1. Introduction
Coronavirus disease 2019 (COVID-19) is a viral respiratory disease that emerged at the end of 2019 in Wuhan, China, and rapidly extended its devastating effects worldwide. COVID-19 is caused by SARS-CoV-2, a large enveloped, positive-strand RNA virus with a genome approximately 30 kb in length, belonging to the genus
Betacoronavirus of the family Coronaviridae [
1]. As of 27
th September 2023, SARS-CoV-2 continues to spread worldwide, with over 770,875,433 total confirmed cases and 6,959,316 deaths worldwide [
2].
SARS-CoV-2 primarily targets the respiratory tract, and the infection begins when the viral spike protein on the surface of the virus binds to the human receptor angiotensin-converting enzyme (ACE2). Cleavage of spike protein by the transmembrane protease serine 2 (TMPRSS2) on the surface of epithelial cells triggers the fusion of the viral and host cell membranes, facilitating the entry of the virus into the host cell [
3,
4]. In addition to causing acute respiratory distress syndrome, SARS-CoV-2 has other pulmonary and extrapulmonary manifestations in the gastrointestinal tract, hepatobiliary system, cardiovascular, neurological, and renal systems, which often lead to multiorgan failure and shock in severe cases [
5,
6]. Furthermore, some survivors experience Long COVID or Post-Acute Sequelae of SARS-CoV-2 (PASC), with cardiovascular, neurological, and pulmonary manifestations [
7,
8,
9,
10]. The exact pathogenesis of acute and chronic disease in extrapulmonary organs in COVID-19 is unknown. It has been suggested that indirect mechanisms such as co-morbidities and/or other pathophysiological conditions may play a role [
11,
12]. Understanding the details of the mechanism of replication and cell tropism of SARS-CoV-2 may provide insights into the pathogenesis and tissue tropism of this virus.
SARS-CoV-2 belongs to coronaviridae, which include enveloped, positive sense, single-stranded RNA viruses. These viruses employ an elaborate mechanism for replicating their genome and for transcribing the coding sequences. The replication cycle of SARS-CoV-2, like other coronaviruses, begins with the entry of the virus into the cell and release of viral RNA into the cytoplasm [
13]. Within the cytoplasm, the first step is the translation of two large open reading frames (ORFs; ORF1a and ORF1b) present at the 5′ end of the positive-strand gRNA for the expression of polyproteins and proteolytic cleavage of these proteins to form 15-16 nonstructural proteins (nsp) of which 15 compose the viral replication and transcription complex (RTC) including RNA-processing and RNA-modifying enzymes [
14]. RTC leads to the generation of negative-strand viral RNA replication intermediates from the positive-strand genomic RNA. Discontinuous transcription of the newly synthesized negative-strand RNAs from the 3′ end leads to the formation of a series of shorter sub-genomic RNAs (sgRNAs), which encode for the structural and accessory proteins [
1]. During its replication, the virus modifies the intracellular host endoplasmic reticulum membrane to generate the replication organelles (ROs), which are the powerhouses consisting of double-membrane vesicles (DMVs) enclosing the viral RNAs [
15,
16]. DMVs are likely to provide a protective environment for the replication of gRNA and sgRNA. Newly synthesized gRNA and sgRNA are thought to translocate from the lumen of the DMVs into the cytoplasm through pores present on DMVs. While the exact composition of DMVs and the pores are not completely understood, it has been established the nsp3 and nsp4 proteins are required for the formation of DMVs and for the biogenesis of DMVs [
17].
Many questions remain unanswered about the early replication events of SARS-CoV-2 such as: i) the time it takes for the viral RNA to complete translation and start the replication process after entry into the cytoplasm; ii) the generation, subcellular localization, and function of the ROs; iii) timing of formation of gRNA, sgRNAs and RO; and iv) the mechanism of vRNA synthesis within ROs [
13]. Addressing these questions is not only important for obtaining insight about the mechanism of SARS-CoV-2 replication but also for the identification of unexplored drug targets that can be used to curb viral replication at a very early stage of the infection cycle.
Most of the studies aimed at understanding the SARS-CoV-2 replication have generally focused on time-points ~4-5 hours post-infection (p.i.), which is when viral replication is at its midpoint, making it easier for monitoring and visualization of the RNA and ROs [
18,
19]. However, understanding the replication at time points earlier than 4-5 hours p.i may be required to investigate the initial stages of vRNA replication and the formation of ROs. Studying early time points requires highly sensitive and specific methods to detect SARS-CoV-2 RNA at a single molecule level as large amounts of RNA are unlikely to be present at these stages. Several reports have utilized single-molecule RNA-FISH (smRNA-FISH) to detect an absolute number of SARS-CoV-2 transcripts [
18,
20]. However, these reports do not investigate the DMVs at early time points before 4-5 p.i.
To detect and visualize SARS-CoV-2 gRNA and sgRNAs with high specificity and sensitivity at early time points, we employed a combination of single molecule RNA-Fluorescence in situ hybridization (smRNA-FISH) using probes for gRNA and sgRNA and immunofluorescence using nsp3. To facilitate the study of replication kinetics of single SARS-CoV-2 RNA molecules after the virus enters a cell, we infected the cells with low m.o.i. of virus. Furthermore, we employed High-Speed High-Resolution Scanning fluorescence microscopy (HSHRS-FM) to scan and visualize the replication of SARS-CoV-2 in a large number of cells to determine the different stages of replication in these cells by using smRNA-FISH analysis. The HSHRS-FM method involves scanning the entire slide to create a single high-resolution digital image by tiling and stitching many high-magnification fields of view together, thus capturing the images of a large number of cells present on the slide. We designed probes to simultaneously detect positive strands of the gRNA and sgRNA and carried out a time course analysis to visualize the cells at time points starting from 0.5 hours to 24 hours p.i. Our analyses have led to the detection of the SARS-CoV-2 gRNA within the infected cells as early as 30 minutes p.i. Subsequent time points indicated that the replicating RNAs were present in distinct spots that contained both gRNA and sgRNAs. We also tested for the formation of DMVs by monitoring nsp3 protein by combining smRNA-FISH with immunofluorescence. Our finding suggests that atleast at the start of the replication (~ 3 hr p.i.) many of the RNA spots were devoid of nsp3 protein, suggesting that some of these RNA spots may represent the direct accumulation of RNA in the cytoplasm without DMV. At later time points, most of the RNA spots became positive for nsp3, suggesting the formation of DMV. We also observed that the replication is asynchronous and cells with various stages of SARS-CoV-2 RNA replication could be found at any given time point. Our studies indicate that combining smRNA-FISH with IF and utilization of HSHRS-FM enables not only the sensitive detection of SARS-CoV-2 RNAs in a large number of infected cells, but also facilitates the studies to address important questions related to the mechanism of early replication events of SARS-CoV-2.
2. Materials and Methods
2.1. Cell Culture
Vero E6 cells (ATCC, CRL-1586) were maintained in Dulbecco’s Modified Eagle Medium (Hyclone; Cat # SH30081.01) supplemented with 10% fetal bovine serum (Atlas Biologicals; Cat # F-0500-A), 2mM L-glutamine (Gibco; Cat # 25030-081), non-essential amino acids, 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco; Cat # 15140-122). All the cell lines were maintained in a standard 5% CO2 in a culture incubator at 37˚ C.
2.2. Infection of Vero E6 Cells with SARS-CoV-2
For infection of Vero E6 cells with SARS-CoV-2 (SARS-CoV-2 USA-WA1/2020), cells were seeded on Nunc™ Lab-Tek™ sterile 4-chambered slides (Thermo Scientific™, Cat # 177399) at a confluency of 50-70% the day before infection. On the day of the infection, medium was replaced with 500 μl of infection medium (DMEM medium supplemented with 2% fetal bovine serum, non-essential amino acids, HEPES and penicillin/streptomycin) and the cells were transferred to the BSL-3 laboratory. The virus was diluted in infection medium and 100 µl/well added to each well to achieve a m.o.i. of 0.5 PFU/cell. After, 0, 0.5, 1, 2, 3, 4, 5, 6,12 and/or 24 hours post-infection, supernatant was removed, and the cells were washed twice with 1 ml of 1x PBS with 5 mM MgCl2. After the second wash, cells were fixed with 4% paraformaldehyde. During fixation, cells were protected from light.
2.3. Immunofluorescence
The fixed cells were washed with 1x PBS, permeabilized with 0.2% Triton X-100 and incubated with a mouse monoclonal antibody that recognizes the NP protein of SARS-CoV-2 (1C7, kindly provided by Dr. Thomas Moran, Icahn School of Medicine at Mount Sinai). Next, cells were washed with 1x PBS and incubated with Alexa fluor 488 conjugated anti-mouse secondary antibody (Invitrogen) and DAPI. The signals were detected using an EVOS M5000 fluorescent microscope.
2.4. smRNA-FISH Probe Design and Specificity Analysis
Forty different 22 nucleotide long smRNA-FISH probes (5’-> 3’) for spike (S) and RdRp (nsp12) genes were generated using LGC Biosearch Technologies’ Stellaris
® RNA FISH Probe Designer version 4.2 [
21]. Each probe for spike gene was tagged with Quasar 570 and for nsp12 gene with Quasar 670 dyes at 3′ ends, respectively. As a target reference sequence, coding sequences (CDS) of Spike and nsp12 region was selected from the SARS-CoV-2 Wuhan-Hu-1 (NC_045512.2) reference sequence. Each probe sequence was subjected to BLAST and was screened against other coronavirus sequences, human transcriptome, and human intron database. To perform in silico probe sequence specificity analysis, all 40 oligonucleotide sequences of the Spike and nsp12 smRNA-FISH probes were aligned against; SARS-CoV-2 (NC_045512.2); SARS-CoV-1 Tor2 (NC_004718.3); MERS-CoV isolate HCoV-EMC/2012 (NC_019843.3); HCoV-HKU1 (NC_006577.2); HCoV-OC43 strain ATCC VR-759 (AY585228.1); HCoV 229E strain 229E/human/USA/933-40/1993 (KF514433.1); HCoV NL63 strain NL63/human/USA/0111-25/2001 (KF530112.1) and Human hg38_mRNA (AF001540.1), RefSeq genome or transcriptome assembly, using ‘bowtie2′ (2.4.4). To get the minimum edit distance of oligonucleotide sequences to target genome/transcriptome, following bowtie2 v2.4.4 arguments were used; --end-to-end, --no-unal, --align-seed-mm 0, --align-seed-length 5, --align-seed-interval 1-1.15, -- effort-extend 15, --effort-repeat 2 [
18]. On the other hand, various variants of concern (VOCs) of SARS-CoV-2 [
22], in addition to the WA1 strain, were also considered for
in silico probe sequence specificity analysis for both Spike and nsp12 smRNA-FISH probes. For this analysis, the genomes of several SARS-CoV-2 VOCs, including Alpha/B.1.1.7 (OW998408.1), Beta/B.1.351 (OX008586.1), Gamma/P.1 (MZ427312.1), Delta/B.1.617.2 (OX014251.1), Omicron/B.1.1.529 (OW996240.1), and four sub-variants of Omicron, such as BA.1 (OP810428.1), BA.2 (OM617939.1), BA.4 (OP093374.1), and BA.5 (OP093373.1), along with SARS-CoV-2 Wuhan-Hu-1 (NC_045512.2), were selected, and the alignment was performed using ‘bowtie2’ version 2.5.1. To get the minimum edit distance of oligonucleotide sequences to target genome/transcriptome, the following arguments were used; bowtie2 --score-min L,-0.6,-3 --end-to-end -N 0 -L 5 -i S,1,1.15 -D 15 -R 2 -x
${PATH_TO_INDEXED_GENOME} -U
${FASTQ} -S
${OUTPUT_SAMFILE}. In order to determine the specificity of the designed Spike and nsp12 smRNA-FISH probes, the minimum edit distance of oligonucleotide sequences was also calculated for the spike gene of SARS-CoV-2 and the codon-optimized spike gene of SARS-CoV-2. The 40 different smRNA-FISH probe sequences of spike and nsp12 genes are shown as
Table 1. The heatmaps were created using R v4.0.2 with Bioconductor package Complex Heatmap v2.9.3 [
23].
2.5. smRNA-FISH Analysis
Vero E6 cells (ATCC, CRL-1586) were seeded on a four-chambered slide and inoculated with SARS-CoV-2 (USA-WA1/2020) at an m.o.i. of 0.5 for various time points (0, 0.5, 1, 2, 3, 4, 5, 6,12, and/or 24 hours). In the case of VOC infection, Omicron BA.1 was used to infect the Vero cells expressing high levels of transmembrane serine protease 2 (TMPRSS2) under similar conditions (at 3, 6, and 12 hours). However, Vero E6 cells mock-infected or infected with heat-inactivated SARS-CoV-2 WA1 or SARS-CoV-2 BA.1 at 12 hours post-infection were considered as negative controls, respectively. The cells were fixed with 4% paraformaldehyde post-infection for 30 minutes, followed by smRNA-FISH analysis. After fixation, each well of the four chambered slide was treated with 0.1 M Glycine/PBSM for 10 min at room temperature followed by cell permeabilization with PBSM/0.1% Triton X-100 for 10 min. After washing with 1x PBSM, cells were pre-hybridized in 2x Sodium Chloride-Sodium Citrate (SSC) buffer (G-Biosciences; Cat # R019), 15% formamide (Acros Organics; Cat # 75-12-7) for 30 min at 37˚ C. The cells were incubated overnight with 300 μl hybridization buffer containing SARS-CoV-2 spike and/or RdRp/nsp12 stellaris probes tagged with Quasar 570 and Quasar 670 dyes, respectively. SARS-CoV-2 spike and RdRp/nsp12 stellaris probes (250 nM final concentration, Biosearch Technologies)) were added to hybridization buffer containing, 10% dextran sulfate (Sigma; Cat # D61001), 1mg/mL competitor tRNA from E. coli MRE 600 (Cat No # 10109541001; Millipore Sigma), 15% formamide (Cat # 75-12-7; Acros Organics), 0.2 mg/mL UltraPure BSA (Cat #: AM2616; Ambion™), 2x Sodium Chloride-Sodium Citrate (SSC) buffer (Cat # R019, G-Biosciences), 2 mM Ribonucleoside Vanadyl Complex (Cat # S1402S; New England Biolabs), and 10 U/mL SUPERase•In RNase Inhibitor (Cat # AM2694; Ambion™). When combining RNA-FISH with IF, the primary antibodies, α-rabbit SARS-CoV-2 nsp3 antibody (1:500 dilution, GeneTex; Cat # GTX135589) were added to the hybridization buffer. The slides were incubated in a humid hybridization chamber overnight at 37˚ C. Post-incubation, the next day, samples were washed with prehybridization buffer (2x SSC, 15% formamide) three times and if combining IF, the slides were incubated two times for 20 min. each with the anti-rabbit Alexa Fluor 488 secondary Ab (1:1000 in the pre-hybridization buffer, Invitrogen; Cat # A-11008). Following incubation with the secondary antibody, cells were washed with pre-hybridization buffer (2x SSC, 15% formamide) three times followed by 3-4 times washing with 2x SSC. The cells were stained with DAPI (1.0 μg/ml in 2x SSC) for 2 min at room temperature. After DAPI staining the cells were washed with 2x SSC and mounted using the ProLong Gold antifade mountant (Cat # P10144, Thermo Scientific).
2.6. Whole-Slide Scanning, Microscope Setup and Image Acquisition
Fluorescent scanning of the slides was done in two stages. First, whole-slide scanning of the entire slide was obtained at 20x using PANNORAMIC 250 Flash III Slide Scanner. Automatic focusing (default factory settings) was used for scanning the whole slide. For fluorescence imaging, we used DAPI, Cy5-Q and TRITC-Dendra fluorescence filters. Pannoramic scanner software were used for image acquisition and scanned slides were visualized using CaseViewer 2.4 (64-bit version). For setting the parameters for image acquisition, mock control was used to set the background level of the fluorescence in the test images. After scanning, the same slides were subjected to single cell imaging. Cells were imaged using an upright, wide-field Olympus BX-63 Microscope equipped with a SuperApochromatic 60×/1.35 NA Olympus Objective (UPLSAPO60XO), a SOLA light engine (Lumencor), an ORCA-R2 Digital Interline CCD Camera (C10600-10B; Hamamatsu), and zero-pixel shift filter sets: DAPI-5060C-Zero, Cy3-4040C-Zero (for Quasar 570 detection), and Cy5-4040C-Zero (for Quasar 670 detection) from Semrock, as described [
24]. The resulting image pixel dimension was 107.5 nm, and the z-step size (along the optical axis) used for all optical sectioning acquisition was 200 nm. Metamorph software (Molecular Devices) was used for controlling microscope automation and image acquisition. Images were analyzed using ImageJ and/or Fiji software [
25].
4. Discussion
Our studies provide a spatial and temporal characterization of early post-entry events of SARS-CoV-2 RNA replication in Vero E6 cells at a single-cell, single-molecule resolution, and shed light on the formation of replication centers at early time points. Our data complements the previous reports, and in addition, provides information and clear images of very early stages of replication. We visualized cytoplasmically localized single gRNA molecules within cell cytoplasm at 0.5 hours p.i., indicating that these molecules likely represent viral RNA from a particle that has just entered the cell. To our knowledge, these represent the earliest time points of SARS-CoV-2 viral RNA detection in an infected cell. Our studies also indicated that within two hours, single distinct RNA spots/clusters are formed within the cytoplasm that are larger than the diffusion limited single molecules, suggesting that these are results of initial replication of single gRNA that entered the cell. By 3 hr. p.i. we find that some but not all of these RNA clusters are associated with nsp3, which is a marker for the formation of ROs that are characterized by the presence of DMVs, suggesting that these RNA clusters/specles are likely to be precursors for the formation of RO. Eventually, most of the RNA speckles are associated with nsp3 protein at later time points. This temporal association of nsp3 with RNA clusters strongly supports the notion that RNA clusters/speckles that are formed without nsp3 might serve as precursors of ROs during the early phases of SARS-CoV-2 replication. Our results are interesting, as in a previous study conducted by Shu Shi
et al., the intracellular localization and timing of expression of the viral nsp3 protein in SARS-CoV-2-infected cells were investigated at multiple time points (2, 3, 4, 6, 8, and 24 hpi), with the first appearance observed at 6 hpi [
36]. In our study we were able to distinctly visualize the presence and co-localization of nsp3 in some of the RNA clusters at 3 hpi, possibly attributed to the heightened sensitivity of our RNA-FISH approach. It is intriguing to note that many of the RNA clusters at 3 hr time point were without nsp3. It is not clear if these RNA spots without nsp3 are aggregates or phase separated RNA clusters in the cytoplasm without a membrane. More research is needed to characterize these RNA spots that lack nsp3. It is also intriguing to note that while gRNA and nsp3 appears to reside in the center of the RNA clusters, the sgRNA appears to surround these centralized structures. One possibility is that if the RNA clusters represent RO, then sgRNA at the periphery of these clusters may represent the those that have migrated out of the ROs.
Our studies also indicated that there is cell-to-cell heterogeneity and asynchrony in the rate of RNA replication, which can be observed by quantitating the number and nature of RNA spots/clusters present within cells at a given time point. When infected with low m.o.i. of 0.5, most of the cells have one or a few RNA spots/clusters that appear to be at an early stage of replication. As the time progresses, the percentages of cells with early stages of RNA replication decreases, and those with late stages of replication increases. Thus, the heterogeneity appears to be a random or stochastic event, as all stages of replication are found at any given time point. The reason for the heterogeneity in replication in different cells is not clear and it could be due to differences in the rate of entry of RNA into the cytoplasm, the establishment of the first RO from the RNA that entered the cell, or it could be due to differences in the rate at which the RNA replication progresses within the cells. It is well known that the stage of cell cycle and other cellular factors affect the rate of viral replication [
37,
38].
Consistent with previous observations [
31], we have also identified SARS-CoV-2 gRNA molecules within the nuclei of certain infected cells. Recent evidence has stirred controversy in the field, suggesting that, at least under certain specific conditions, SARS-CoV-2 sequences can undergo retrotranscription and integrate into the host genome as DNA, leading to the formation of chimeric genes [
31,
39,
40]. At this point we do not have any information regarding the integration of SARS-CoV-2 nucleic acid in the nucleus, but we clearly find both gRNA and sgRNA within the nucleus. Interestingly, we did not find the presence of nsp3 protein in the nucleus, suggesting that the replication centers are limited to the cytoplasm. More experiments are needed to establish the role of the viral components in the nucleus during replication of the virus.
In summary, high-resolution early kinetic analysis of SARS-CoV-2 RNA replication provides an understanding of the timing of the formation and arrangement of gRNA, sgRNA and possibly formation of RO and sets the stage for further analysis of these unique organelles in the future. While our manuscript was in preparation, several groups have reported the use of smFISH to study the infection of SARS-CoV-2 in human cell lines [
18,
20,
41,
42]. While our data is consistent with these studies, we were able to identify viral RNA within the cells at earlier time points (0.5 hours, and 1 hours p.i.) and were able to determine that the RNA spots could be the precursors to ROs or viral RNA factories at much higher resolution, where we identified the presence of a central region within these ROs filled with gRNAs and sgRNAs that are surrounded by migrating sgRNAs. This central region colocalized with the nsp3 protein, suggesting it is an RO. Interestingly, at early time points p.i. (3 hr p.i.), many RNA spots, which were larger than single RNA molecules, did not colocalize with nsp3, suggesting that either these are aggregates of RNA or represent phase separated RNAs due to the accumulation of newly replicated RNA molecules. Because of this reason, we find that quantitating individual gRNAs or sgRNA after 2 hours p.i. becomes challenging. More studies need to be done to establish the nature of these RNA spots that do not co-localize with nsp3. Our findings open avenues for further investigation into the precise nature of RNA speckles during viral replication and the molecular mechanisms governing their interaction with viral proteins. Understanding the dynamics of these early events in SARS-CoV-2 replication may offer valuable insights for the development of antiviral strategies and the elucidation of the virus’s pathogenesis.
Author Contributions
Conceptualization, GK; Funding acquisition, GK, AG-S. and CE; Investigation, GK, RP, CE, IM, AG-S. and RS; Methodology, RP, CE, IM and AC; Supervision: GK, RS, CE, and AG-S; Visualization and imaging: RP and CE; Writing-original draft, GK, RP with contribution from other authors, CE, IM, AG-S, AC and RS; Writing-review and editing, GK and RP.
Conflicts of Interest
The A.G.-S. laboratory has received research support from GSK, Pfizer, Senhwa Biosciences, Kenall Manufacturing, Blade Therapeutics, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma, Applied Biological Laboratories and Merck, outside of the reported work. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Castlevax, Amovir, Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar, CureLab Oncology, CureLab Veterinary, Synairgen and Pfizer, outside of the reported work. A.G.-S. has been an invited speaker in meeting events organized by Seqirus, Janssen, Abbott and Astrazeneca. A.G.-S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside of the reported work.
Figure 1.
Design and optimization of SARS-CoV-2 specific smRNA-FISH probes to detect SARS-CoV-2 infection. (A) Schematic representation of SARS-CoV-2 gRNA and sgRNA-S, indicating the position of the smRNA-FISH probes, P1 (in magenta) and P2 (in green) probes that hybridize to Spike and nsp12 ORF sequences, respectively. (B) Heatmap representing the probe sequence alignment against various coronavirus genomes (including SARS-CoV-1, SARS-CoV-2, MERS-CoV, HCoV-OC43, HCoV-NL63, HCoV-229E) and human transcriptomes (hg38-mrna). Each column represents individual 22 nt spike gene probe sequences from Spike gene (S1-S40, upper panel, represented in the shades of red) and from nsp12 gene (N1-N40, middle panel, represented in shades of green). The lower panel representing the heatmap of the spike probe sequence alignment against spike gene of the SARS-CoV-2 and codon-optimized spike gene of the SARS-CoV-2 (S1-S40, lower panel, represented in the shades of red), showing the specificity of the probe. The minimum edit distance represents mismatch score between the SARS-CoV-2 sequence and the other genome sequences, where ‘0′ indicates a perfect match (in white) and >10 represents the most mismatch (in dark red or green). (C) Specificity of SARS-CoV-2 spike RNA Probe P1 in detecting SARS-CoV-2 viral genome. Vero E6 cells mock- or SARS-CoV-2- infected, at 12 hours p.i. (m.o.i.: 0.5 PFU/cell), were probed with α-NP antibody (green, panels 1 and 2) as positive control for infection and spike RNA probe P1 (magenta, panels 3 and 4), respectively, to test the specificity of the probe. Panel 5 and 6 shows the specificity of spike RNA probe P1 against Vero E6 cells mock- or infected with heat-inactivated SARS-CoV-2 at 12 hours p.i. (magenta, panels 3 and 4), respectively. Blue color indicates nuclei upon DAPI staining. Scale bar, 150 µm. (D) Vero E6 cells, mock-infected or infected with VSV-Spike virus containing codon-optimized spike gene open reading frame, were probed with spike RNA probe P1 after 24 hours p.i. The cells were imaged under FITC channel for expression of GFP (shown in green, panels 1 and 2) or under TritC channel for detecting hybridization with P1 probe (magenta, panels 3 and 4). GFP expression demonstrates the infection of cells (green, panel 2), whereas the absence of a P1 probe signal (panel 4) with cells infected with VSV-Spike, indicates the specificity of SARS-CoV-2 spike RNA probe P1. Scale bar, 150 µm. (E) Photomicrographs of scanned images of infected cells to determine the extent of SARS-CoV-2 infection and plaque formation. Vero E6 cells infected with SARS-CoV-2 virus at 0.5 m.o.i., were probed with spike RNA probe P1 at 24 hours p.i. The entire slide was scanned at high speed and high resolution. The panels 1-3 represent images at 2x, 10x, and 40x magnifications, respectively. The white arrow in the panel 3 represents the dead center of a plaque. Blue color represents DAPI staining of nuclei and Magenta, SARS-CoV-2 RNA. Scale bars, 1000 μm, 200 μm and 50 μm for 2x, 10x, and 40x images, respectively.
Figure 1.
Design and optimization of SARS-CoV-2 specific smRNA-FISH probes to detect SARS-CoV-2 infection. (A) Schematic representation of SARS-CoV-2 gRNA and sgRNA-S, indicating the position of the smRNA-FISH probes, P1 (in magenta) and P2 (in green) probes that hybridize to Spike and nsp12 ORF sequences, respectively. (B) Heatmap representing the probe sequence alignment against various coronavirus genomes (including SARS-CoV-1, SARS-CoV-2, MERS-CoV, HCoV-OC43, HCoV-NL63, HCoV-229E) and human transcriptomes (hg38-mrna). Each column represents individual 22 nt spike gene probe sequences from Spike gene (S1-S40, upper panel, represented in the shades of red) and from nsp12 gene (N1-N40, middle panel, represented in shades of green). The lower panel representing the heatmap of the spike probe sequence alignment against spike gene of the SARS-CoV-2 and codon-optimized spike gene of the SARS-CoV-2 (S1-S40, lower panel, represented in the shades of red), showing the specificity of the probe. The minimum edit distance represents mismatch score between the SARS-CoV-2 sequence and the other genome sequences, where ‘0′ indicates a perfect match (in white) and >10 represents the most mismatch (in dark red or green). (C) Specificity of SARS-CoV-2 spike RNA Probe P1 in detecting SARS-CoV-2 viral genome. Vero E6 cells mock- or SARS-CoV-2- infected, at 12 hours p.i. (m.o.i.: 0.5 PFU/cell), were probed with α-NP antibody (green, panels 1 and 2) as positive control for infection and spike RNA probe P1 (magenta, panels 3 and 4), respectively, to test the specificity of the probe. Panel 5 and 6 shows the specificity of spike RNA probe P1 against Vero E6 cells mock- or infected with heat-inactivated SARS-CoV-2 at 12 hours p.i. (magenta, panels 3 and 4), respectively. Blue color indicates nuclei upon DAPI staining. Scale bar, 150 µm. (D) Vero E6 cells, mock-infected or infected with VSV-Spike virus containing codon-optimized spike gene open reading frame, were probed with spike RNA probe P1 after 24 hours p.i. The cells were imaged under FITC channel for expression of GFP (shown in green, panels 1 and 2) or under TritC channel for detecting hybridization with P1 probe (magenta, panels 3 and 4). GFP expression demonstrates the infection of cells (green, panel 2), whereas the absence of a P1 probe signal (panel 4) with cells infected with VSV-Spike, indicates the specificity of SARS-CoV-2 spike RNA probe P1. Scale bar, 150 µm. (E) Photomicrographs of scanned images of infected cells to determine the extent of SARS-CoV-2 infection and plaque formation. Vero E6 cells infected with SARS-CoV-2 virus at 0.5 m.o.i., were probed with spike RNA probe P1 at 24 hours p.i. The entire slide was scanned at high speed and high resolution. The panels 1-3 represent images at 2x, 10x, and 40x magnifications, respectively. The white arrow in the panel 3 represents the dead center of a plaque. Blue color represents DAPI staining of nuclei and Magenta, SARS-CoV-2 RNA. Scale bars, 1000 μm, 200 μm and 50 μm for 2x, 10x, and 40x images, respectively.
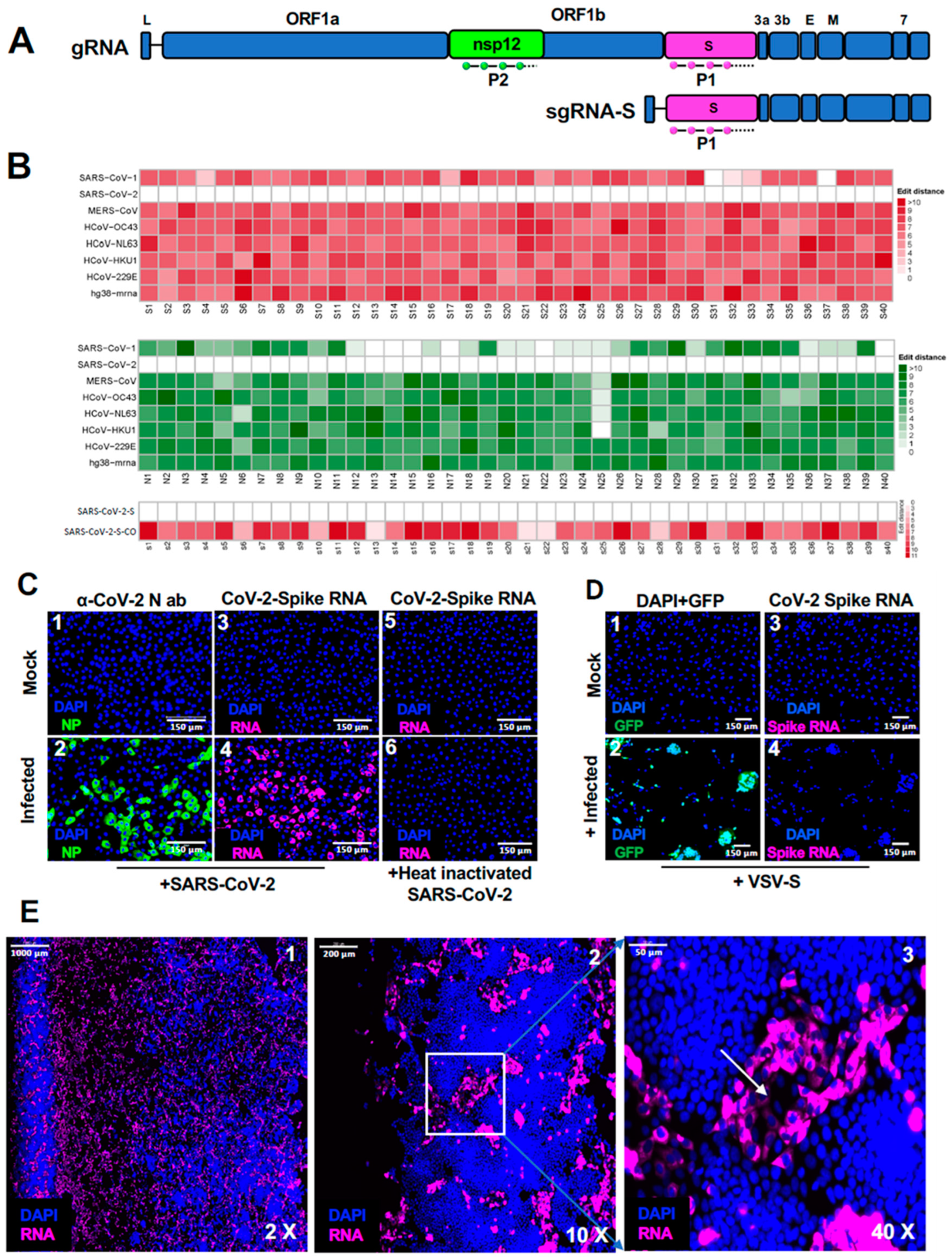
Figure 2.
Time-course analysis of SARS-CoV-2 replication using smRNA-FISH. Cells were infected and hybridized with Quasar 570-labeled P1 probes (magenta) against spike RNA at 0, 0.5, 2, 6, 12, and 24 hours p.i. (A) Photomicrographs representing the images acquired using high resolution-high speed scanning microscopy are represented at 2x (upper panels) and 40x (middle panels) magnifications, respectively. Scale bars, 1000 μm and 50 μm for 2x and 40x images, respectively. Graphical representation showing the quantitation of total percentage of positive cells infected with SARS-CoV-2 at 0, 6, 12, and 24 hours p.i. (lower panel). (B) Photomicrographs representing images of infected cells at single molecule resolution, probed with P1 at 0.5, 2, 6, 12 and 24 hours p.i., acquired using a wide-field Olympus BX-63 Microscope. White arrows point to single molecules of SARS-CoV-2 RNA seen at early time points, 0.5 and 2 hours p.i. DAPI (nucleus, blue). Scale bar, 10 μm.
Figure 2.
Time-course analysis of SARS-CoV-2 replication using smRNA-FISH. Cells were infected and hybridized with Quasar 570-labeled P1 probes (magenta) against spike RNA at 0, 0.5, 2, 6, 12, and 24 hours p.i. (A) Photomicrographs representing the images acquired using high resolution-high speed scanning microscopy are represented at 2x (upper panels) and 40x (middle panels) magnifications, respectively. Scale bars, 1000 μm and 50 μm for 2x and 40x images, respectively. Graphical representation showing the quantitation of total percentage of positive cells infected with SARS-CoV-2 at 0, 6, 12, and 24 hours p.i. (lower panel). (B) Photomicrographs representing images of infected cells at single molecule resolution, probed with P1 at 0.5, 2, 6, 12 and 24 hours p.i., acquired using a wide-field Olympus BX-63 Microscope. White arrows point to single molecules of SARS-CoV-2 RNA seen at early time points, 0.5 and 2 hours p.i. DAPI (nucleus, blue). Scale bar, 10 μm.
Figure 3.
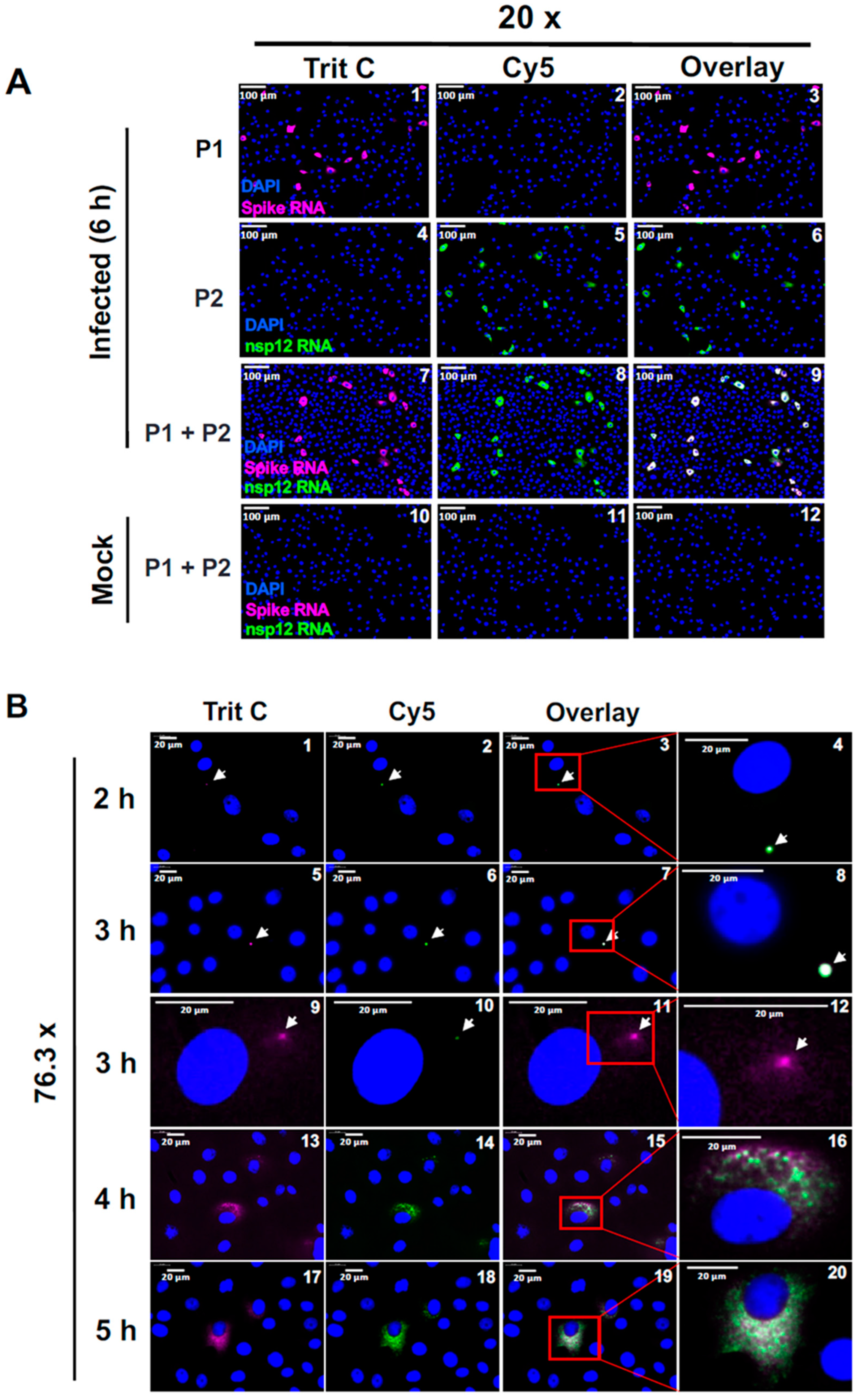
Simultaneous detection of SARS-CoV-2 gRNA and sgRNA-S in a time-course analysis. (A) Validation of spike RNA probe P1 and nsp12 RNA probe P2 to simultaneously detect gRNA and sgRNA-S in the infected cells. Cells were infected for 6 hours and hybridized with probes P1 and P2 alone or together and subjected to smRNA-FISH. Images are represented at 20x magnification. Scale bar at 100 μm. (B) A time-course analysis to detect the replication of gRNA and sgRNA in infected cells: The infected cells at 2, 3, 4 and 5 hours p.i. were hybridized with both spike RNA probe P1 and nsp12 RNA probe P2 and subjected to high-speed, high-resolution scanning. The images are represented at 76.3x magnifications. Panels 4, 8, 12,16 and 20 are the zoomed images of insets for indicated time-points. White arrows point to RNA spots that are surrounds by potentially single molecule sgRNA-S. Blue indicates DAPI stained nuclei. Scale bar at 20 μm.
Figure 3.
Simultaneous detection of SARS-CoV-2 gRNA and sgRNA-S in a time-course analysis. (A) Validation of spike RNA probe P1 and nsp12 RNA probe P2 to simultaneously detect gRNA and sgRNA-S in the infected cells. Cells were infected for 6 hours and hybridized with probes P1 and P2 alone or together and subjected to smRNA-FISH. Images are represented at 20x magnification. Scale bar at 100 μm. (B) A time-course analysis to detect the replication of gRNA and sgRNA in infected cells: The infected cells at 2, 3, 4 and 5 hours p.i. were hybridized with both spike RNA probe P1 and nsp12 RNA probe P2 and subjected to high-speed, high-resolution scanning. The images are represented at 76.3x magnifications. Panels 4, 8, 12,16 and 20 are the zoomed images of insets for indicated time-points. White arrows point to RNA spots that are surrounds by potentially single molecule sgRNA-S. Blue indicates DAPI stained nuclei. Scale bar at 20 μm.
Figure 4.
Single molecule analysis to determine the kinetics of SARS-CoV-2 gRNA and sgRNA replication. Transcript specific visualization of gRNA and sgRNA-S in SARS-CoV-2 infected Vero E6 cells using P1 (spike, represented in magenta) and P2 (nsp12, represented in green) probes. When spike RNA probe P1 and nsp12 RNA probe P2 both hybridize to the same molecule, it is shown in white (overlay). Blue indicates DAPI stained nuclei. Images represent the cells infected with SARS-CoV-2 and hybridized with P1 and P2 probes at 0, 2, 3, 4, 5 and 12 hours p.i. White arrows in panels 6-16 point to replication centers. Yellow arrow heads point to single molecules of sgRNA-S. Panels 13-16 show the magnified area of the corresponding overlay images at 3 hours p.i., illustrating two replication centers (white arrows). The larger RNA spots in these panels harbors gRNA in the center surrounded by the sgRNA-S. Single molecule RNA are indicated by yellow arrow heads (panels 14-16). The white arrows in panels 22-24 point to RNA in the nucleus. The images were acquired using a wide-field microscope to detect the diffraction limited single molecules. Scale bar at 10 μm.
Figure 4.
Single molecule analysis to determine the kinetics of SARS-CoV-2 gRNA and sgRNA replication. Transcript specific visualization of gRNA and sgRNA-S in SARS-CoV-2 infected Vero E6 cells using P1 (spike, represented in magenta) and P2 (nsp12, represented in green) probes. When spike RNA probe P1 and nsp12 RNA probe P2 both hybridize to the same molecule, it is shown in white (overlay). Blue indicates DAPI stained nuclei. Images represent the cells infected with SARS-CoV-2 and hybridized with P1 and P2 probes at 0, 2, 3, 4, 5 and 12 hours p.i. White arrows in panels 6-16 point to replication centers. Yellow arrow heads point to single molecules of sgRNA-S. Panels 13-16 show the magnified area of the corresponding overlay images at 3 hours p.i., illustrating two replication centers (white arrows). The larger RNA spots in these panels harbors gRNA in the center surrounded by the sgRNA-S. Single molecule RNA are indicated by yellow arrow heads (panels 14-16). The white arrows in panels 22-24 point to RNA in the nucleus. The images were acquired using a wide-field microscope to detect the diffraction limited single molecules. Scale bar at 10 μm.
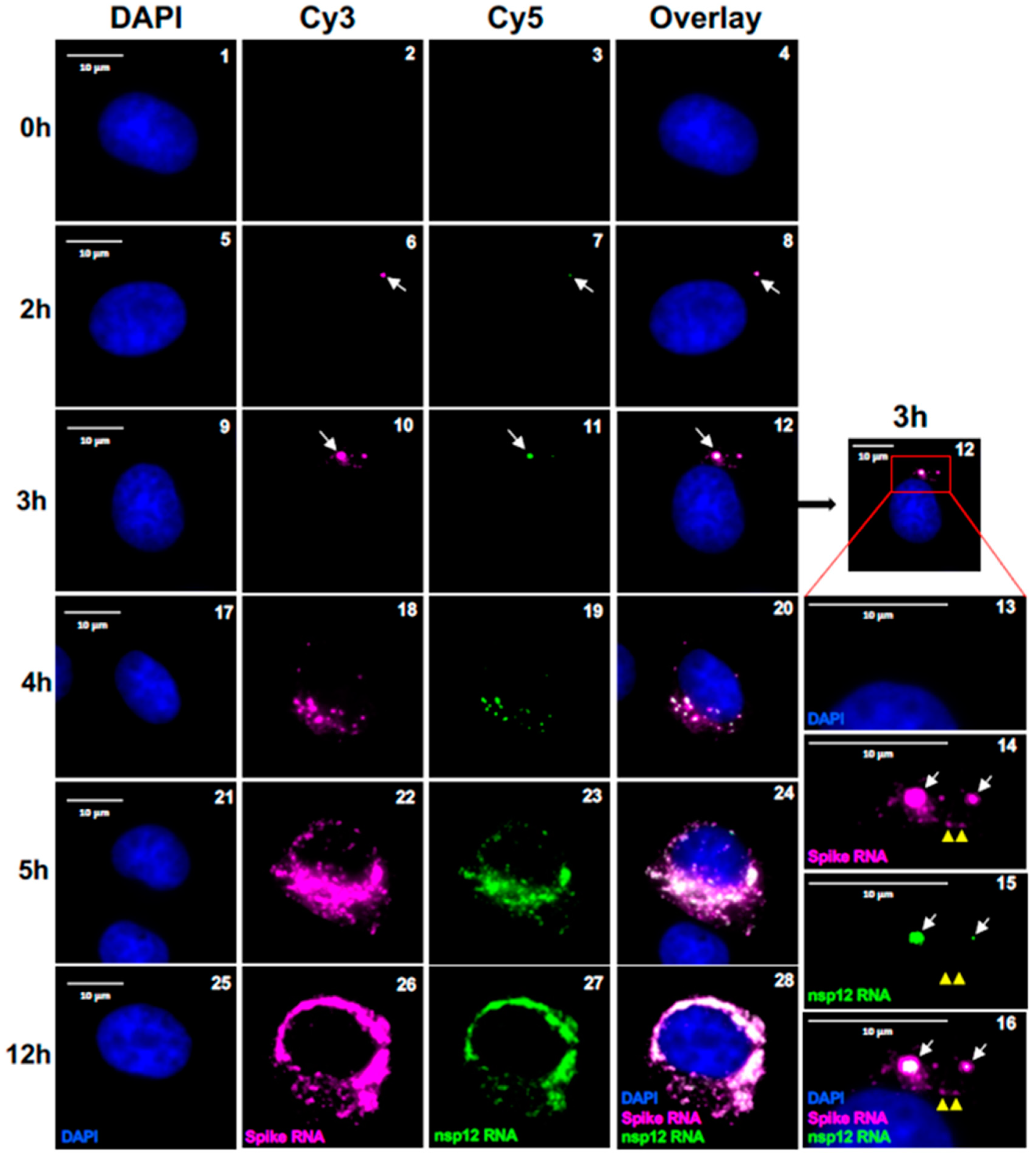
Figure 5.
Images of cells infected with SARS-CoV-2 at 5 hours p.i. (A) Representative cells infected with SARS-CoV-2 at 5 hours p.i. Panels 1-4 represent a cell filled with ROs at 5 hours p.i. Panels 5-8 represent a single cell with migrating gRNA and sgRNA-S to the distal end of the cell (yellow arrow heads). (B) Presence of SARS-CoV-2 RNA in the nucleus. The images from a few of the Z-stack of cell are represented to demonstrate the presence of the positive RNA spot inside the nucleus. The Z-stack images were acquired starting from the bottom of the cell moving upwards. The panel numbers represent the stack number in a total of 41 optical sections (each with a 200 nm Z step size). In both A and B, images were acquired using a wide-field microscope to detect the diffraction limited single molecules. The spike RNA P1 probe signals are colored in magenta (sgRNA-S), nsp12 RNA P2 probe signals are colored in green (gRNA) and when P1 and P2 both hybridize to the same molecule, it is shown in white (overlay). Scale bar, 10 μm. DAPI (nucleus, blue).
Figure 5.
Images of cells infected with SARS-CoV-2 at 5 hours p.i. (A) Representative cells infected with SARS-CoV-2 at 5 hours p.i. Panels 1-4 represent a cell filled with ROs at 5 hours p.i. Panels 5-8 represent a single cell with migrating gRNA and sgRNA-S to the distal end of the cell (yellow arrow heads). (B) Presence of SARS-CoV-2 RNA in the nucleus. The images from a few of the Z-stack of cell are represented to demonstrate the presence of the positive RNA spot inside the nucleus. The Z-stack images were acquired starting from the bottom of the cell moving upwards. The panel numbers represent the stack number in a total of 41 optical sections (each with a 200 nm Z step size). In both A and B, images were acquired using a wide-field microscope to detect the diffraction limited single molecules. The spike RNA P1 probe signals are colored in magenta (sgRNA-S), nsp12 RNA P2 probe signals are colored in green (gRNA) and when P1 and P2 both hybridize to the same molecule, it is shown in white (overlay). Scale bar, 10 μm. DAPI (nucleus, blue).
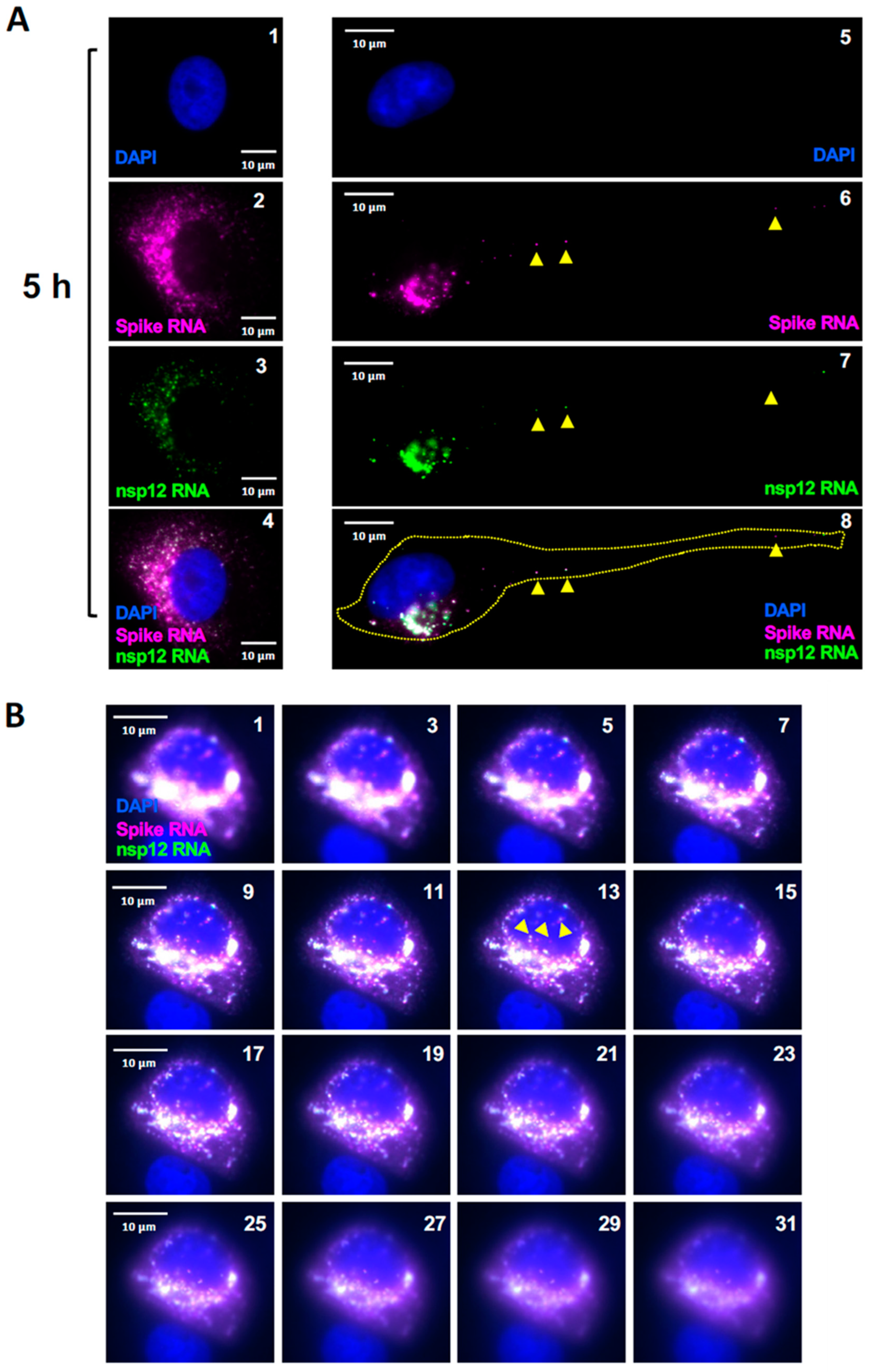
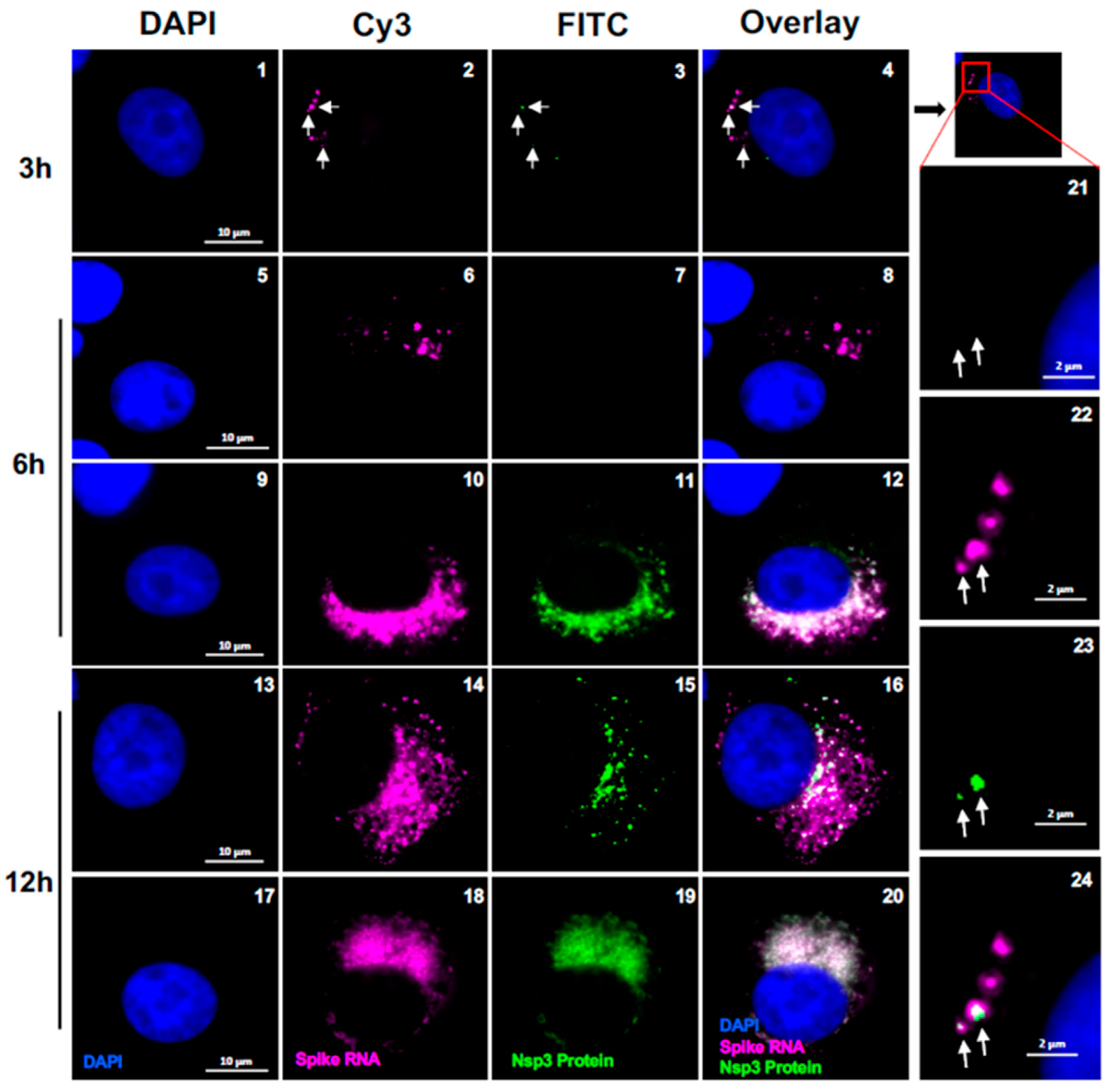
Figure 6.
Co-localization of SARS-CoV-2 RNA speckles with Nsp3 protein. Representative cells showing the co-localization of spike probe P1 and Nsp3 protein within the infected Vero E6 cells with SARS-CoV-2 at 3, 6 and 12 hours p.i., respectively. The images were acquired using a wide-field microscope to detect the diffraction limited single molecules. Blue color represents DAPI nuclear staining; Green color represents Nsp3 protein, magenta color represents SARS-CoV-2 spike RNA. Scale bar, 10 μm. Panels 21-24 show the magnified area of the corresponding images at 3 hours p.i. (Scale bar at 2 μm). White arrows indicate RNA speckles that co-localize with nsp3.
Figure 6.
Co-localization of SARS-CoV-2 RNA speckles with Nsp3 protein. Representative cells showing the co-localization of spike probe P1 and Nsp3 protein within the infected Vero E6 cells with SARS-CoV-2 at 3, 6 and 12 hours p.i., respectively. The images were acquired using a wide-field microscope to detect the diffraction limited single molecules. Blue color represents DAPI nuclear staining; Green color represents Nsp3 protein, magenta color represents SARS-CoV-2 spike RNA. Scale bar, 10 μm. Panels 21-24 show the magnified area of the corresponding images at 3 hours p.i. (Scale bar at 2 μm). White arrows indicate RNA speckles that co-localize with nsp3.
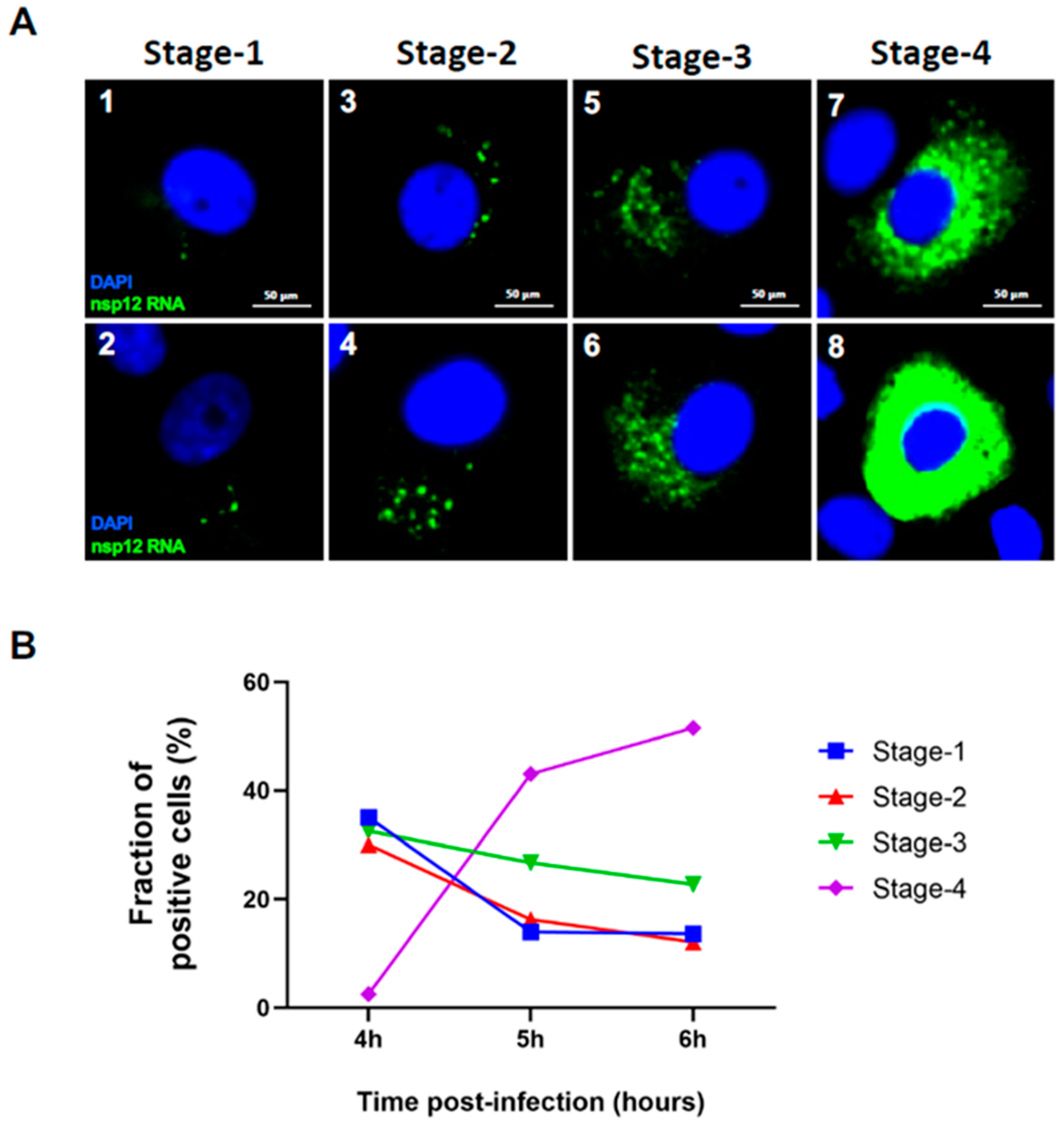
Figure 7.
Quantitation of cells containing viral RNA at different stages of replication. (A) Examples of images of cells with four different stages of viral replication at Stage-1 (panels 1 and 2), Stage-2 (panels 3 and 4), Stage-3 (panels 5 and 6), and Stage-4 (panels 7 and 8). Blue color represents DAPI nuclear staining; and nsp12 RNA (P2 probe) signals are colored in green which represents replication centers containing genomic RNA. Scale bar, 50 μm. (B) Graphical representation of quantitation of percentage of positive cells harboring four different stages of replication at 4, 5 and 6 hours p.i.
Figure 7.
Quantitation of cells containing viral RNA at different stages of replication. (A) Examples of images of cells with four different stages of viral replication at Stage-1 (panels 1 and 2), Stage-2 (panels 3 and 4), Stage-3 (panels 5 and 6), and Stage-4 (panels 7 and 8). Blue color represents DAPI nuclear staining; and nsp12 RNA (P2 probe) signals are colored in green which represents replication centers containing genomic RNA. Scale bar, 50 μm. (B) Graphical representation of quantitation of percentage of positive cells harboring four different stages of replication at 4, 5 and 6 hours p.i.
Figure 8.
Ability of SARS-CoV-2 probes to detect variants of concern (VOCs) of SARS-CoV-2. (A) Heatmap representing the probe sequence alignment against various SARS-CoV-2 variants of concern (VOCs) genomes (SARS-CoV-2 B.1.1.7, B.1.351, P.1, B.1.617.2, B.1.1.529, BA.1, BA.2, BA.4, and BA.5) along with SARS-CoV-2 Wuhan-Hu-1. Each column represents individual 22 nt spike gene probe sequences from Spike gene (S1-S40, upper panel, represented in the shades of red) and from nsp12 gene (N1-N40, lower panel, represented in shades of green). The minimum edit distance represents mismatch score between the SARS-CoV-2 Wuhan-Hu-1 gene sequence and the other SARS-CoV-2 VOC’s genome sequences, where ‘0′ indicates a perfect match (in white) and >10 represents the most mismatch (in dark red or green). (B and C) Validation of spike RNA probe P1 and nsp12 RNA probe P2 to simultaneously detect gRNA and sgRNA-S in the Vero-TMPRSS2 cells infected with SARS-CoV-2 BA.1 VOC. Cells were infected at 3h, 6h and 12h, respectively. (B). Panels 2, 4 and 6 indicates infected cells and panels 1, 3, and 5, represent mock infected cells respectively shown at a20x magnification. Panel 7 indicates Vero-TMPRSS2 cells infected with heat-inactivated SARS-CoV-2 BA.1 at 12 hours p.i. under similar conditions. Scale bar at 50 μm. White arrows point to infected cells. (C). Visualization of representative single Vero-TMPRSS2 cells infected with SARS-CoV-2 BA.1 VOCs at 6 hours (Panels 1-8) and 12 hours (Panels: 9-16) post-infection. Scale bar at 20 μm. Magenta = Spike RNA probe P1; Green = nsp12 RNA probe P2; and Blue =DAPI staining.
Figure 8.
Ability of SARS-CoV-2 probes to detect variants of concern (VOCs) of SARS-CoV-2. (A) Heatmap representing the probe sequence alignment against various SARS-CoV-2 variants of concern (VOCs) genomes (SARS-CoV-2 B.1.1.7, B.1.351, P.1, B.1.617.2, B.1.1.529, BA.1, BA.2, BA.4, and BA.5) along with SARS-CoV-2 Wuhan-Hu-1. Each column represents individual 22 nt spike gene probe sequences from Spike gene (S1-S40, upper panel, represented in the shades of red) and from nsp12 gene (N1-N40, lower panel, represented in shades of green). The minimum edit distance represents mismatch score between the SARS-CoV-2 Wuhan-Hu-1 gene sequence and the other SARS-CoV-2 VOC’s genome sequences, where ‘0′ indicates a perfect match (in white) and >10 represents the most mismatch (in dark red or green). (B and C) Validation of spike RNA probe P1 and nsp12 RNA probe P2 to simultaneously detect gRNA and sgRNA-S in the Vero-TMPRSS2 cells infected with SARS-CoV-2 BA.1 VOC. Cells were infected at 3h, 6h and 12h, respectively. (B). Panels 2, 4 and 6 indicates infected cells and panels 1, 3, and 5, represent mock infected cells respectively shown at a20x magnification. Panel 7 indicates Vero-TMPRSS2 cells infected with heat-inactivated SARS-CoV-2 BA.1 at 12 hours p.i. under similar conditions. Scale bar at 50 μm. White arrows point to infected cells. (C). Visualization of representative single Vero-TMPRSS2 cells infected with SARS-CoV-2 BA.1 VOCs at 6 hours (Panels 1-8) and 12 hours (Panels: 9-16) post-infection. Scale bar at 20 μm. Magenta = Spike RNA probe P1; Green = nsp12 RNA probe P2; and Blue =DAPI staining.
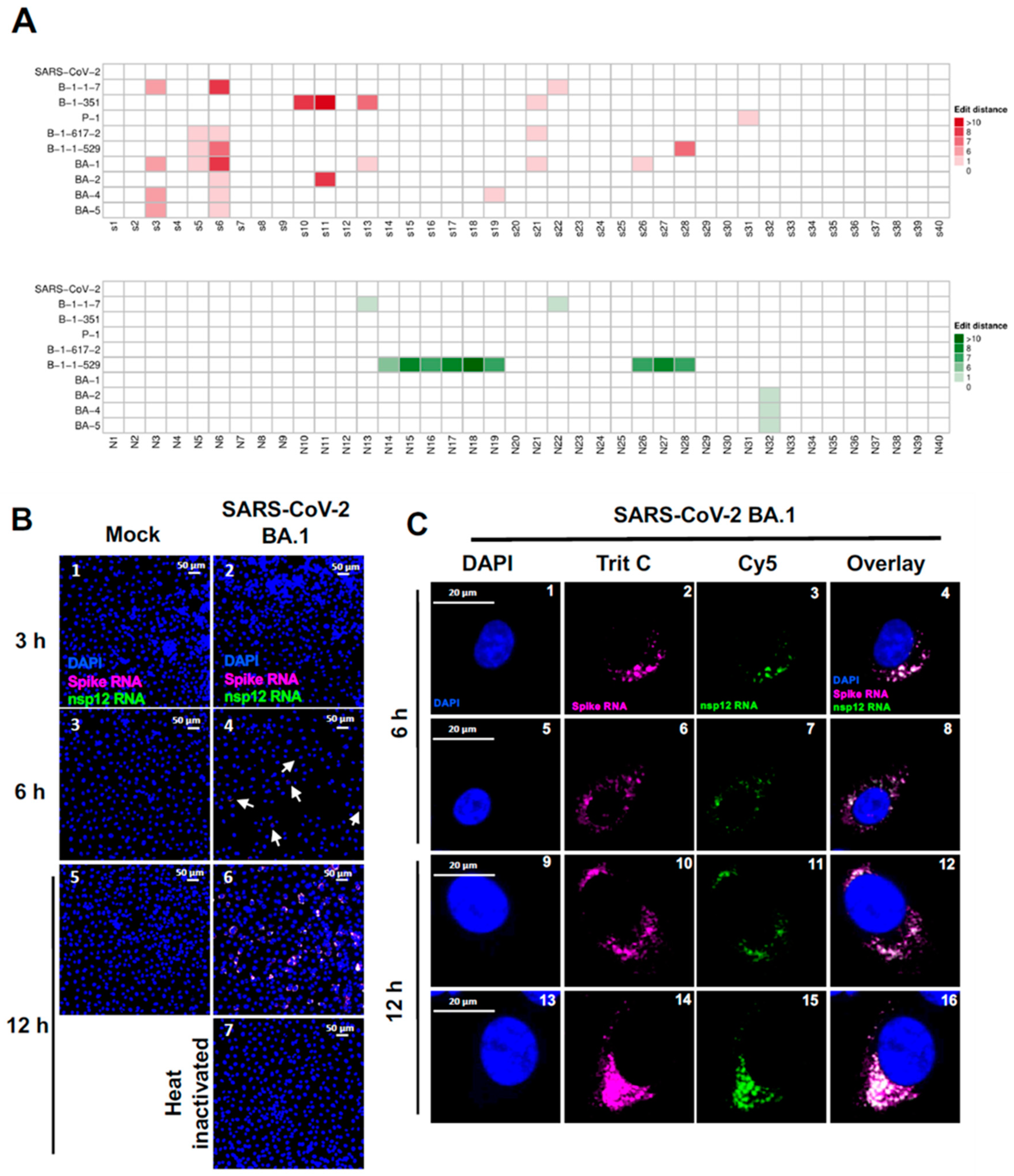
Table 1.
Sequences of SARS-CoV-2 Spike RNA (P1) and nsp12 RNA (P2) probes used for smRNA-FISH analyses.
Table 1.
Sequences of SARS-CoV-2 Spike RNA (P1) and nsp12 RNA (P2) probes used for smRNA-FISH analyses.
| Probe number |
smRNA-FISH Spike RNA probe (P1)
(5′-> 3′) |
smRNA-FISH nsp12 RNA probe (P2)
(5′-> 3′) |
| 1 |
tgactagagactagtggcaata |
gctactttatcattgtagatgt |
| 2 |
tttgtcagggtaataaacacca |
agttagagaaagtgtgtctctt |
| 3 |
atgtatagcatggaaccaagta |
aattaccttcatcaaaatgcct |
| 4 |
accatcattaaatggtaggaca |
ttctacaaaatcataccagtcc |
| 5 |
tatgttagacttctcagtggaa |
ttttaacaaagcttggcgtaca |
| 6 |
tttttgtggtaataaacaccca |
agttaccattgagatcttgatt |
| 7 |
gtgcaattattcgcactagaat |
agaatctacaacaggaactcca |
| 8 |
aataggcgtgtgcttagaatat |
acatgtgactctgcagttaaag |
| 9 |
gcaaatctaccaatggttctaa |
ctgtcatccaaacagttaacac |
| 10 |
taggttgaagataacccacata |
cagcagcatacacaagtaattc |
| 11 |
ctacagtgaaggatttcaacgt |
gtaatagattaccagaagcagc |
| 12 |
agaattccaagctataacgcag |
taagtgcagctactgaaaagca |
| 13 |
tataattaccaccaaccttaga |
aaaattaccgggtttgacagtt |
| 14 |
ggcagaaactttttgttagact |
caacagaacttccttccttaaa |
| 15 |
tgacaccaccaaaagaacatgg |
atcctgagcaaagaagaagtgt |
| 16 |
tatttgttcctggtgttataac |
acgatagtagtcataatcgctg |
| 17 |
gttgatctgcatgaatagcaac |
ttgtctgatatcacacattgtt |
| 18 |
aataaacacgccaagtaggagt |
acgatgacttggttagcattaa |
| 19 |
cactcatatgagttgttgacat |
gaaaaccagctgatttgtctag |
| 20 |
atagtgtaggcaatgatggatt |
atgcgaaaagtgcatcttgatc |
| 21 |
gtaagcaactgaattttctgca |
tagtagggatgacattacgttt |
| 22 |
agtaaaatttgtgggtatggca |
tattctttgcactaatggcata |
| 23 |
ggtagaatttctgtggtaacac |
ctattggtcatagtactacaga |
| 24 |
acttgtgcaaaaacttcttggg |
cttgttccaattactacagtag |
| 25 |
tggtggtgttttgtaaatttgt |
tcacatttaggataatcccaac |
| 26 |
aacagtaaggccgttaaacttt |
acaagtgaggccataattctaa |
| 27 |
agaacattctgtgtaactccaa |
gaaacggtgtgacaagctacaa |
| 28 |
acttgctgtggaagaaagtgag |
atgaccatttcactcaatactt |
| 29 |
ggagctaagttgtttaacaagc |
acatacttatcggcaattttgt |
| 30 |
ttgagtcacatatgtctgcaaa |
cactcataaagtctgtgttgta |
| 31 |
gacattttagtagcagcaagat |
tcacaaagtctgtgtcaacatc |
| 32 |
cctttccacaaaaatcaactct |
gtcagagagtatcatcattgag |
| 33 |
ctgactgagggaaggacataag |
actagaccttgagatgcataag |
| 34 |
agtcacatgcaagaagactaca |
gaaggtacacataatcatcacc |
| 35 |
gttgttgacaattcctattaca |
atatcatctacaaaacagccgg |
| 36 |
tgaagcattaatgccagagatg |
gacacgaaccgttcaatcataa |
| 37 |
gcggtcaatttctttttgaatg |
tagtaagtgggtaagcatctat |
| 38 |
taaattcttggcaacctcattg |
atgtagctttcttatgtattgt |
| 39 |
ttggagatcgatgagagattca |
aatacatgtctaacatgtgtcc |
| 40 |
gctataaaacctagccaaatgt |
gtgtacatagcctcataaaact |