Submitted:
29 January 2024
Posted:
30 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Prevalence of Enterococci
2.2. Virulence of Enterococci
2.3. Antibiotic resistance
| Antibiotic Group | Antibiotics | Enterococcus Species | |||||
|---|---|---|---|---|---|---|---|
|
E. faecalis n=11 |
E. hirae n=34 |
E. faecium n=14 |
E.casseliflavus n=30 |
E. gallinarum n=1 |
Total n=90 |
||
| n(%) | n(%) | n(%) | n(%) | n(%) | n(%) | ||
| Penicillins | AM | - | 5 (5.5) | 8(8.8) | 4(4.4) | 1(1.1) | 18(20) |
| Penicillins | P | - | - | 2(2.2) | - | - | 2(2.2) |
| Lipoglycopeptides | TEC | - | - | - | - | - | - |
| Macrolides | E | * | 3(3.3) | * | * | * | 3(3.3) |
| Tetracyclines | TE | 6(6.6) | 4(4.4) | 3(3.3) | 6(6.6) | - | 19(21.1) |
| Fluoroquinolones | CIP | 1(1.1) | - | 5(5.5) | - | - | 6(6.6) |
| Fluoroquinolones | LEV | 1(1.1) | - | - | - | - | 1(1.1) |
| Nitrofurans | F | - | 4(4.4) | - | - | - | 4(4.4) |
| Ansamycins | RA | 2(2.2) | 4(4.4) | 4(4.4) | 3(3.3) | 1(1.1) | 14(15.5) |
| Fosfomycins | FF | - | - | 2(2.2) | 2(2.2) | - | 4(4.4) |
| Phenicols | C | 2(2.2) | - | 1(1.1) | 1(1.1) | - | 4(4.4) |
| Streptogramins | QD | * | 19(21.1) | 7(7.7) | * | * | 26(28.8) |
| Oxazolidinones | LNZ | - | - | - | - | - | - |
| Tetracyclines | TIG | - | - | - | 1(1.1) | - | 1(1.1) |
| Carbapenems | IPM | - | - | - | - | - | - |
| Glycopeptides | VA | 3(3.3) | 3(3.3) | 1(1.1) | - | - | 7(7.7) |
| Aminoglycosides | HSLR | 1(1.1) | - | 2(2.2) | 2(2.2) | - | 5(5.5) |
| Aminoglycosides | HGLR | - | - | - | - | - | - |
| MDR | 4(36.3) | 4(11.7) | 7(30) | 2(6.6) | 17(18.8) | ||
| Strain | Animal species | Carcass part | Antibiotic resistance | Virulence factor | |
|---|---|---|---|---|---|
| Phenotype | Genotype | ||||
| EFM-4 | Sheep | Hind leg | CIP, RA, QD | ||
| EFM-7 | Sheep | Hind leg | RA,P,AM | ||
| EC-39 | Goat | Rectal | HLSR, TE, AM | ||
| EFM-45 | Sheep | Rectal | CIP, RA, TE | ||
| EH-48 | Sheep | Rectal | E, AM, QD | ||
| EH-49 | Sheep | Brisket | RA, AM, QD | ermB | |
| EFM-57 | Sheep | Rectal | HLSR, FF, AM, | ||
| EH-66 | Goat | Hind leg | RA, E, QD | ||
| EFS-76 | Sheep | Hind leg | C, CIP, LEV, RA, TE, AM | aac(6’)Ie-aph(2'')-la, Isa, efrA, tetM | efaA |
| EFM-88 | Goat | Flank | VA, AM, QD | VanC1 | |
| EFS-97 | Goat | Rectal | VA, C, HLSR, TE | Isa, efrA, emeA, tetM | efaA |
| EC-98 | Sheep | Brisket | C, HLSR, AM, TE | ||
| EFM-99 | Goat | Rectal | C, HLSR, TE, FF, QD | Isa, efrA, tetM | |
| EFS-106 | Cattle | Brisket | VA, TE, AM | Isa, tetM | |
| EFS-108 | Sheep | Flank | VA, RA, TE, AM, QD | Isa, tetM | |
| EFM-113 | Sheep | Hind leg | CIP, RA, TE, P, AM | ||
| EH-116 | Goat | Hind leg | F, E, AM | ||
2.4. Genotyping of antibiotic resistance
3. Discussion
4. Materials and Methods
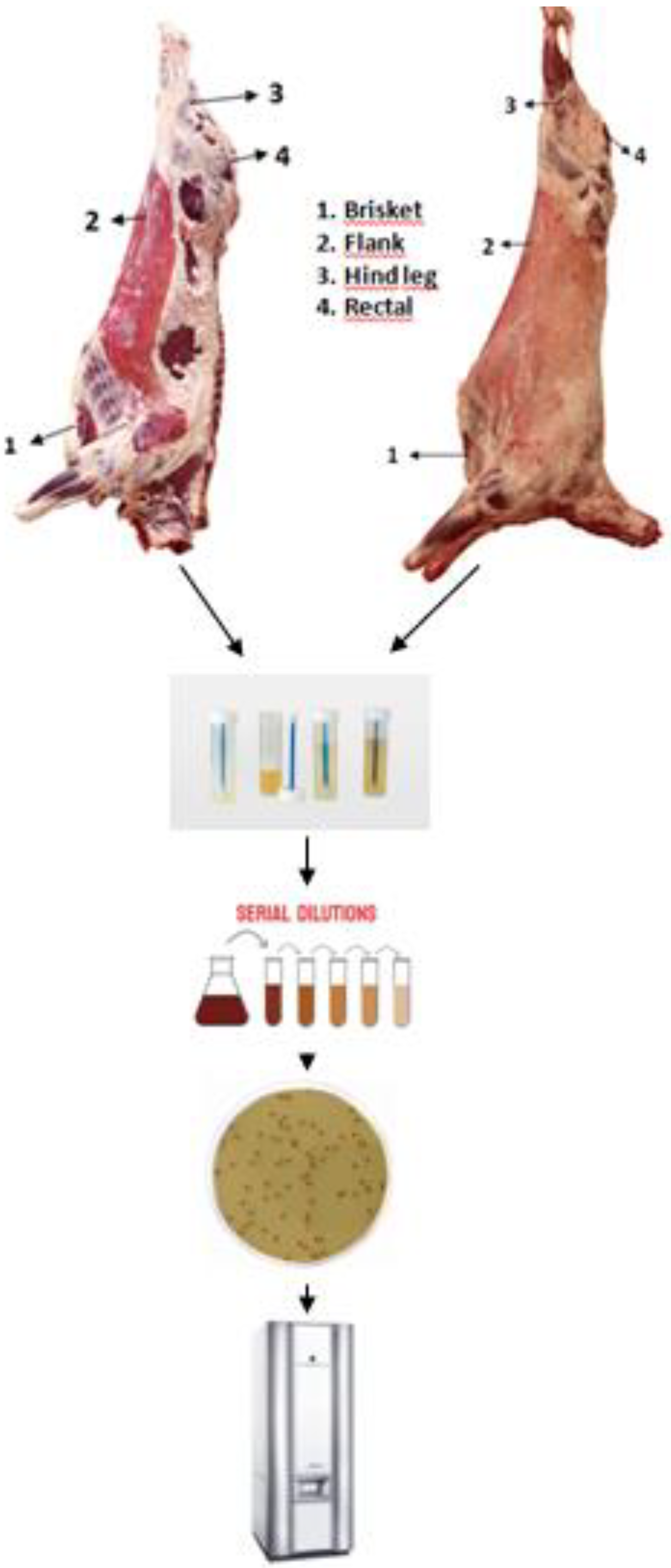
4.1. Sample collection

4.2. Enterococus species isolation
4.3. DNA isolation protocols
4.4. Screening for confirmation and virulence genes
| Target gene | Primer sequence (5'-3') | Fragment size (pb) |
|---|---|---|
| Enterococcus spp. (16S rRNA) | F: AGCGCAGGCGGTTTCTTAA R: CTCGTTGTACTTCCCATTGT |
941 |
| Enterococcus faecalis | F: ATCAAGTACAGTTAGTCTTTATTAG R:ACGATTCAAAGCTAACTGAATCAGT |
658 |
| Enterococcus faecium | F: TTGAGGCAGACCAGATTGACG R: GCTGCTAAAGCTGCGCTT |
822 |
| Enterococcus gallinarum | F: GGTATCAAGGAAACCTC R: CTTCCGCCATCATAGCT |
484 |
| Enterococcus casseliflavus | F: CGGGGAAGATGGCAGTAT R: CGCAGGGACGGTGATTTT |
521 |
| Enterococcus hirae | F: GCATATTTATCCAGCACTAG R: CTCTGGATCAAGTCCATAAGTGG |
639 |
| asa1 | F: CACGCTATTACGAACTATGA R: TAAGAAAGAACATCACCACGA |
375 |
| ace | F: GGAATGACCGAGAACGATGGC R: GCTTGATGTTGGCCTGCTTCCG |
616 |
| cylA | F: ACTCGGGGATTGATAGGC R: GCTGCTAAAGCTGCGCTT |
688 |
| efaA | F: CGTGAGAAAGAAATGGAGGA R: CTACTAACACGTCACGAATG |
499 |
| esp | F: AGATTTCATCTTTGATTCTTG R: AATTGATTCTTTAGCATCTGG |
510 |
| gelE | F: TATGACAATGCTTTTTGGGAT R: AGATGCACCCGAAATAATATA |
213 |
| hyl | F: ACAGAAGAGCTGCAGGAAATG R: GACTGACGTCCAAGTTTCCAA |
276 |
2.5. Antimicrobial susceptibility testing
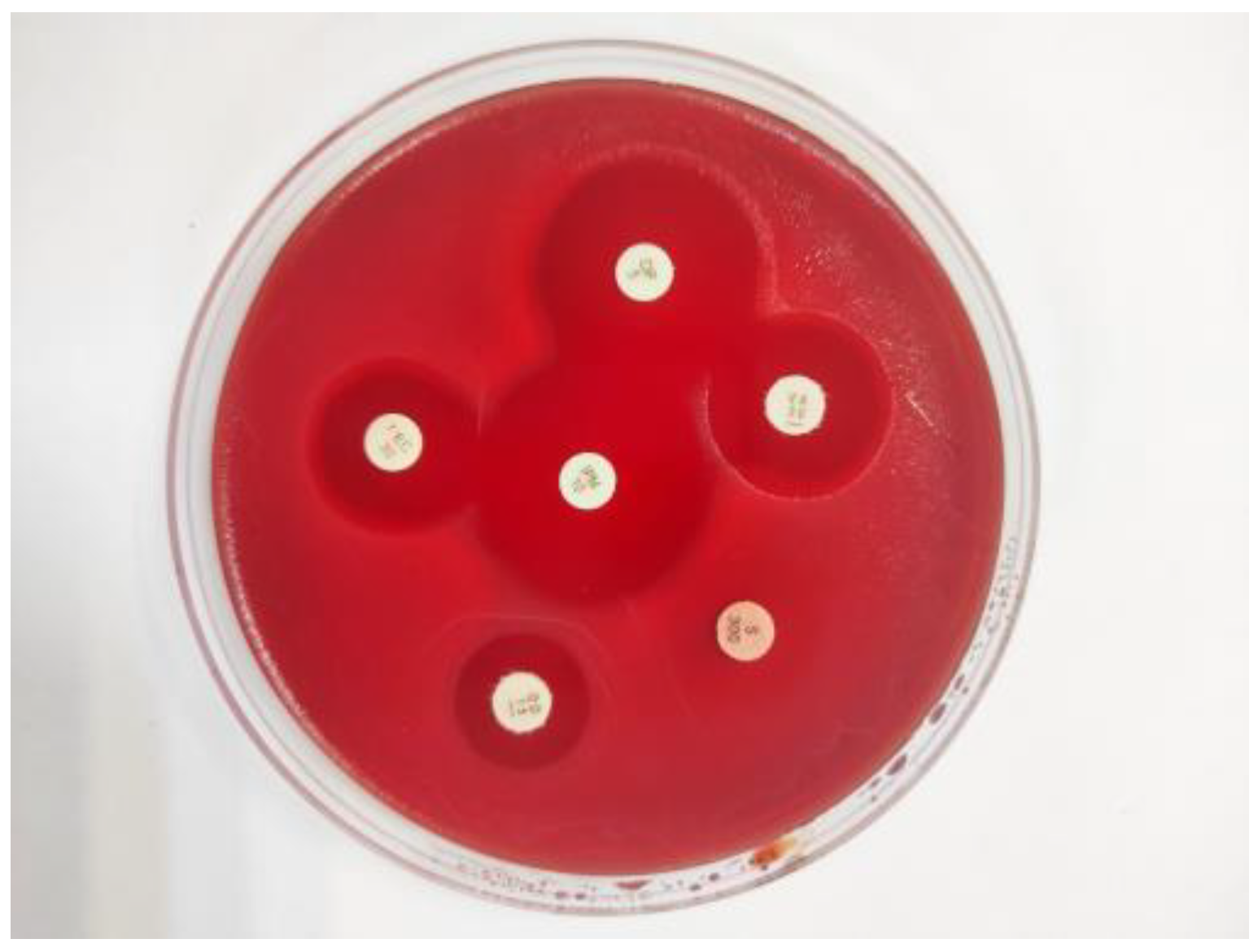
2.6. PCR detection of genes for antimicrobial resistance
| Antimicrobial agent | Target gene | Primer sequence (5'-3') | Fragment size (pb) |
|---|---|---|---|
| Macrolides | ermA | F: TAACATCAGTACGGATATTG R: AGTCTACACTTGGCTTAGG |
200 |
| Macrolides | ermB | F: CCGAACACTAGGGTTGCTC R: ATCTGGAACATCTGTGGTATG |
139 |
| Macrolides | mef | F: AGTATCATTAATCACTAGTGC R: TTCTTCTGGTACTAAAAGTGG |
348 |
| Tetracyclines | tet(L) | F: ATAAATTGTTTCGGGTCGGTAAT R: AACCAGCCAACTAATGACAATGAT |
1077 |
| Tetracyclines | tet(M) | F: GTTAAATAGTGTTCTTGGAG R: CTAAGATATGGCTCTAACAA |
657 |
| Tetracyclines | tet(O) | F: GATGGCATACAGGCACAGAC R: CAATATCACCAGAGCAGGCT |
614 |
| Phenicols | cfr | F: TGAAGTATAAAGCAGGTTGGGAGTCA R: ACCATATAATTGACCACAAGCAGC |
746 |
| Phenicols | fexA | F: GTACTTGTAGGTGCAATTACGGCTGA R: CGCATCTGAGTAGGACATAGCGTC |
1272 |
| Phenicols | optrA | F: AGGTGGTCAGCGAACTAA R: ATCAACTGTTCCCATTCA |
1379 |
| Efflux pump | eme(A) | F: AGCCCAAGCGAAAAGCGGTTT R: CCATCGCTTTCGGACGTTCA |
123 |
| Efflux pump | efr(A) | F: GTCTGTTTCGTTTAATGGCAGCAGCC R: CGAATAGCTGGTTCATGTCTAAGGC |
258 |
| Efflux pump | lsa | F: GTGACTTCTTTTGAACAGTGGGA R: TTCAGCCACTTGTTGTCTGCC |
232 |
| Aminoglycoside modifying enzyme |
aac(6’)Ie-aph(2")-la | F: CAGAGCCTTGGGAAGATGAAG R: CCTCGTGTAATTCATGTTCTGGC |
348 |
| Aminoglycoside modifying enzyme |
aph(2")-Ib | F: CTTGGACGCTGAGATATATGAGCAC R: GTTTGTAGCAATTCAGAAACACCCTT |
867 |
| Aminoglycoside modifying enzyme |
aph(2")-Ic | F: CCACAATGATAATGACTCAGTTCCC R: CCACAGCTTCCGATAGCAAGAG |
641 |
| Aminoglycoside modifying enzyme |
aph(2")-Id | F: GTGGTTTTTACAGGAATGCCATC R: CCCTCTTCATACCAATCCATATAACC |
284 |
| Aminoglycoside modifying enzyme |
ant(3")-Ia | F: TGATTTGCTGGTTACGGTGAC R: CGCTATGTTCTCTTGCTTTTG |
284 |
| Aminoglycoside modifying enzyme |
aph(6)-Ia | F: ACTGGCTTAATCAATTTGGG R: GCCTTTCCGCCACCTCACCG |
596 |
| Glycopeptıdes | vanA | F: ATTGCTATTCAGCTGTACTC R: GGCTCGACTTCCTGATGAAT |
559 |
| Glycopeptıdes | vanB | F: AACGGCGTATGGAAGCTATG R: CCATCATATTGTCCTGCTGC |
467 |
| Glycopeptıdes | vanC1 | F: GGCATCGCACCAACAATGGA R: TCCTCTGCCAGTGCAATCAA |
902 |
| Glycopeptıdes | vanC2 | F: TTCAGCAACTAGCGCAATCG R: TCACAAGCACCGACAGTCAA |
663 |
2.7. Statistical analysis
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ramos, S.; Igrejas, G.; Capelo-Martinez, J. L.; Poeta, P. Antibiotic resistance and mechanisms implicated in fecal enterococci recovered from pigs, cattle and sheep in a Portuguese slaughterhouse. Annals of microbiology 2012, 62, 1485–1494. [Google Scholar] [CrossRef]
- Angulo, F. J.; Heuer, O. E.; Hammerum, A. M.; Collignon, P.; Wegener, H. C. Human health hazard from antimicrobial-resistant enterococci in animals and food. Clinical Infectious Diseases 2006, 43, 911–916. [Google Scholar]
- Na, S. H.; Moon, D. C.; Choi, M. J.; Oh, S. J.; Jung, D. Y.; Kang, H. Y.; Hyun, S. J.; Lim, S. K. Detection of oxazolidinone and phenicol resistant enterococcal isolates from duck feces and carcasses. International journal of food microbiology 2019, 293, 53–59. [Google Scholar] [CrossRef]
- Kim, M. H.; Moon, D. C.; Kim, S. J.; Mechesso, A. F.; Song, H. J.; Kang, H. Y.; Choi, J. Y.; Yoon, S. S.; Lim, S. K. Nationwide surveillance on antimicrobial resistance profiles of Enterococcus faecium and Enterococcus faecalis isolated from healthy food animals in South Korea, 2010 to 2019. Microorganisms 2021, 9, 925. [Google Scholar] [CrossRef]
- Smoglica, C.; Vergara, A.; Angelucci, S.; Festino, A. R.; Antonucci, A.; Marsilio, F.; Di Francesco, C. E. Evidence of linezolid resistance and virulence factors in Enterococcus spp. isolates from wild and domestic ruminants, Italy. Antibiotics 2022, 11, 223. [Google Scholar] [CrossRef]
- Desire, O. E.; Larson, B.; Richard, O.; Rolande, M. M.; Serge, K. B. Investigating antibiotic resistance in enterococci in Gabonese livestock. Veterinary World 2022, 15, 714. [Google Scholar] [CrossRef]
- Strateva, T.; Atanasova, D.; Savov, E.; Petrova, G.; Mitov, I. Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Brazilian Journal of Infectious Diseases 2016, 20, 127–133. [Google Scholar] [CrossRef]
- Song, H.; Bae, Y.; Jeon, E.; Kwon, Y.; Joh, S. Multiplex PCR analysis of virulence genes and their influence on antibiotic resistance in Enterococcus spp. isolated from broiler chicken. Journal of veterinary science 2019, 20. [Google Scholar] [CrossRef]
- Alzahrani, O. M.; Fayez, M.; Alswat, A. S.; Alkafafy, M.; Mahmoud, S. F.; Al-Marri, T.; Almuslem, A.; Ashfaq, H.; Yusuf, S. Antimicrobial resistance, biofilm formation, and virulence genes in Enterococcus species from small backyard chicken flocks. Antibiotics 2022, 11, 380. [Google Scholar] [CrossRef]
- Beukers, A. G.; Zaheer, R.; Goji, N.; Amoako, K. K.; Chaves, A. V.; Ward, M. P.; McAllister, T. A. Comparative genomics of Enterococcus spp. isolated from bovine feces. BMC microbiology 2017, 17, 1–18. [Google Scholar] [CrossRef]
- Ribeiro, J.; Silva, V.; Monteiro, A.; Vieira-Pinto, M.; Igrejas, G.; Reis, F. S.; Barros, L.; Poeta, P. Antibiotic Resistance among Gastrointestinal Bacteria in Broilers: A Review Focused on Enterococcus spp. and Escherichia coli. Animals 2023, 13, 1362. [Google Scholar] [CrossRef]
- Klibi, N.; Aouini, R.; Borgo, F.; Ben Said, L.; Ferrario, C.; Dziri, R.; Boudabous, A.; Torres, C.; Ben Slama, K. Antibiotic resistance and virulence of faecal enterococci isolated from food-producing animals in Tunisia. Annals of Microbiology 2015, 65, 695–702. [Google Scholar] [CrossRef]
- Zaheer, R.; Cook, S. R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R. O.; Thymensen, L.; Stamm, C.; Song, J.; Hannon, S.; Jones, T.; Church, D.; Booker, C. W.; Amoako, K.; Domselaar, G.; Read, R. R.; McAllister, T. A. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020, 10, 1–16. [Google Scholar]
- Dafale, N.A.; Srivastava, S.; Purohit, H.J. Zoonosis: an emerging link to antibiotic resistance under “one health approach”. Indian journal of microbiology 2020, 60, 139–152. [Google Scholar] [CrossRef]
- Bortolaia, V.; Guardabassi, L. Zoonotic Transmission of Antimicrobial-Resistant Enterococci: A Threat to Public Health or an Overemphasized Risk? In Zoonoses: Infections Affecting Humans and Animals, Sing, A. Ed.; Cham: Springer International Publishing, Germany, 2023; pp. 1–33. [Google Scholar]
- Wambui, J.; Tasara, T.; Njage, P. M. K.; Stephan, R. Species distribution and antimicrobial profiles of Enterococcus spp. isolates from Kenyan small and medium enterprise slaughterhouses. Journal of food protection 2018, 81, 1445–1449. [Google Scholar] [CrossRef]
- European Food Safety Authority Panel on Biological Hazards-BIOHAZ. Scientific Opinion on the public health hazards to be covered by inspection of meat (bovine animals). EFSA Journal 2013, 11, 3266. [Google Scholar] [CrossRef]
- European Food Safety Authority Panel on Biological Hazards-BIOHAZ. Scientific Opinion on the public health hazards to be covered by inspection of meat from sheep and goats. EFSA Journal 2013, 11, 3265. [Google Scholar] [CrossRef]
- Wardhana, D. K. Risk factors for bacterial contamination of bovine meat during slaughter in ten Indonesian abattoirs. Veterinary medicine international 2019. [Google Scholar]
- Microbiology of the food chain − Carcass sampling for microbiological analysis, ISO 17604; International Organization for Standardization: Geneva, Switzerland, 2015.
- Cebeci, T. Listeria monocytogenes in Ruminants at an Abattoir: Prevalence, Virulence Characteristics, Serotypes and Antibiotic Resistance in Eastern Türkiye. Israel Journal of Veterinary Medicine 2022, 77, 4. [Google Scholar]
- Pesavento, G.; Calonico, C.; Ducci, B.; Magnanini, A.; Nostro, A. L. Prevalence and antibiotic resistance of Enterococcus spp. isolated from retail cheese, ready-to-eat salads, ham, and raw meat. Food microbiology 2014, 41, 1–7. [Google Scholar] [CrossRef]
- Quintela-Baluja, M.; Böhme, K.; Fernández-No, I. C.; Morandi, S.; Alnakip, M. E.; Caamaño-Antelo, S.; Barros-Velázquez, J.; Calo-Mata, P. Characterization of different food-isolated Enterococcus strains by MALDI-TOF mass fingerprinting. Electrophoresis 2013, 34, 2240–2250. [Google Scholar] [CrossRef]
- Chotinantakul, K.; Chansiw, N.; Okada, S. Antimicrobial resistance of Enterococcus spp. isolated from Thai fermented pork in Chiang Rai Province, Thailand. Journal of Global Antimicrobial Resistance 2018, 12, 143–148. [Google Scholar] [CrossRef]
- Billström, H.; Lund, B.; Sullivan, Å.; Nord, C. E. Virulence and antimicrobial resistance in clinical Enterococcus faecium. International journal of antimicrobial agents 2008, 32, 374–377. [Google Scholar] [CrossRef]
- Performance standards for antimicrobial susceptibility testing, CLSI supplement M100, 33th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2023.
- The European Committee on Antimicrobial Susceptibility Testing, EUCAST supplement 2023, version 13; Breakpoint tables for interpretation of MICs and zone diameters. The European Committee: Växjö, Sweden, 2023.
- Kim, Y. B.; Seo, K. W.; Son, S. H.; Noh, E. B.; Lee, Y. J. Genetic characterization of high-level aminoglycoside-resistant Enterococcus faecalis and Enterococcus faecium isolated from retail chicken meat. Poultry Science 2019, 98, 5981–5988. [Google Scholar] [CrossRef]
- Ben Braiek, O.; Smaoui, S. Enterococci: between emerging pathogens and potential probiotics. BioMed research international 2019, 2019, 5938210. [Google Scholar] [CrossRef]
- Efstratiou, A.; Lamagni, T.; Turner, C. E. Streptococci and Enterococci. In Infectious Diseases, 4th ed.; Cohen, J., Powderly, W. G., Opal, S. M., Eds.; Elsevier: China, 2017; Volume 2, pp. 1523–1536. [Google Scholar]
- Holman, D. B.; Gzyl, K. E.; Zaheer, R.; Jones, T. H.; McAllister, T. A. Draft genome sequences of 43 Enterococcus faecalis and Enterococcus faecium isolates from a commercial beef processing plant and retail ground beef. Microbiology Resource Announcements 2019, 8, 10–1128. [Google Scholar] [CrossRef]
- Holman, D. B.; Klima, C. L.; Gzyl, K. E.; Zaheer, R.; Service, C.; Jones, T. H.; McAllister, T. A. A Longitudinal Study of Antimicrobial Resistance in Enterococcus spp. Isolated from a Beef Processing Plant and Retail Ground Beef. bioRxiv 2021, 2021-05. [Google Scholar]
- Telli, N.; Telli, A. E.; Biçer, Y.; Turkal, G.; Uçar, G. Isolation and antimicrobial resistance of vancomycin resistant Enterococcus spp.(VRE) and methicillin-resistant S. aureus (MRSA) on beef and chicken meat, and workers hands from slaughterhouses and retail shops in Turkey. Journal of the Hellenic Veterinary Medical Society 2021, 72, 3345–3354. [Google Scholar] [CrossRef]
- Messele, Y. E.; Hasoon, M. F.; Trott, D. J.; Veltman, T.; McMeniman, J. P.; Kidd, S. P.; Low, W. Y.; Petrovski, K. R. Longitudinal Analysis of Antimicrobial Resistance among Enterococcus Species Isolated from Australian Beef Cattle Faeces at Feedlot Entry and Exit. Animals 2022, 12, 2690. [Google Scholar] [CrossRef]
- Guzman Prieto, A. M.; van Schaik, W.; Rogers, M. R.; Coque, T. M.; Baquero, F.; Corander, J.; Willems, R. J. Global emergence and dissemination of enterococci as nosocomial pathogens: attack of the clones? Frontiers in microbiology 2016, 7, 788. [Google Scholar] [CrossRef]
- Igbinosa, E. O.; Beshiru, A. Antimicrobial resistance, virulence determinants, and biofilm formation of Enterococcus species from ready-to-eat seafood. Frontiers in Microbiology 2019, 10, 728. [Google Scholar] [CrossRef]
- Golob, M.; Pate, M.; Kušar, D.; Dermota, U.; Avberšek, J.; Papić, B.; Zdovc, I. Antimicrobial resistance and virulence genes in Enterococcus faecium and Enterococcus faecalis from humans and retail red meat. BioMed research international 2019.
- Fiore, E.; Van Tyne, D.; Gilmore, M. S. Pathogenicity of enterococci. Microbiology spectrum 2019, 7, 7-4. [Google Scholar] [CrossRef]
- Zhang, F., Jiang, M., Wan, C., Chen, X., Chen, X., Tao, X., Shah, N. P.; Wei, H. Screening probiotic strains for safety: Evaluation of virulence and antimicrobial susceptibility of enterococci from healthy Chinese infants. Journal of dairy science 2016, 99, 4282–4290. [CrossRef]
- Mohanty, S.; Behera, B. Antibiogram Pattern and Virulence Trait Characterization of Enterococcus Species Clinical Isolates in Eastern India: A Recent Analysis. Journal of Laboratory Physicians 2022, 14, 237–246. [Google Scholar] [CrossRef]
- Ahmed, M. O.; Baptiste, K. E. Vancomycin-resistant enterococci: a review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microbial Drug Resistance 2018, 24, 590–606. [Google Scholar] [CrossRef]
- Papich, M. G. Oxytetracycline. In Saunders Handbook of Veterinary Drug Small and Large Animal, 4th ed.; Papich, M. G., Ed.; Sounders: USA, 2016; pp. 595–598. [Google Scholar]
- Ayeni, F. A.; Odumosu, B. T.; Oluseyi, A. E.; Ruppitsch, W. Identification and prevalence of tetracycline resistance in enterococci isolated from poultry in Ilishan, Ogun State, Nigeria. Journal of pharmacy & bioallied sciences 2016, 8, 69. [Google Scholar]
- Smith, M. V. Therapeutics. In Textbook of Rabbit Medicine, 3rd ed.; Smith, M. V., Ed.; Elsevier: Poland, 2023; pp. 100–137. [Google Scholar]
- Ngbede, E. O.; Raji, M. A.; Kwanashie, C. N.; Kwaga, J. K. P. Antimicrobial resistance and virulence profile of enterococci isolated from poultry and cattle sources in Nigeria. Tropical animal health and production 2017, 49, 451–458. [Google Scholar] [CrossRef]
- Barlow, R. S.; McMillan, K. E.; Duffy, L. L.; Fegan, N.; Jordan, D.; Mellor, G. E. Antimicrobial resistance status of Enterococcus from Australian cattle populations at slaughter. PLoS One 2017, 12, e0177728. [Google Scholar] [CrossRef]
- Xuan, H.; Yao, X.; Pan, R.; Gao, Y.; Wei, J.; Shao, D.; Liu, K.; Li, Z.; Qiu, Y.; Ma, Z.; Li, B.; Xia, L. Antimicrobial resistance in Enterococcus faecium and Enterococcus faecalis isolates of swine origin from eighteen provinces in China. Journal of Veterinary Medical Science 2021, 83, 1952–1958. [Google Scholar] [CrossRef]
- Yu, L.; Liu, Y.; Liu, M.; Li, Z.; Li, L.; Wang, F. Research Note: Molecular characterization of antimicrobial resistance and virulence gene analysis of Enterococcus faecalis in poultry in Tai'an, China. Poultry Science 2022, 101, 101763. [Google Scholar] [CrossRef]
- Hollenbeck, B. L.; Rice, L. B. Intrinsic and acquired resistance mechanisms in Enterococcus. Virulence 2012, 3, 421–569. [Google Scholar] [CrossRef]
- Jian, Z., Zeng, L., Xu, T., Sun, S., Yan, S., Yang, L., Huang, Y.; Jia, J.; Dou, T. Antibiotic resistance genes in bacteria: Occurrence, spread, and control. Journal of basic microbiology 2021, 61, 1049–1070. [CrossRef]
- Li, W.; Li, J.; Wei, Q.; Hu, Q.; Lin, X.; Chen, M.; Ye, R.; Lv, H. Characterization of aminoglycoside resistance and virulence genes among Enterococcus spp. isolated from a hospital in China. International journal of environmental research and public health 2015, 12, 3014–3025. [Google Scholar] [CrossRef]
- Kristich, C. J.; Rice, L. B.; Arias, C. A. Enterococcal infection—treatment and antibiotic resistance. In Enterococci: from commensals to leading causes of drug resistant infection [Internet], Gilmore, M. S., Ed.; Massachusetts Eye and Ear Infirmary: Boston, USA, 2014; pp. 1–47. [Google Scholar]
- Channaiah, L. H.; Subramanyam, B.; Zurek, L. Molecular characterization of antibiotic resistant and potentially virulent enterococci isolated from swine farms and feed mills. Journal of Stored Products Research 2018, 77, 189–196. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Blanco, P.; Alcalde-Rico, M.; Corona, F.; Reales-Calderón, J. A.; Sánchez, M. B.; Martínez, J. L. Multidrug efflux pumps as main players in intrinsic and acquired resistance to antimicrobials. Drug Resistance Updates 2016, 28, 13–27. [Google Scholar] [CrossRef]
- Sanchez Valenzuela, A.; Lavilla Lerma, L.; Benomar, N.; Gálvez, A.; Perez Pulido, R.; Abriouel, H. Phenotypic and molecular antibiotic resistance profile of Enterococcus faecalis and Enterococcus faecium isolated from different traditional fermented foods. Foodborne pathogens and disease 2013, 10, 143–149. [Google Scholar] [CrossRef]
- Rehman, M.A., Yin, X., Zaheer; R., Goji; N., Amoako; K.K., McAllister, T., Pritchard, J.; Diarra, M.S. Genotypes and phenotypes of Enterococci isolated from broiler chickens. Frontiers in Sustainable Food Systems 2018, 2, 83. [CrossRef]
| Animal species |
Carcass surface point |
Sample number |
E. faecalis |
E. hirae |
E. faecium |
E. casseliflavus |
E. gallinarum |
Total |
|---|---|---|---|---|---|---|---|---|
| n(%) | n(%) | n(%) | n(%) | n(%) | n(%) | |||
| Sheep | Brisket | 80 | 0(0) | 4(11.7) | 0(0) | 7(23.3) | 0(0) | 11(12.2) |
| Flank | 80 | 1(9.09) | 5(14.7) | 2(14.2) | 1(3.3) | 0(0) | 9(10) | |
| Hind leg | 80 | 2(18.1) | 4(11.7) | 6(42.8) | 4(13.3) | 0(0) | 16(17.7) | |
| Rectal | 80 | 4(36.3) | 8(23.5) | 2(14.2) | 4(13.3) | 0(0) | 18(20) | |
| Goat | Brisket | 50 | 1(9.09) | 1(2.9) | 0(0) | 3(10) | 0(0) | 5(5.5) |
| Flank | 50 | 0(0) | 1(2.9) | 1(7.1) | 2(6.6) | 0(0) | 4(4.4) | |
| Hind leg | 50 | 0(0) | 6(17.6) | 0(0) | 4(13.3) | 0(0) | 10(11.1) | |
| Rectal | 50 | 1(9.09) | 5(14.7) | 3(21.4) | 5(16.6) | 0(0) | 14(15.5) | |
| Cattle | Brisket | 20 | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) |
| Flank | 20 | 2(18.1) | 0(0) | 0(0) | 0(0) | 0(0) | 2(2.2) | |
| Hind leg | 20 | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | |
| Rectal | 20 | 0(0) | 0(0) | 0(0) | 0(0) | 1(100) | 1(1.1) | |
| Total | 600 | 11(12.2) | 34(37.7) | 14(15.5) | 30(33.3) | 1(1.1) | 90 |
| Virulence genotypes | Number (%) of Enterococcus virulence factor genotypes | |||||
|---|---|---|---|---|---|---|
|
E. faecalis n=11 |
E. hirae n=34 |
E. faecium n=14 |
E.casseliflavus n=30 |
E.gallinarum n=1 |
Total (n = 90) |
|
| n(%) | n(%) | n(%) | n(%) | n(%) | n(%) | |
| ace | 1(9.09) | 0(0) | 0(0) | 1(3.3) | 0(0) | 2(2.2) |
| gelE | 1(9.09) | 2(18.8) | 0(0) | 0(0) | 0(0) | 3(3.3) |
| efaA | 6(54.5) | 2(18.8) | 3(21.4) | 0(0) | 0(0) | 11(12.2) |
| esp | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) |
| asaI | 2(18.8) | 1(2.9) | 0(0) | 0(0) | 0(0) | 3(3.3) |
| cylA | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) |
| hyl | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) |
| Antibiotic resistance genes | Enterococcus Species | |||||
|---|---|---|---|---|---|---|
|
E. faecalis n=11 |
E. hirae n=34 |
E. faecium n=14 |
E.casseliflavus n=30 |
E. gallinarum n=1 |
Total (n = 90) |
|
| n(%) | n(%) | n(%) | n(%) | n(%) | n(%) | |
| ermA | * | 1(1.1) | * | * | * | 1(1.1) |
| ermB | * | 2(2.2) | * | * | * | 2(2.2) |
| mef | * | - | * | * | * | - |
| tet(L) | - | - | - | - | - | - |
| tet(M) | 5(5.5) | - | - | 1(1.1) | - | 6(6.6) |
| tet(O) | - | - | - | - | - | - |
| cfr | - | - | - | - | - | - |
| fexA | - | - | - | - | - | - |
| optrA | - | - | - | - | - | - |
| aac(6’)Ie-aph(2")-la | 1(1.1) | - | - | - | - | 1(1.1) |
| aph(2")-Ib | - | - | - | - | - | - |
| aph(2")-Ic | - | - | - | - | - | - |
| aph(2")-Id | - | - | - | - | - | - |
| ant(3")-Ia | - | - | - | - | - | - |
| aph(6)-Ia | - | - | - | - | - | - |
| vanA | - | - | - | * | * | - |
| vanB | - | - | - | * | * | - |
| vanC1 | - | - | 1(1.1) | * | * | 1(1.1) |
| vanC2 | - | 1(1.1) | - | * | * | 1(1.1) |
| efr(A) | 5(5.5) | 2(2.2) | 1(1.1) | 1(1.1) | - | 9(10) |
| lsa | 9(10) | 2(2.2) | 1(1.1) | 1(1.1) | - | 13(14.4) |
| eme(A) | 6(6.6) | 1(1.1) | - | 2(2.2) | - | 9(10) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




