Submitted:
29 January 2024
Posted:
29 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Progress in Genome Sequencing and Assembly of Jujube Genomes
3. Genomics Improves the Construction of High-Density Genetic Linkage Maps of Jujube
4. Domestication-Related Genes Identified Based on Jujube Genomics
5. Molecular Regulation Identification of Jujube Biology Traits Based on Multi-Omics
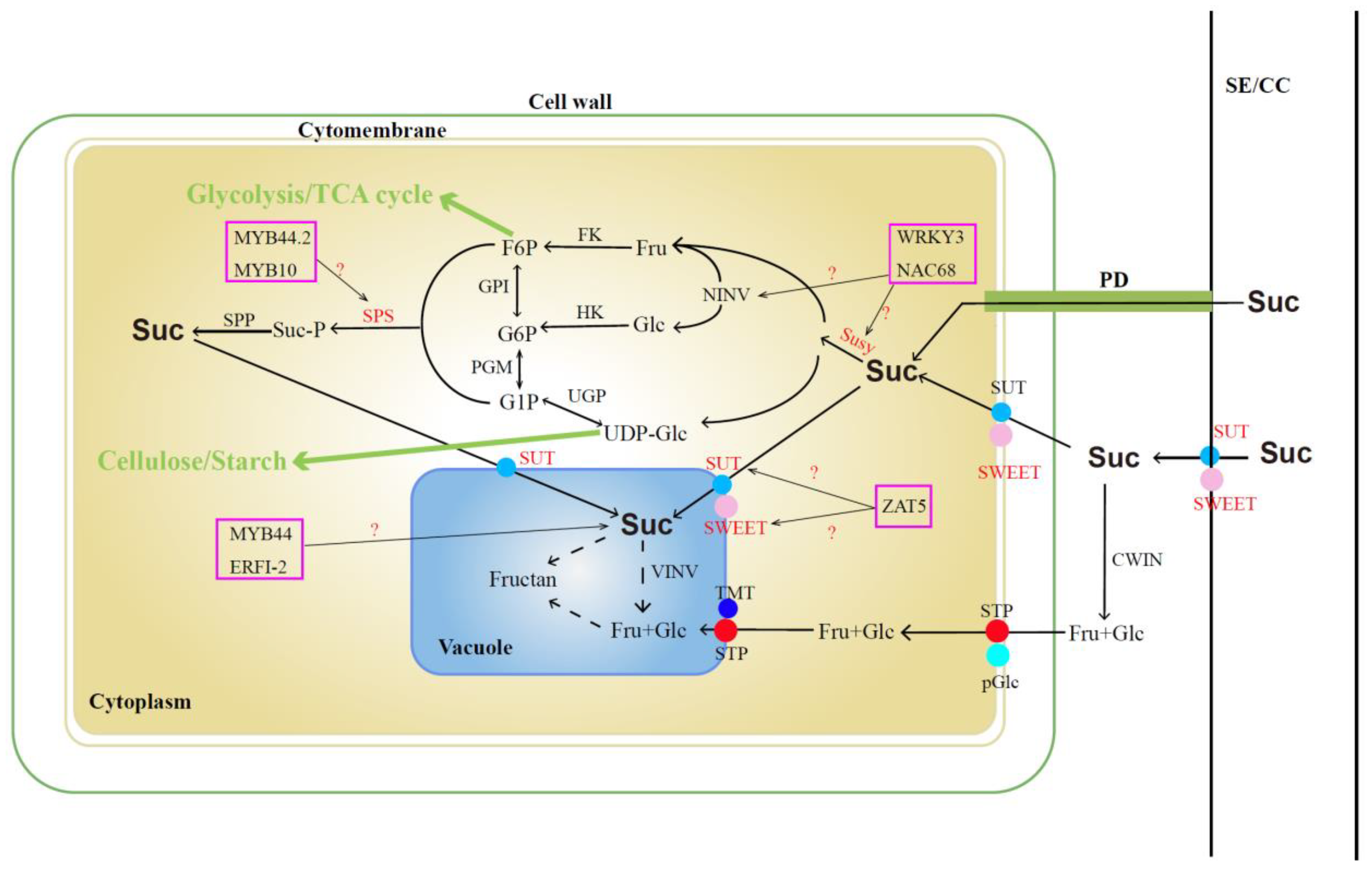
5.1. The Accumulation and Metabolism of Sugar and Acid in Jujube
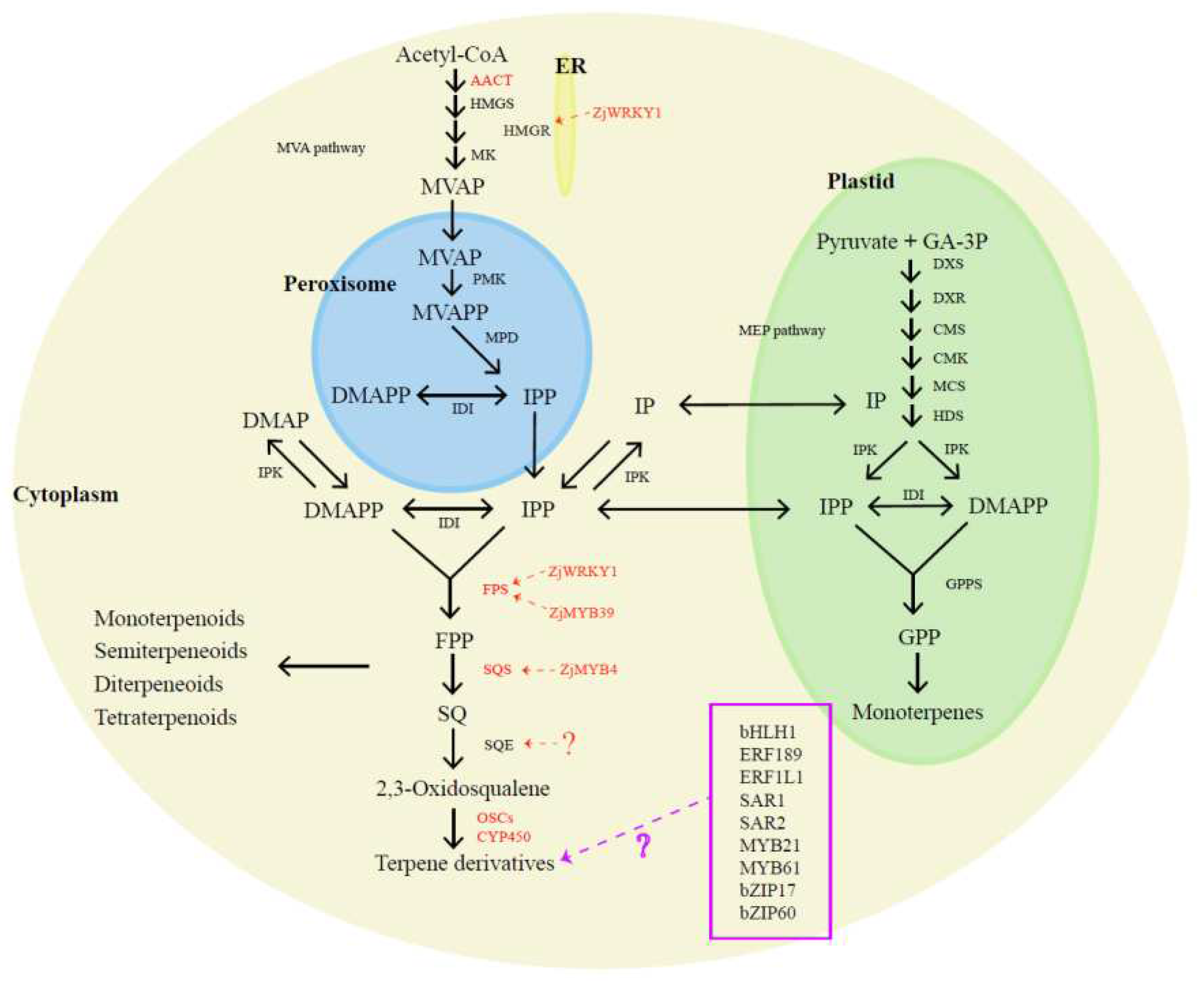
5.2. Biosynthesis of Terpenoids in Jujube
5.3. Biosynthesis of Jujube Flavonoids
5.4. Abiotic Stress Response of Jujube
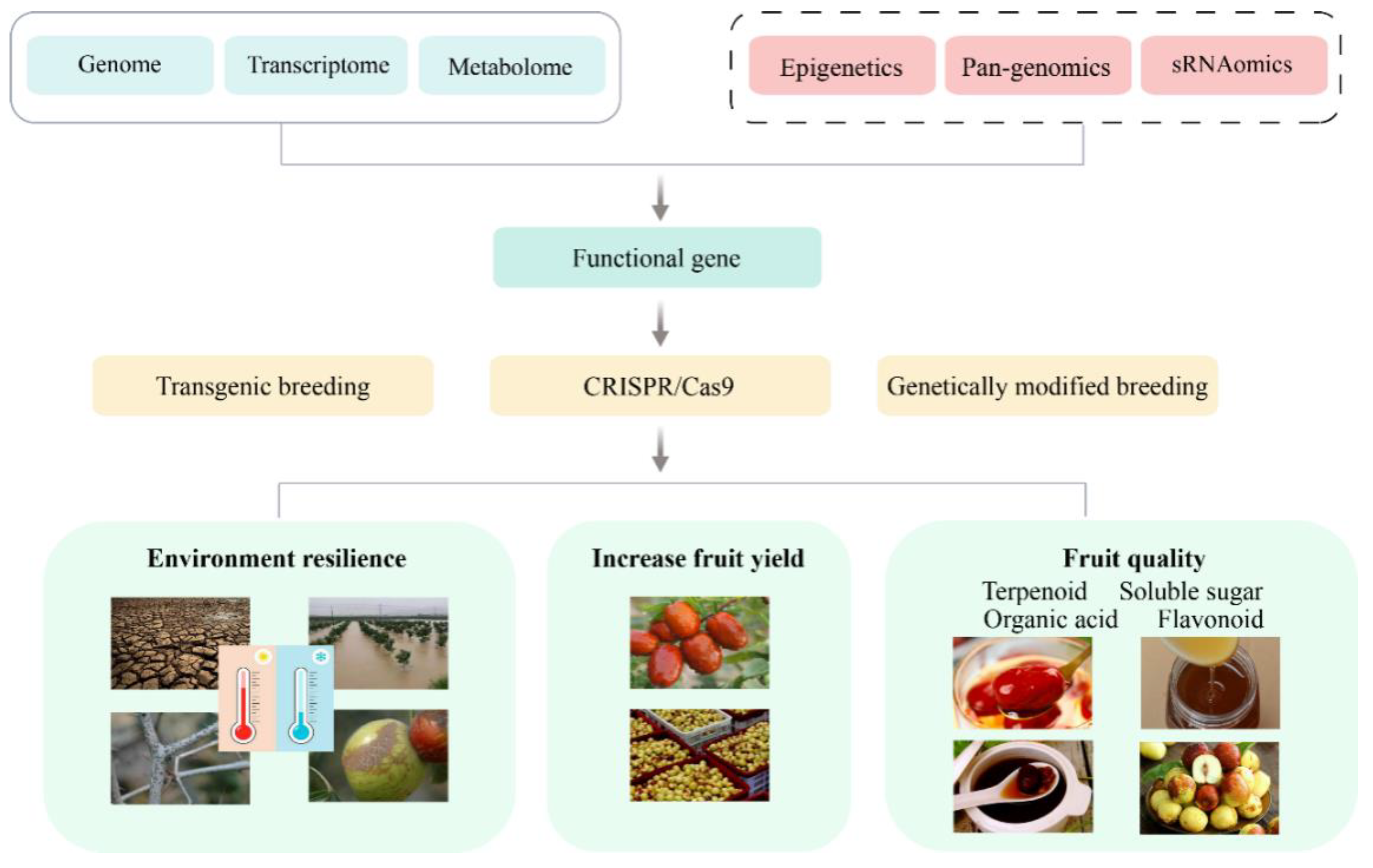
6. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mengjun, L. Advances in taxonomy study on the genus Ziziphus. Acta Hortic. 1999, 390, 161–166. [Google Scholar]
- Xu, T.; Zhou, X.; Degen, A.; Yin, J.; Zhang, S.; Chen, N. The inclusion of jujube by-products in animal feed: A review. Sustainability 2022, 14, 7882. [Google Scholar] [CrossRef]
- Dou, J.; Kou, X.; Wu, C.; Fan, G.; Li, T.; Li, X. Recent advances and development of postharvest management research for fresh jujube fruit: A review. Sci Hortic. 2023, 310. [Google Scholar] [CrossRef]
- Liu, M.; Wang, J.; Wang, L.; Liu, P.; Zhao, J.; Zhao, Z.; et al. The historical and current research progress on jujube-a superfruit for the future. Hort Res. 2020, 7, 119. [Google Scholar] [CrossRef]
- Shen, D.; Kou, X.; Wu, C.; Fan, G.; Li, T.; Dou, J.; et al. Cocktail enzyme-assisted alkaline extraction and identification of jujube peel pigments. Food Chem. 2021, 357, 129747. [Google Scholar] [CrossRef]
- Liu, M. Advances of research on germplasm resources of Chinese Jujube. Acta Hortic. 2013, 993, 15–20. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, H.; Jung, S.; Main, D.; Guo, L. Developmental mechanisms of fleshy fruit diversity in rosaceae. Annu Rev Plant Biol. 2020, 71, 547–573. [Google Scholar] [CrossRef]
- Chen, P.; Chen, L.; Ye, X.; Tan, B.; Zheng, X.; Cheng, J.; et al. Phytoplasma effector Zaofeng6 induces shoot proliferation by decreasing the expression of ZjTCP7 in Ziziphus jujuba. Hortic Res. 2022, 9, uhab032. [Google Scholar] [CrossRef]
- Yuan, L.; Lao, F.; Shi, X.; Zhang, D.; Wu, J. Effects of cold plasma, high hydrostatic pressure, ultrasound, and high-pressure carbon dioxide pretreatments on the quality characteristics of vacuum freeze-dried jujube slices. Ultrason Sonochem. 2022, 90, 106219. [Google Scholar] [CrossRef]
- Choi, S.; Ahn, J.; Kozukue, N.; Levin, C.; Friedman, M. Distribution of free amino acids, flavonoids, total phenolics, and antioxidative activities of Jujube (Ziziphus jujuba) fruits and seeds harvested from plants grown in Korea. J Agric Food Chem. 2011, 59, 6594–6604. [Google Scholar] [CrossRef] [PubMed]
- Bao, T.; Hao, X.; Shishir, M.R.I.; Karim, N.; Chen, W. Cold plasma: An emerging pretreatment technology for the drying of jujube slices. Food Chem. 2021, 337, 127783. [Google Scholar] [CrossRef] [PubMed]
- Arslan, M.; Zareef, M.; Tahir, H. Comparative analyses of phenolic compounds and antioxidant properties of Chinese jujube as affected by geographical region and drying methods (Puff-drying and convective hot air-drying systems). Food Meas. 2021, 15, 933–943. [Google Scholar] [CrossRef]
- Ahmed, K.; Naymul, K.; Mohammad, R.; Tao, B.; Yang, L.; Wei, C. Jujube fruit: A potential nutritious fruit for the development of functional food products. J. Funct. Foods 2020, 75, 104205. [Google Scholar]
- Ahudoukayoumu, A.; Fan, D.; Yue, W.; Zhan, J.; Hao, Q. Changes of nutrients, endogenous hormones and antioxidant enzymes activities during flower bud differentiation process of Ziziphus jujuba. Acta Botanica Boreali-Occidentalia Sinica 2021, 41, 142–150. [Google Scholar]
- Abudoukayoumu, A. Observation study of the process of flower bud differentiation and flower development of Jujube. Xinjiang Agric. Sci. 2020, 57, 798–805. [Google Scholar]
- Lu, D.; Wu, Y.; Zhang, J.; Qi, Y.; Zhang, Y.; Pan, Q. Visualizing the distribution of Jujube metabolites at different maturity stages using matrix-assisted laser desorption/ionization mass spectrometry imaging. Foods. 2023, 12, 3795. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Saand, M.; Huang, L.; Abdelaal, W.; Zhang, J.; Wu, Y.; et al. Applications of multi-omics technologies for crop improvement. Front Plant Sci. 2021, 12, 563953. [Google Scholar] [CrossRef]
- Sun, L.; Wang, J.; Cui, Y.; Cui, R.; Kang, R.; Zhang, Y.; et al. Changes in terpene biosynthesis and submergence tolerance in cotton. BMC Plant Biol. 2023, 23, 330. [Google Scholar] [CrossRef]
- Owusu, A.; Lv, Y.; Liu, M.; Wu, Y.; Li, C.; Guo, N.; et al. Transcriptomic and metabolomic analyses reveal the potential mechanism of waterlogging resistance in cotton (Gossypium hirsutum L.). Front Plant Sci. 2023, 14, 1088537. [Google Scholar] [CrossRef]
- Yan, F.; Luo, Y.; Bao, J.; Pan, Y.; Wang, J.; Liu, M.; et al. Construction of a highly saturated genetic map and identification of quantitative trait loci for leaf traits in jujube. Front Plant Sci. 2022, 13, 1001850. [Google Scholar] [CrossRef]
- Liu, M.; Zhao, J.; Cai, Q.; Liu, G.; Wang, J.; Zhao, Z.; et al. The complex jujube genome provides insights into fruit tree biology. Nat Commun. 2014, 5, 5315. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, C.; Zhao, X.; Fei, Z.; Wan, K.; Zhang, Z.; et al. The jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. PLoS Genet. 2016, 12, e1006433. [Google Scholar] [CrossRef]
- Shen, L.; Luo, H.; Wang, X.; Wang, X.; Qiu, X.; Liu, H.; et al. Chromosome-scale genome assembly for chinese sour jujube and insights into its genome evolution and domestication signature. Front Plant Sci. 2021, 12, 773090. [Google Scholar] [CrossRef]
- Wang, L.; Luo, Z.; Liu, Z.; Liu, P.; Liu, M. Genome size variation within species of chinese jujube (Ziziphus jujuba Mill.) and its wild ancestor sour jujube (Z. acidojujuba Cheng et Liu). Forests 2019, 10, 460. [Google Scholar] [CrossRef]
- Yang, M.; Han, L.; Zhang, S.; Dai, L.; Li, B.; Han, S.; et al. Insights into the evolution and spatial chromosome architecture of jujube from an updated gapless genome assembly. Plant Commun. 2023, 4, 100662. [Google Scholar] [CrossRef]
- Zhao, J.; Jian, J.; Liu, G.; Wang, J.; Lin, M.; Ming, Y.; et al. Rapid SNP discovery and a RAD-based high-density linkage map in Jujube (Ziziphus Mill.). Plos One 2014, 9, e109850. [Google Scholar] [CrossRef]
- Zhang, Z.; Wei, T.; Zhong, Y.; Li, X.; Huang, J. Construction of a high-density genetic map of Ziziphus jujuba Mill. using genotyping by sequencing technology. Tree Genet. Genomes 2016, 12, 76. [Google Scholar] [CrossRef]
- Zhang, Z. Optimization of a high-density genetic map for chinese jujube and QTL mapping for several important traits. Beijing Forestry University 2016. [Google Scholar]
- Wang, Z.; Zhang, Z.; Tang, H.; Zhang, Q.; Zhou, G.; Li, X. High-density genetic map construction and QTL mapping of leaf and needling traits in Ziziphus jujuba Mill. Front Plant Sci. 2019, 10, 1424. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Zhang, Z.; Li, S.; Lian, Q.; Fu, P.; He, Y.; et al. Genomic analyses of diverse wild and cultivated accessions provide insights into the evolutionary history of jujube. Plant Biotechnol J. 2021, 19, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Luo, H.; Wang, X.; Wang, X.; Qiu, X.; Liu, H.; et al. Chromosome-scale genome assembly for chinese sour jujube and insights into its genome evolution and domestication signature. Front Plant Sci. 2021, 12, 773090. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Zhang, Z.; Cheng, Y.; Li, S.; Shao, P.; Yu, Q.; et al. Comparative population genomics dissects the genetic basis of seven domestication traits in jujube. Hortic Res. 2020, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Fabres, P.; Collins, C.; Cavagnaro, T.; Rodríguez López, C. A concise review on multi-omics data integration for terroir analysis in vitis vinifera. Front Plant Sci. 2017, 8, 1065. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y. Research progress in chemical constituents of Lycium Ruthenicum Murr. Prog Pharm Sci. 2015, 39, 351–356. [Google Scholar]
- Zhang, C. Molecular mechanism related to the metabolism of sugar,acid and domestication for Ziziphus Jujuba Mill. Northwest A&F University, 2017. [Google Scholar]
- Zhao, A. Characteristic analysis of sugars and organic acids components and contents of chinese Jujube and wild jujube fruits. Journal of Tarim University. 2016, 28, 29–36. [Google Scholar]
- Zhang, C.; Bian, Y.; Hou, S.; Li, X. Sugar transport played a more important role than sugar biosynthesis in fruit sugar accumulation during Chinese jujube domestication. Planta. 2018, 248, 1187–1199. [Google Scholar] [CrossRef]
- Patrick, J.; Offler, C. Post-sieve element transport of photoassimilates in sink regions. J Exp Bot. 1996, 47, 1165–1177. [Google Scholar] [CrossRef]
- Patrick, J. PHLOEM UNLOADING: Sieve element unloading and post-sieve element transport. Annu Rev Plant Physiol Plant Mol Biol. 1997, 48, 191–222. [Google Scholar] [CrossRef]
- Chen, X. Comparative proteomics analysis of the difference in fruit size,sugar and acid content between jujube and wild jujube. Northwest A&F University, 2019. [Google Scholar]
- Yang, C.; Zhao, X.; Luo, Z.; Wang, L.; Liu, M. Genome-wide identification and expression profile analysis of SWEET genes in Chinese jujube. PeerJ. 2023, 11, e14704. [Google Scholar] [CrossRef]
- Wei, L.; Mao, W.; Jia, M.; Xing, S.; Ali, U.; Zhao, Y.; et al. FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10. J Exp Bot. 2018, 69, 4805–4820. [Google Scholar] [CrossRef]
- Wei, W.; Cheng, M.; Ba, L.; Zeng, R.; Luo, D.; Qin, Y.; et al. Pitaya HpWRKY3 is associated with fruit sugar accumulation by transcriptionally modulating sucrose metabolic genes HpINV2 and HpSuSy1. Int J Mol Sci. 2019, 20, 1890. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Zhang, J.; Ren, Y.; Li, M.; Tian, S.; et al. The NAC transcription factor ClNAC68 positively regulates sugar content and seed development in watermelon by repressing ClINV and ClGH3.6. Hortic Res. 2021, 8, 265. [Google Scholar] [PubMed]
- Fang, H.; Shi, Y.; Liu, S.; Jin, R.; Sun, J.; Grierson, D.; et al. The transcription factor CitZAT5 modifies sugar accumulation and hexose proportion in citrus fruit. Plant Physiol. 2023, 192, 1858–1876. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Yang, F.; Wang, C.; Duan, X.; Li, M.; Ma, Y.; et al. The transcription factor CmERFI-2 represses CmMYB44 expression to increase sucrose levels in oriental melon fruit. Plant Physiol. 2023, 192, 1378–1395. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, H.; Liu, C. Analysis of nutritional quality of different jujube cultivars. Ningxia J. of Agri. Fores. Sci. Tech. 2019, 60, 25–27. [Google Scholar]
- Ma, Q. Study on the changes of main organic acid content and acid-metabolism during the development of jujube fruits. Tarim University 2017. [Google Scholar]
- Tong, P. Acid accumulation pattern and related gene mining in jujube fruit. Tarim University 2021. [Google Scholar]
- Zhang, C.; Geng, Y.; Liu, H.; Wu, M.; Bi, J.; Wang, Z.; et al. Low-acidity ALUMINUM-DEPENDENT MALATE TRANSPORTER4 genotype determines malate content in cultivated jujube. Plant Physiol. 2023, 191, 414–427. [Google Scholar] [CrossRef]
- Pan, F.; Zhao, X.; Liu, F.; Luo, Z.; Chen, S.; Liu, Z.; et al. Triterpenoids in jujube: A Review of composition, content diversity, pharmacological effects, synthetic pathway, and variation during domestication. Plant 2023, 12, 1501. [Google Scholar] [CrossRef]
- Guo, S.; Duan, J.; Qian, D.; Tang, Y.; Wu, D.; Su, S.; et al. Content variations of triterpenic acid, nucleoside, nucleobase, and sugar in jujube (Ziziphus jujuba) fruit during ripening. Food Chem. 2015, 167, 468–474. [Google Scholar] [CrossRef]
- Sakna, S.; Yasmin, R.; Mohamed, S.; Mohamed, A. Phytochemical diversity and pharmacological effects of triterpenes from genus Ziziphus: a comprehensive review. Phytochem Rev. 2022, 22, 1611–1636. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, C. Biosynthesis of plant triterpenoid saponins in microbial cell factories. J Agric Food Chem. 2018, 66, 12155–12165. [Google Scholar] [CrossRef] [PubMed]
- Thimmappa, R.; Geisler, K.; Louveau, T.; O'Maille, P.; Osbourn, A. Triterpene biosynthesis in plants. Annu Rev Plant Biol. 2014, 65, 225–257. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Zhang, Z.; Shi, Q.; Yue, R.; Li, X. Metabolite and gene expression analysis underlying temporal and spatial accumulation of pentacyclic triterpenoids in Jujube. Genes 2022, 13, 823. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Zhang, Z.; Shi, Q.; Duan, X.; Du, J.; Wu, C.; et al. Methyl jasmonate- and salicylic acid-induced transcription factor ZjWRKY18 regulates triterpenoid accumulation and salt stress tolerance in Jujube. Int J Mol Sci. 2023, 24, 3899. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Zhang, Z.; Shi, Q.; Niu, R.; Duan, X.; Shen, B.; et al. Transcription factors ZjMYB39 and ZjMYB4 regulate farnesyl diphosphate synthase- and squalene synthase-mediated triterpenoid biosynthesis in Jujube. J Agric Food Chem. 2023, 71, 4599–4614. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Miyamoto, K.; Ogawa, S.; Masuda, Y.; Shimizu, T.; Kishi-Kaboshi, M.; et al. Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS One 2014, 9, e98737. [Google Scholar] [CrossRef]
- Wang, C.; Hao, X.; Wang, Y.; Maoz, I.; Zhou, W.; Zhou, Z.; et al. Identification of WRKY transcription factors involved in regulating the biosynthesis of the anti-cancer drug camptothecin in Ophiorrhiza pumila. Hortic Res. 2022, 9, uhac099. [Google Scholar] [CrossRef]
- Zhang, X.; Ge, F.; Deng, B.; Shah, T.; Huang, Z.; Liu, D.; et al. Molecular cloning and characterization of PnbHLH1 transcription factor in Panax notoginseng. Molecules 2017, 22, 1268. [Google Scholar] [CrossRef]
- Aslam, M.; Lin, X.; Li, X.; Yang, N.; Chen, L. Molecular cloning and functional characterization of CpMYC2 and CpBHLH13 transcription factors from Wintersweet (Chimonanthus praecox L.). Plants 2020, 9, 785. [Google Scholar] [CrossRef]
- Yin, J.; Li, X.; Zhan, Y.; Li, Y.; Qu, Z.; Sun, L.; et al. Cloning and expression of BpMYC4 and BpbHLH9 genes and the role of BpbHLH9 in triterpenoid synthesis in birch. BMC Plant Biol. 2017, 17, 214. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Bai, Z.; Wu, J.; Huang, W.; Jiao, J.; Zhang, C.; Hou, Z.; et al. The ethylene response factor SmERF8 regulates the expression of SmKSL1 and is involved in tanshinone biosynthesis in Saliva miltiorrhiza hairy roots. J Plant Physiol. 2020, 244, 153006. [Google Scholar] [CrossRef]
- Wei, C.; Li, M.; Cao, X.; Jin, Z.; Zhang, C.; Xu, M.; et al. Linalool synthesis related PpTPS1 and PpTPS3 are activated by transcription factor PpERF61 whose expression is associated with DNA methylation during peach fruit ripening. Plant Sci. 2022, 317, 111200. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, L.; Zheng, X.; Zhang, J.; Yang, L.; et al. Overexpression of SmMYB9b enhances tanshinone concentration in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 2017, 36, 1297–1309. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Han, L.; Zhu, X.; Zhang, C.; Li, Y.; Xue, X.; et al. SmMYB4 is a R2R3-MYB transcriptional repressor regulating the Biosynthesis of phenolic acids and tanshinones in Salvia miltiorrhiza. Metabolites 2022, 12, 968. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Pu, Z.; Cao, G.; You, D.; Zhou, Y.; Deng, C.; et al. Tanshinone and salvianolic acid biosynthesis are regulated by SmMYB98 in Salvia miltiorrhiza hairy roots. J Adv Res. 2020, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, Y.; Gao, F.; Jin, W.; Li, S.; Kimani, S.; et al. MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J Exp Bot. 2020, 71, 4140–4158. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Li, L.; Liu, H.; Yan, X.; Ma, Y.; Li, Y.; et al. AaMYB15, an R2R3-MYB TF in Artemisia annua, acts as a negative regulator of artemisinin biosynthesis. Plant Sci. 2021, 308, 110920. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, B.; Erffelinck, M.; Lacchini, E.; Ceulemans, E.; Colinas, M.; Williams, C.; et al. Interference between ER stress-related bZIP-type and jasmonate-inducible bHLH-type transcription factors in the regulation of triterpene saponin biosynthesis in Medicago truncatula. Front Plant Sci. 2022, 13, 903793. [Google Scholar] [CrossRef]
- Hao, X.; Zhong, Y.; Nï Tzmann, H.; Fu, X.; Yan, T.; Shen, Q.; et al. Light-induced artemisinin biosynthesis is regulated by the bZIP transcription factor AaHY5 in Artemisia annua. Plant Cell Physiol. 2019, 60, 1747–1760. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Z.; Ji, A.; Luo, H.; Song, J. Genomic survey of bZIP transcription factor genes related to tanshinone biosynthesis in Salvia miltiorrhiza. Acta Pharm Sin B. 2018, 8, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, P.; Sonawane, P.; Pollier, J.; Vanden Bossche, R.; Dewangan, V.; Weithorn, E.; et al. GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat Commun. 2016, 7, 10654. [Google Scholar] [CrossRef] [PubMed]
- Thagun, C.; Imanishi, S.; Kudo, T.; Nakabayashi, R.; Ohyama, K.; Mori, T.; et al. Jasmonate-responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in Tomato. Plant Cell Physiol. 2016, 57, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Sun, M.; Yuan, T.; Wang, Y.; Shi, M.; Lu, S.; et al. The AP2/ERF transcription factor SmERF1L1 regulates the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Food Chem. 2019, 274, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Mertens, J.; Pollier, J.; Vanden Bossche, R.; Lopez-Vidriero, I.; Franco-Zorrilla, J.; Goossens, A. The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in medicago truncatula. Plant Physiol. 2016, 170, 194–210. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Sun, L.; Li, Y.; Xiao, J.; Wang, S.; Yang, J.; et al. Functional identification of BpMYB21 and BpMYB61 transcription factors responding to MeJA and SA in birch triterpenoid synthesis. BMC Plant Biol. 2020, 20, 374. [Google Scholar] [CrossRef] [PubMed]
- Pawlowska, A. Flavonoids of Zizyphus jujuba L. and Zizyphus spina-christi (L.) Willd (Rhamnaceae) fruits. Food Chem. 2009, 112, 858–862. [Google Scholar] [CrossRef]
- Wang, C.; Cheng, D.; Cao, J.; Jiang, W. Antioxidant capacity and chemical constituents of Chinese jujube (Ziziphus jujuba Mill.) at different ripening stages. Food Sci. Biotechnol. 2013, 22, 639–644. [Google Scholar] [CrossRef]
- Li, X. The patterns of flavonoids accumulation and the expression of biosynthesis related genes during the course of maturation of the Chinese jujube fruit. J. Fruit Sci. 2020, 37, 1464–1474. [Google Scholar]
- Shi, Q.; Du, J.; Zhu, D.; Li, X.; Li, X. Metabolomic and transcriptomic analyses of anthocyanin biosynthesis mechanisms in the color mutant Ziziphus jujuba cv. Tailihong. J Agric Food Chem. 2020, 68, 15186–15198. [Google Scholar] [CrossRef]
- Li, S.; Shen, Y.; Zheng, S.; Zhu, Q.; Cai, L.; Wang, Y.; et al. ZjFAS2 is involved in the fruit coloration in Ziziphus jujuba Mill. by regulating anthocyanin accumulation. Front Plant Sci. 2023, 14, 1142757. [Google Scholar] [CrossRef]
- Muhammad, N.; Luo, Z.; Zhao, X.; Yang, M.; Liu, Z.; Liu, M. Transcriptome-wide expression analysis of MYB gene family leads to functional characterization of flavonoid biosynthesis in fruit coloration of Ziziphus Mill. Front Plant Sci. 2023, 14, 1171288. [Google Scholar] [CrossRef]
- Wang, W.; Pu, Y.; Wen, H.; Lu, D.; Yan, M.; Liu, M.; et al. Transcriptome and weighted gene co-expression network analysis of jujube (Ziziphus jujuba Mill.) fruit reveal putative genes involved in proanthocyanin biosynthesis and regulation. FSHW 2023, 12, 1557–1570. [Google Scholar] [CrossRef]
- Wang, L.; Li, M.; Liu, Z.; Dai, L.; Zhang, M.; Wang, L.; et al. Genome-wide identification of CNGC genes in Chinese jujube (Ziziphus jujuba Mill.) and ZjCNGC2 mediated signalling cascades in response to cold stress. BMC Genom. 2020, 21, 191. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Han, Y.; Feng, X.; Gao, H.; Cao, B.; Song, L. Genome-wide identification of BAM (β-amylase) gene family in jujube (Ziziphus jujuba Mill.) and expression in response to abiotic stress. BMC Genom. 2022, 23, 438. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Wang, L.; Luo, Z.; Zhao, R.; Liu, Z.; Liu, P.; et al. Molecular characteristics of CML genes in Chinese jujube and their expression patterns in resistance to cold stress. J. Beijing For. Univ. 2023, 45, 58–67. [Google Scholar]
- Zhou, H. Comparative transcriptome analysis of Jujube under freezing stress and functional studies of related genes. Beijing Forestry University 2020. [Google Scholar]
- Chen, X.; Chen, R.; Wang, Y.; Wu, C.; Huang, J. Genome-wide identification of WRKY transcription factors in Chinese jujube (Ziziphus jujuba Mill.) and their involvement in fruit developing, ripening, and abiotic stress. Genes 2019, 10, 360. [Google Scholar] [CrossRef] [PubMed]
- He, A.; Ma, Z.; Li, Y.; Huang, C.; Yong, J.; Huang, J. Spatiotemporal, physiological and transcriptomic dynamics of wild jujube seedlings under saline conditions. Tree Physiol. 2023, 43, 832–850. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hou, L.; Liu, S.; Zhang, C.; Yang, W.; Pang, X.; et al. Genome-wide identification and expression analysis of NAC transcription factors in Ziziphus jujuba Mill. reveal their putative regulatory effects on tissue senescence and abiotic stress responses. Ind Crops Prod. 2021, 173, 114093. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, S.; Liu, B. Research progress on pangenome and its application in plant functional genetics. J. Plant Genet. Resour. 2021, 22, 7–15. [Google Scholar]
- Wang, J.; Yang, W.; Zhang, S.; Hu, H.; Yuan, Y.; Dong, J.; et al. A pangenome ana1ysis pipe1ine provides insights into functiona1 gene identification in rice. Genome Biol. 2023, 24, 19. [Google Scholar] [CrossRef]
- Wang, B.; Hou, M.; Shi, J.; Ku, L.; Song, W.; Li, C.; et al. De novo genome assembly and analyses of 12 founder inbred lines provide insights into maize heterosis. Nat Genet. 2023, 55, 312–323. [Google Scholar] [CrossRef]
- Wang, M.; Li, J.; Qi, Z.; Long, Y.; Pei, L.; Huang, X.; et al. Genomic innovation and regulatory rewiring during evolution of the cotton genus Gossypium. Nat Genet. 2022, 54, 1959–1971. [Google Scholar] [CrossRef]
- Xia, R.; Chen, C.; Pokhrel, S.; Ma, W.; Huang, K.; Patel, P.; et al. 24-nt reproductive phasiRNAs are broadly present in angiosperms. Nat Commun. 2019, 10, 627. [Google Scholar] [CrossRef]
- Zhan, J.; Meyers, B. Plant small RNAs: their biogenesis, regulatory roles, and functions. Annu Rev Plant Biol. 2023, 74, 21–51. [Google Scholar] [CrossRef]
- Kong, X.; Yang, M.; Le, B.H.; He, W.; Hou, Y. The master role of siRNAs in plant immunity. Mol Plant Pathol. 2022, 23, 1565–1574. [Google Scholar] [CrossRef]
- Meers, C.; Le, H.; Pesari, S.; Hoffmann, F.; Walker, M.; Gezelle, J.; et al. Transposon-encoded nucleases use guide RNAs to promote their selfish spread. Nature 2023, 622, 863–871. [Google Scholar] [CrossRef]
| Infomation | Dongzao | Junzao | Suanzao | Dongzao |
|---|---|---|---|---|
| Sequencing platform | Illumina | Illumina | PacBio+ Illumina | Nanopore+ PacBio |
| Assembly strategy | WGS+BAC | WGS | Hi-C | HiFi+ONT+Hi-C |
| Total length of scaffolds (bp) | 437,645,007 | 351,115,537 | 406,163,984 | 393,332,932 |
| Contig N50 length (bp) | 33,948 | 34,020 | 2,144,872 | 32,986,920 |
| sequences anchored to chromosomes (%) | 73.6% | 83.6% | 93.7% | 100% |
| BUSCO genes (%) | 89.0% | 93.2% | 95.56% | 98.50% |
| Number of protein-coding genes | 27,443 | 31,067 | 25,089 | 29,633 |
| Transposable elements (bp) | 136.33 | 204.92 | 215.93 | 220.88 |
| Domesticated traits | Candidate gene name | Candidate gene name ID | Validated method | Reference |
|---|---|---|---|---|
| Sugar-and acid-related metabolism | NADP-dependent malic enzym | Zj.jz006119090 | Expression profiling | Huang et al. 2016 |
| 6-phosphofructokinase | Zj.jz010621015 | |||
| Phosphoglucomutase | Zj.jz021807003 | |||
| Sugar transporters | Zj.jz042571026 | |||
| Zj.jz036789032 | ||||
| Zj.jz034227050 | ||||
| Zj.jz002249011 | ||||
| Zj.jz002249010 | ||||
| Pyruvate kinase | Zj.jz006429010 | |||
| ERD6-like Sugar transporter | Zj.jz001627070 | |||
| Zj.jz007429007 | ||||
| Zj.jz007429005 | ||||
| Zj.jz007429006 | ||||
| sucrose synthase | Zj.jz031941019 | |||
| Pyruvate kinase | Zj.jz006429010 | |||
| Fruit shape and kernel shape | FS3 | Zj.jz044531027 | GWAS,qPCR and Transgenic | Guo et al. 2020 |
| Bearing shoots | NLBS | Zj.jz003639032 | GWAS | |
| Prickles on bearing shoots | HDG2 | Zj.jz044447010 | GWAS | |
| BLT1 | Zj.jz040945037 | |||
| Seed-setting rate | OVA4 | Zj.jz006119092 | GWAS | |
| MIK1 | Zj.jz007373151 | |||
| RAD51D | Zj.jz001293012 | |||
| Fruit softening | Polygalacturonase | Zj.jz044553003 | NO | |
| Flowering time | Early flowering 3 | Zj.jz000799141 | NO | Guo et al. 2021 |
| Seed-setting rate | POD1 | Zj.jz015743041 | GWAS,qPCR | |
| Fruit weight | DA3/UBP14 | Zj.jz038707057 | GWAS,qPCR and Transgenic | |
| Fruit size | FW2.2/CNR1 | Zj.jz029849045 | NO | |
| Environmental adaptation | Histidine kinase 4 | Zijuj10G0113500 | NO | Shen et al. 2021 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





