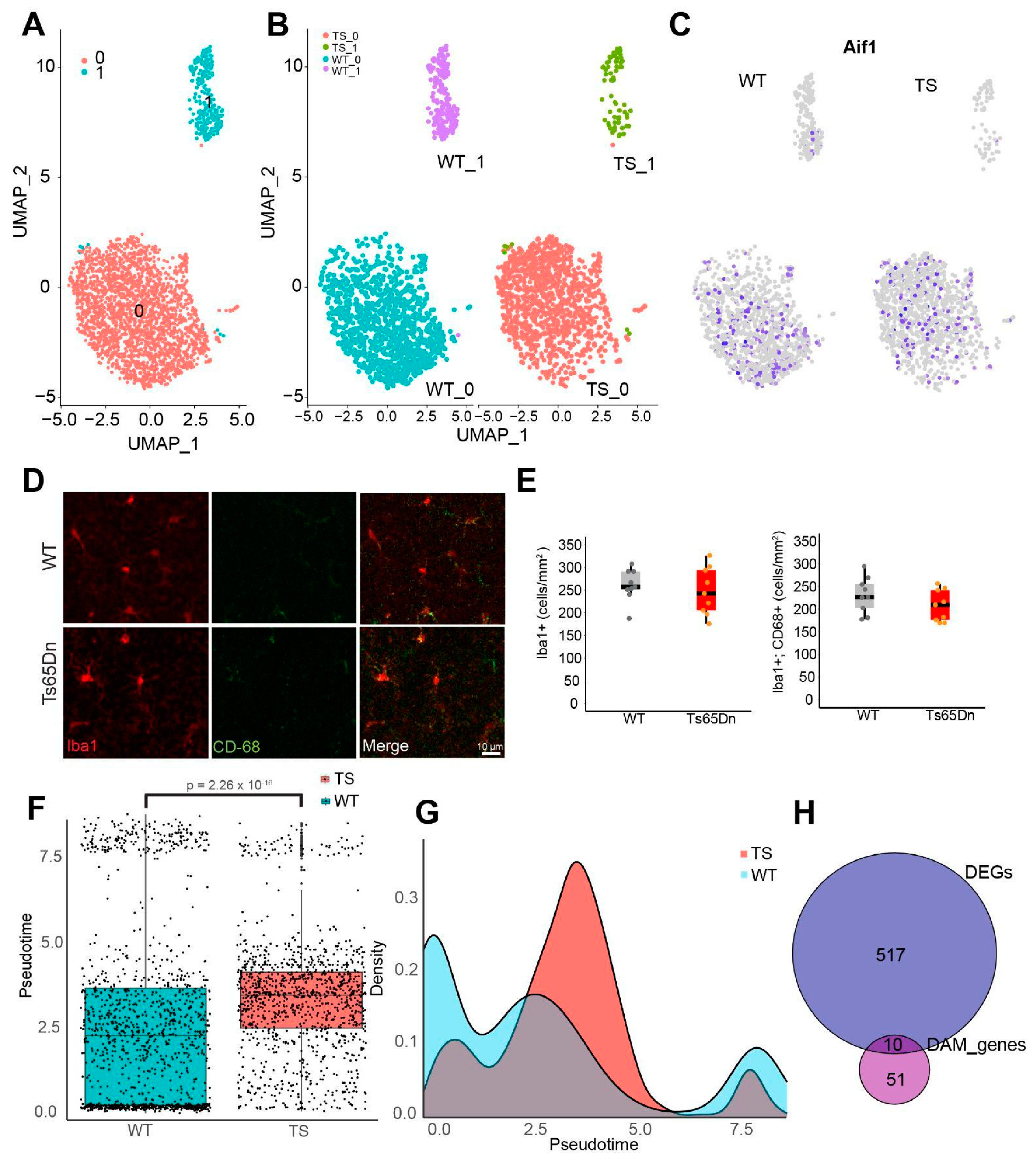
- More than 60% of trisomic microglial cells are in an intermediate reactive state
3. Discussion
4. Materials and Methods
- Animals
- Histology
- Immunohistochemistry
- Cell counting
- Single Nucleus RNA sequencing
- Nucleus isolation
- 10x single-cell barcoding, library preparation and sequencing
- 10x data pre-processing
- Batch Correction and scaling data matrix
- Dimensionality reduction, clustering and visualization
- Identification of marker genes within every cluster
- Identification of differentially expressed genes between WT and Ts65Dn
- Gene set enrichment
- Cellular proportion
- Disease-associated microglia (DAM) markers
- Pseudotime analysis
- Statistical analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ross MH, Galaburda AM, Kemper TL. Down’s syndrome: Is there a decreased population of neurons? Neurology. 1984;34(7):909–16. [CrossRef]
- Guidi S, Bonasoni P, Ceccarelli C, Santini D, Gualtieri F, Ciani E, et al. Neurogenesis impairment and increased cell death reduce total neuron number in the hippocampal region of fetuses with Down syndrome. Brain Pathol. 2008. [CrossRef]
- Pinto B, Morelli G, Rastogi M, Savardi A, Fumagalli A, Petretto A, et al. Rescuing Over-activated Microglia Restores Cognitive Performance in Juvenile Animals of the Dp(16) Mouse Model of Down Syndrome. Neuron . 2020 Dec 9 ;108(5):887-904.e12. Available from: 10.1016/J.NEURON.2020.09.010. [CrossRef]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Neuroscience: Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science (80- ) . 2005 May 27 ;308(5726):1314–8. Available from: 10.1126/SCIENCE.1110647/SUPPL_FILE/1110647S9.MOV. [CrossRef]
- Paolicelli RC, Sierra A, Stevens B, Tremblay ME, Aguzzi A, Ajami B, et al. Microglia states and nomenclature: A field at its crossroads. Neuron . 2022 Nov 2 ;110(21):3458–83. Available from: 10.1016/J.NEURON.2022.10.020. [CrossRef]
- Sarlus H, Heneka MT. Microglia in Alzheimer’s disease. J Clin Invest . 2017 Sep 1 ;127(9):3240–9. Available from: 10.1172/JCI90606. [CrossRef]
- Cserép C, Pósfai B, Lénárt N, Fekete R, László ZI, Lele Z, et al. Microglia monitor and protect neuronal function through specialized somatic purinergic junctions. Science (80- ) . 2020 Jan 31 ;367(6477):528–37. Available from: 10.1126/SCIENCE.AAX6752/SUPPL_FILE/AAX6752_S7.MOV. [CrossRef]
- Prinz M, Jung S, Priller J. Microglia Biology: One Century of Evolving Concepts. Cell . 2019 Oct 3 ;179(2):292–311. Available from: 10.1016/J.CELL.2019.08.053. [CrossRef]
- Cornell J, Salinas S, Huang HY, Zhou M. Microglia regulation of synaptic plasticity and learning and memory. Neural Regen Res . 2022 Apr 1 ;17(4):705–16. Available from: 10.4103/1673-5374.322423. [CrossRef]
- Woodburn SC, Bollinger JL, Wohleb ES. The semantics of microglia activation: neuroinflammation, homeostasis, and stress. J Neuroinflammation 2021 181 . 2021 Nov 6 ;18(1):1–16. Available from: 10.1186/S12974-021-02309-6. [CrossRef]
- Provenzano F, Pérez MJ, Deleidi M. Redefining Microglial Identity in Health and Disease at Single-Cell Resolution. Trends Mol Med . 2021 Jan 1 ;27(1):47–59. Available from: 10.1016/J.MOLMED.2020.09.001 . [CrossRef]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell . 2017 Jun 15 ;169(7):1276-1290.e17. Available from: 10.1016/J.CELL.2017.05.018. [CrossRef]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity . 2017 Sep 19 ;47(3):566-581.e9. Available from: 10.1016/J.IMMUNI.2017.08.008. [CrossRef]
- Sala Frigerio C, Wolfs L, Fattorelli N, Thrupp N, Voytyuk I, Schmidt I, et al. The Major Risk Factors for Alzheimer’s Disease: Age, Sex, and Genes Modulate the Microglia Response to Aβ Plaques. Cell Rep . 2019 Apr 23 ;27(4):1293-1306.e6. Available from: 10.1016/J.CELREP.2019.03.099. [CrossRef]
- Srinivasan K, Friedman BA, Etxeberria A, Huntley MA, van der Brug MP, Foreman O, et al. Alzheimer’s Patient Microglia Exhibit Enhanced Aging and Unique Transcriptional Activation. Cell Rep . 2020 Jun 30 ;31(13). Available from: 10.1016/J.CELREP.2020.107843. [CrossRef]
- Absinta M, Maric D, Gharagozloo M, Garton T, Smith MD, Jin J, et al. A lymphocyte-microglia-astrocyte axis in chronic active multiple sclerosis. Nature . 2021 Sep 30 ;597(7878):709–14. Available from: 10.1038/S41586-021-03892-7. [CrossRef]
- Marschallinger J, Iram T, Zardeneta M, Lee SE, Lehallier B, Haney MS, et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat Neurosci . 2020 Feb 1 ;23(2):194–208. Available from: 10.1038/S41593-019-0566-1. [CrossRef]
- Wierzba-Bobrowicz T, Lewandowska E, Schmidt-Sidor B, Gwiazda E. The comparison of microglia maturation in CNS of normal human fetuses and fetuses with Down’s syndrome. Folia Neuropathol . 1999 Jan 1 ;37(4):227–34.
- Xue QS, Streit WJ. Microglial pathology in Down syndrome. Acta Neuropathol . 2011 Oct 17 ;122(4):455–66. Available from: 10.1007/S00401-011-0864-5/METRICS. [CrossRef]
- Flores-Aguilar L, Iulita MF, Kovecses O, Torres MD, Levi SM, Zhang Y, et al. Evolution of neuroinflammation across the lifespan of individuals with Down syndrome. Brain . 2020 Dec 1 ;143(12):3653–71. Available from: 10.1093/BRAIN/AWAA326. [CrossRef]
- Martini AC, Helman AM, McCarty KL, Lott IT, Doran E, Schmitt FA, et al. Distribution of microglial phenotypes as a function of age and Alzheimer’s disease neuropathology in the brains of people with Down syndrome. Alzheimer’s Dement Diagnosis, Assess Dis Monit . 2020 Jan 1 ;12(1):e12113. Available from: 10.1002/DAD2.12113. [CrossRef]
- Colton CA, Yao J, Taffs RE, Keri JE, Oster-Granite ML. Abnormal production of interleukin-1 by microglia from trisomy 16 mice. Neurosci Lett. 1991 Nov 11;132(2):270–4. [CrossRef]
- Palmer CR, Liu CS, Romanow WJ, Lee MH, Chun J. Altered cell and RNA isoform diversity in aging down syndrome brains. Proc Natl Acad Sci U S A . 2021 Nov 23 ;118(47):e2114326118. Available from: 10.1073/PNAS.2114326118/SUPPL_FILE/PNAS.2114326118.SD10.XLSX. [CrossRef]
- Wilcock DM, Hurban J, Helman AM, Sudduth TL, McCarty KL, Beckett TL, et al. Down syndrome individuals with Alzheimer’s disease have a distinct neuroinflammatory phenotype compared to sporadic Alzheimer’s disease. Neurobiol Aging. 2015 Sep 1;36(9):2468–74. [CrossRef]
- Rueda N, Vidal V, García-Cerro S, Narcís JO, Llorens-Martín M, Corrales A, et al. Anti-IL17 treatment ameliorates Down syndrome phenotypes in mice. Brain Behav Immun. 2018 Oct 1;73:235–51. [CrossRef]
- Illouz T, Madar R, Biragyn A, Okun E. Restoring microglial and astroglial homeostasis using DNA immunization in a Down Syndrome mouse model. Brain Behav Immun. 2019 Jan 1;75:163–80. [CrossRef]
- Lockrow J, Boger H, Bimonte-Nelson H, Granholm AC. Effects of long-term memantine on memory and neuropathology in Ts65Dn mice, a model for Down syndrome. Behav Brain Res. 2011 Aug 10;221(2):610–22. [CrossRef]
- Hamlett ED, Hjorth E, Ledreux A, Gilmore A, Schultzberg M, Granholm AC. RvE1 treatment prevents memory loss and neuroinflammation in the Ts65Dn mouse model of Down syndrome. Glia . 2020 Jul 1 ;68(7):1347–60 Available from: 10.1002/GLIA.23779. [CrossRef]
- Gupta M, Dhanasekaran AR, Gardiner KJ. Mouse models of Down syndrome: gene content and consequences. Mamm Genome 2016 2711 . 2016 Aug 18 ;27(11):538–55. Available from: 10.1007/S00335-016-9661-8. [CrossRef]
- Rueda N, Flórez J, Martínez-Cué C. Mouse models of down syndrome as a tool to unravel the causes of mental disabilities. Neural Plast. 2012;2012.
- Olmos-Serrano JL, Kang HJ, Tyler WA, Silbereis JC, Cheng F, Zhu Y, et al. Down Syndrome Developmental Brain Transcriptome Reveals Defective Oligodendrocyte Differentiation and Myelination. Neuron . 2016 Mar 16 ;89(6):1208–22. Available from: 10.1016/J.NEURON.2016.01.042. [CrossRef]
- Zhou Z, Zhi C, Chen D, Cai Z, Jiang X. Single-nucleus RNA sequencing reveals cell type-specific transcriptome alterations of Down syndrome hippocampus using the Dp16 mouse model. Genes Genomics . 2023 Oct 1 ;45(10):1305–15. Available from: 10.1007/S13258-023-01433-2. [CrossRef]
- Qiu JJ, Liu YN, Wei H, Zeng F, Yan J Bin. Single-cell RNA sequencing of neural stem cells derived from human trisomic iPSCs reveals the abnormalities during neural differentiation of Down syndrome. Front Mol Neurosci . 2023 ;16. Available from: 10.3389/FNMOL.2023.1137123. [CrossRef]
- Ponroy Bally B, Farmer WT, Jones E V., Jessa S, Kacerovsky JB, Mayran A, et al. Human iPSC-derived Down syndrome astrocytes display genome-wide perturbations in gene expression, an altered adhesion profile, and increased cellular dynamics. Hum Mol Genet. 2020;29(5):785–802. [CrossRef]
- Sierra C, Sabariego-Navarro M, Fernández-Blanco Á, Cruciani S, Zamora-Moratalla A, Novoa EM, et al. The lncRNA Snhg11, a new candidate contributing to neurogenesis, plasticity and memory deficits in Down syndrome. Res Sq . 2023 Sep 25 ;rs.3.rs-3184329. Available from: 10.21203/RS.3.RS-3184329/V1. [CrossRef]
- Jones RE, Andrews R, Holmans P, Hill M, Taylor PR. Modest changes in Spi1 dosage reveal the potential for altered microglial function as seen in Alzheimer’s disease. Sci Reports 2021 111 . 2021 Jul 22 [cited 2024 Jan 22];11(1):1–14. Available from: 10.1038/s41598-021-94324-z. [CrossRef]
- Reifschneider A, Robinson S, Van Lengerich B, Gnörich J, Logan T, Heindl S, et al. Loss of TREM2 rescues hyperactivation of microglia, but not lysosomal deficits and neurotoxicity in models of progranulin deficiency. EMBO J . 2022 Feb 15 [cited 2024 Jan 26];41(4). Available from: 10.15252/EMBJ.2021109108. [CrossRef]
- Sobue A, Komine O, Hara Y, Endo F, Mizoguchi H, Watanabe S, et al. Microglial gene signature reveals loss of homeostatic microglia associated with neurodegeneration of Alzheimer’s disease. Acta Neuropathol Commun 2020 91 . 2021 Jan 5 [cited 2024 Jan 26];9(1):1–17. Available from: 10.1186/S40478-020-01099-X. [CrossRef]
- Gally F, Rao DM, Schmitz C, Colvin KL, Yeager ME, Perraud AL. The TRPM2 ion channel contributes to cytokine hyperproduction in a mouse model of Down Syndrome. Biochim Biophys Acta - Mol Basis Dis. 2018 Jan 1;1864(1):126–32. [CrossRef]
- Wilcock DM, Griffin WST. Down’s syndrome, neuroinflammation, and Alzheimer neuropathogenesis. J Neuroinflammation . 2013 Jul 16 ;10. Available from: 10.1186/1742-2094-10-84. [CrossRef]
- Seol S, Kwon J, Kang HJ. Cell type characterization of spatiotemporal gene co-expression modules in Down syndrome brain. iScience . 2022 Jan 20 ;26(1). Available from: 10.1016/J.ISCI.2022.105884. [CrossRef]
- Wolf Y, Shemer A, Levy-Efrati L, Gross M, Kim JS, Engel A, et al. Microglial MHC class II is dispensable for experimental autoimmune encephalomyelitis and cuprizone-induced demyelination. Eur J Immunol . 2018 Aug 1 ;48(8):1308–18. Available from: 10.1002/EJI.201847540. [CrossRef]
- Vázquez-Oliver A, Pérez-García S, Pizarro N, Molina-Porcel L, Torre R de la, Maldonado R, et al. Long-term decreased cannabinoid type-1 receptor activity restores specific neurological phenotypes in the Ts65Dn mouse model of Down syndrome. bioRxiv . 2021 Nov 22 ;2021.11.22.469296. Available from: 10.1101/2021.11.22.469296. [CrossRef]
- Safaiyan S, Besson-Girard S, Kaya T, Cantuti-Castelvetri L, Liu L, Ji H, et al. White matter aging drives microglial diversity. Neuron . 2021 Apr 7 ;109(7):1100-1117.e10. Available from: 10.1016/J.NEURON.2021.01.027. [CrossRef]
- Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, et al. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity . 2019 Jan 15 ;50(1):253-271.e6. Available from: 10.1016/J.IMMUNI.2018.11.004. [CrossRef]
- Erny D, De Angelis ALH, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci . 2015 Jun 25 ;18(7):965–77. Available from: 10.1038/NN.4030. [CrossRef]
- Thion MS, Low D, Silvin A, Chen J, Grisel P, Schulte-Schrepping J, et al. Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner. Cell . 2018 Jan 25 ;172(3):500-516.e16. Available from: 10.1016/J.CELL.2017.11.042. [CrossRef]
- Deczkowska A, Keren-Shaul H, Weiner A, Colonna M, Schwartz M, Amit I. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell . 2018 May 17 ;173(5):1073–81. Available from: 10.1016/j.cell.2018.05.003. [CrossRef]
- Song WM, Colonna M. The identity and function of microglia in neurodegeneration. Nat Immunol 2018 1910 . 2018 Sep 24 ;19(10):1048–58. Available from: 10.1038/s41590-018-0212-1. [CrossRef]
- Song W, Li Q, Wang T, Li Y, Fan T, Zhang J, et al. Putative complement control protein CSMD3 dysfunction impairs synaptogenesis and induces neurodevelopmental disorders. Brain Behav Immun . 2022 May 1 ;102:237–50. Available from: 10.1016/J.BBI.2022.02.027. [CrossRef]
- Kálmán J, Juhász A, Laird G, Dickens P, Járdánházy T, Rimanóczy Á, et al. Serum interleukin-6 levels correlate with the severity of dementia in Down syndrome and in Alzheimer’s disease. Acta Neurol Scand . 1997 Oct 1 ;96(4):236–40. Available from: 10.1111/J.1600-0404.1997.TB00275.X. [CrossRef]
- Zhang Y, Che M, Yuan J, Yu Y, Cao C, Qin XY, et al. Aberrations in circulating inflammatory cytokine levels in patients with Down syndrome: a meta-analysis. Oncotarget . 2017 Oct 10 ;8(48):84489. Available from: 10.18632/ONCOTARGET.21060. [CrossRef]
- Wilcock, DM. Neuroinflammation in the aging down syndrome brain; Lessons from Alzheimer’s disease. Curr Gerontol Geriatr Res. 2012;2012.
- García O, Flores-Aguilar L. Astroglial and microglial pathology in Down syndrome: Focus on Alzheimer’s disease. Front Cell Neurosci . 2022 Sep 20 [cited 2022 Dec 1];16. Available from: 10.3389/FNCEL.2022.987212. [CrossRef]
- Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates - George Paxinos, Keith B.J. Franklin - Google Libros . [cited 2022 Dec 11].
- Martelotto LG. ‘Frankenstein’ protocol for nuclei isolation from fresh and frozen tissue for snRNAseq. 2020 Mar 6 [cited 2022 Dec 11]; Available from: 10.17504/PROTOCOLS.IO.3FKGJKW. [CrossRef]
- Hochgerner H, Zeisel A, Lönnerberg P, Linnarsson S. Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat Neurosci . 2018 Feb 1 [cited 2022 Dec 1];21(2):290–9. Available from: 10.1038/S41593-017-0056-2. [CrossRef]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019 1612 . 2019 Nov 18 ;16(12):1289–96. Available from: 10.1038/s41592-019-0619-0. [CrossRef]
- Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019 202 . 2019 Jan 14 ;20(2):163–72. Available from: 10.1038/s41590-018-0276-y. [CrossRef]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, et al. Molecular Architecture of the Mouse Nervous System. Cell . 2018 Aug 9 ;174(4):999-1014.e22. Available from: 10.1016/j.cell.2018.06.021. [CrossRef]
- Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020 Apr 15;36(8):2628–9. [CrossRef]
- Cao Y, Lin Y, Ormerod JT, Yang P, Yang JYH, Lo KK. scDC: single cell differential composition analysis. BMC Bioinformatics. 2019 Dec 24;20(S19):721. [CrossRef]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 2017 143 . 2017 Jan 23 ;14(3):309–15. Available from: 10.1038/nmeth.4150. [CrossRef]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014 324 . 2014 Mar 23 ;32(4):381–6. Available from: 10.1038/nbt.2859. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




