Submitted:
15 January 2024
Posted:
16 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Patient’s recruitment
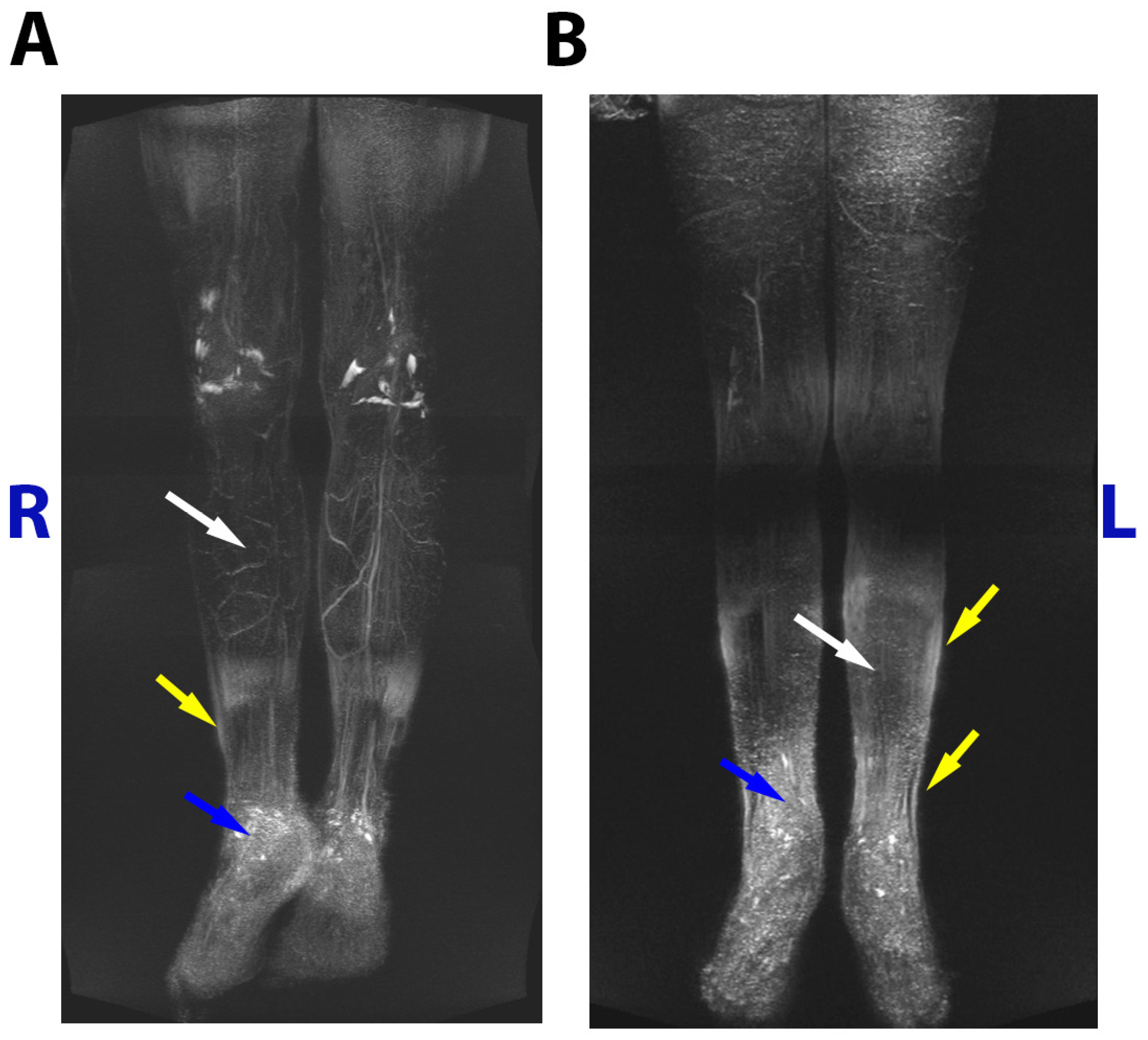
MRI
Exome sequencing and analysis
Sanger sequencing
3. Results
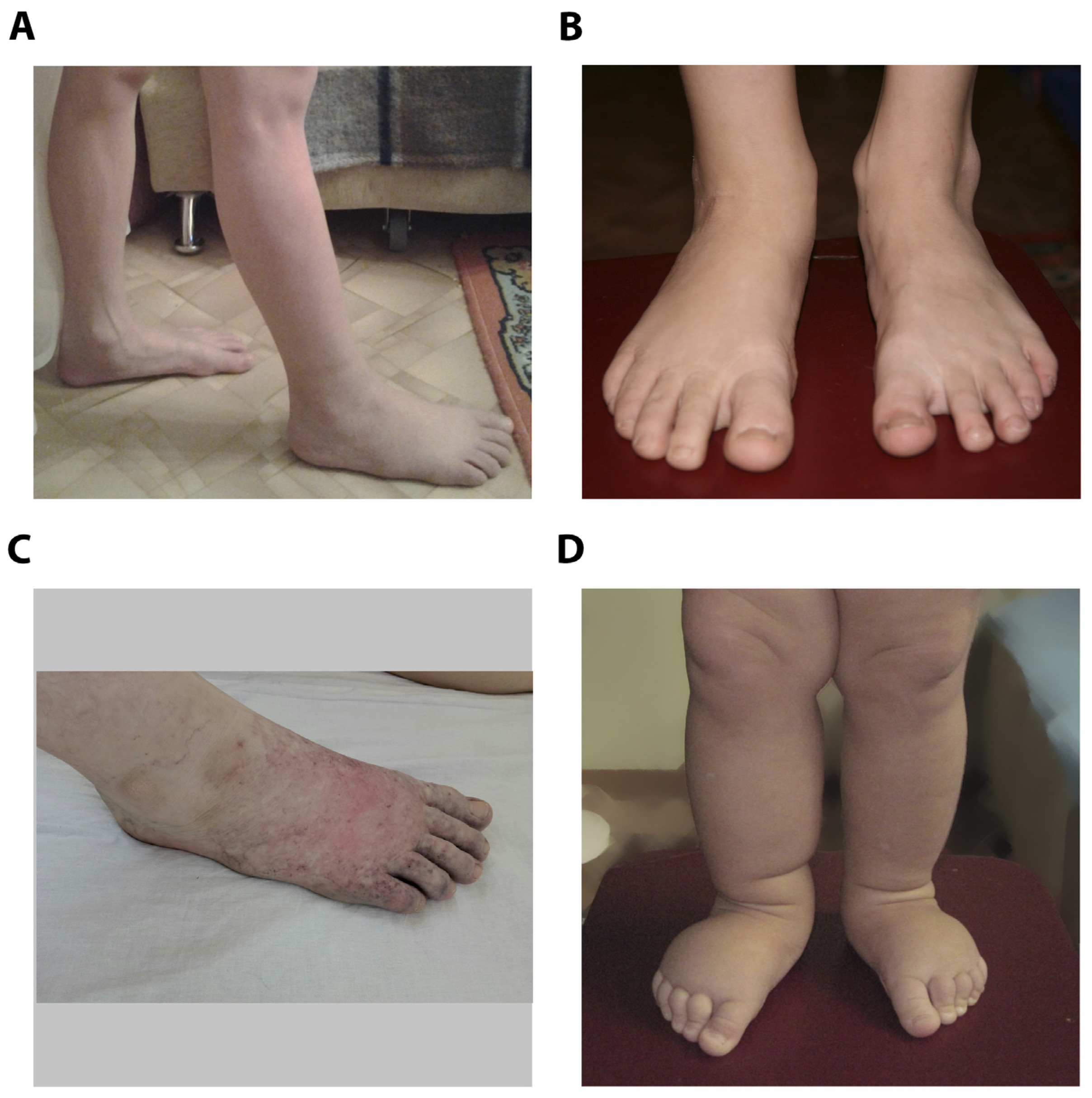
3.1. Family case
3.2. Family case
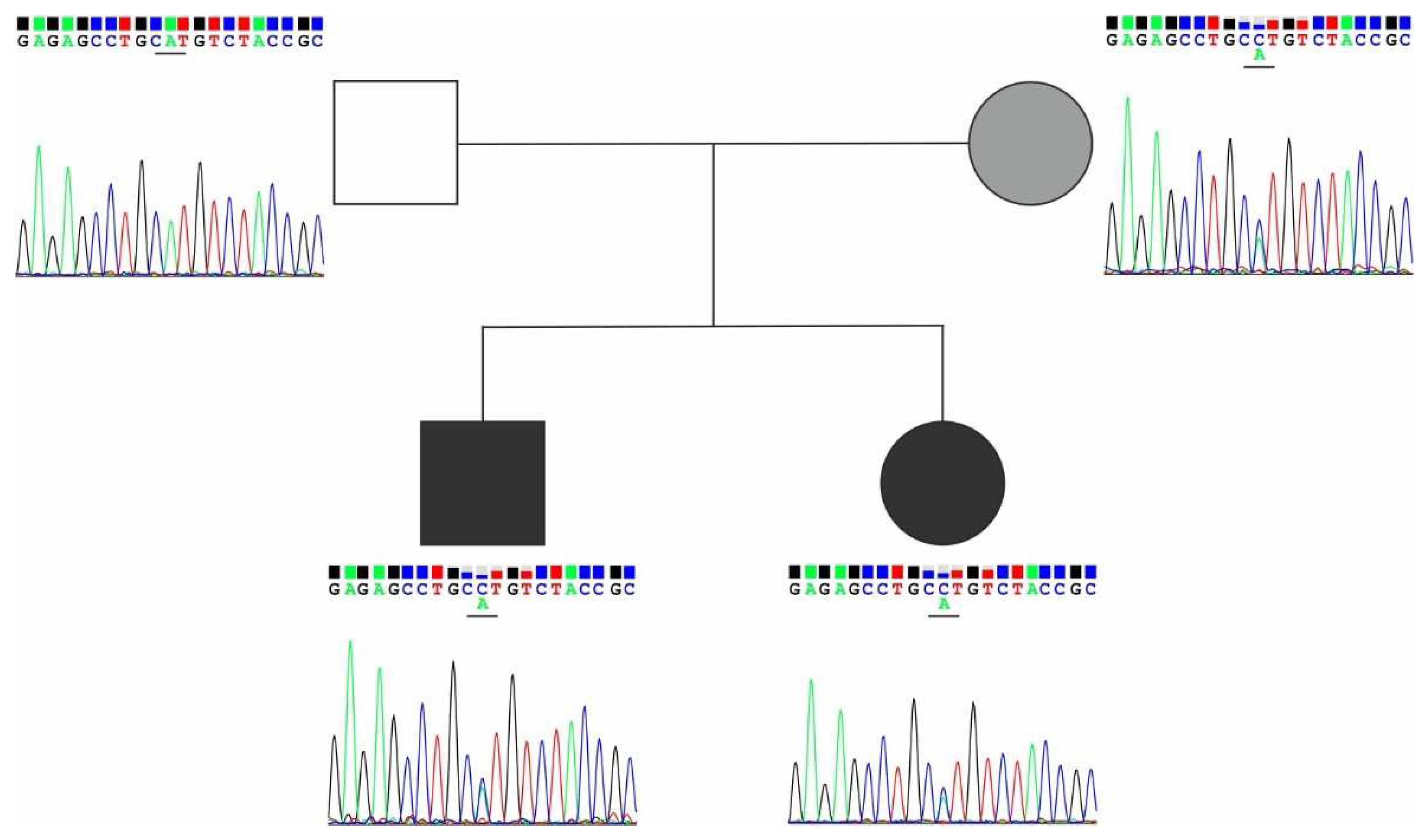
3.3. Exome analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rockson, S. G., and K. K. Rivera. Estimating the Population Burden of Lymphedema. Ann N Y Acad Sci 1131 (2008): 147-54. [CrossRef]
- Greene, Arin K. Epidemiology and Morbidity of Lymphedema. In Lymphedema: Presentation, Diagnosis, and Treatment, edited by Arin K. Greene, Sumner A. Slavin and Håkan Brorson, 33-44. Cham: Springer International Publishing, 2015. [CrossRef]
- Brouillard, P., L. Boon, and M. Vikkula. Genetics of Lymphatic Anomalies. J Clin Invest 124, no. 3 (2014): 898-904. [CrossRef]
- Mendola, A., M. J. Schlögel, A. Ghalamkarpour, A. Irrthum, H. L. Nguyen, E. Fastré, A. Bygum, C. van der Vleuten, C. Fagerberg, E. Baselga, I. Quere, J. B. Mulliken, L. M. Boon, P. Brouillard, and M. Vikkula. Mutations in the Vegfr3 Signaling Pathway Explain 36% of Familial Lymphedema. Mol Syndromol 4, no. 6 (2013): 257-66. [CrossRef]
- Bonetti, G., S. Paolacci, M. Samaja, P. E. Maltese, S. Michelini, S. Michelini, S. Michelini, M. Ricci, M. Cestari, A. Dautaj, M. C. Medori, and M. Bertelli. Low Efficacy of Genetic Tests for the Diagnosis of Primary Lymphedema Prompts Novel Insights into the Underlying Molecular Pathways. Int J Mol Sci 23, no. 13 (2022). [CrossRef]
- Li, H., and R. Durbin. Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform. Bioinformatics 26, no. 5 (2010): 589-95. [CrossRef]
- McKenna, A., M. Hanna, E. Banks, A. Sivachenko, K. Cibulskis, A. Kernytsky, K. Garimella, D. Altshuler, S. Gabriel, M. Daly, and M. A. DePristo. The Genome Analysis Toolkit: A Mapreduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res 20, no. 9 (2010): 1297-303. [CrossRef]
- Van der Auwera, G. A., M. O. Carneiro, C. Hartl, R. Poplin, G. Del Angel, A. Levy-Moonshine, T. Jordan, K. Shakir, D. Roazen, J. Thibault, E. Banks, K. V. Garimella, D. Altshuler, S. Gabriel, and M. A. DePristo. From Fastq Data to High Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr Protoc Bioinformatics 43, no. 1110 (2013): 11 10 1-11 10 33. [CrossRef]
- Danecek, P., A. Auton, G. Abecasis, C. A. Albers, E. Banks, M. A. DePristo, R. E. Handsaker, G. Lunter, G. T. Marth, S. T. Sherry, G. McVean, R. Durbin, and Group Genomes Project Analysis. The Variant Call Format and Vcftools. Bioinformatics 27, no. 15 (2011): 2156-8. [CrossRef]
- Wang, K., M. Li, and H. Hakonarson. Annovar: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res 38, no. 16 (2010): e164. [CrossRef]
- Karczewski, K. J., L. C. Francioli, G. Tiao, B. B. Cummings, J. Alfoldi, Q. Wang, R. L. Collins, K. M. Laricchia, A. Ganna, D. P. Birnbaum, L. D. Gauthier, H. Brand, M. Solomonson, N. A. Watts, D. Rhodes, M. Singer-Berk, E. M. England, E. G. Seaby, J. A. Kosmicki, R. K. Walters, K. Tashman, Y. Farjoun, E. Banks, T. Poterba, A. Wang, C. Seed, N. Whiffin, J. X. Chong, K. E. Samocha, E. Pierce-Hoffman, Z. Zappala, A. H. O'Donnell-Luria, E. V. Minikel, B. Weisburd, M. Lek, J. S. Ware, C. Vittal, I. M. Armean, L. Bergelson, K. Cibulskis, K. M. Connolly, M. Covarrubias, S. Donnelly, S. Ferriera, S. Gabriel, J. Gentry, N. Gupta, T. Jeandet, D. Kaplan, C. Llanwarne, R. Munshi, S. Novod, N. Petrillo, D. Roazen, V. Ruano-Rubio, A. Saltzman, M. Schleicher, J. Soto, K. Tibbetts, C. Tolonen, G. Wade, M. E. Talkowski, Consortium Genome Aggregation Database, B. M. Neale, M. J. Daly, and D. G. MacArthur. The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. Nature 581, no. 7809 (2020): 434-43. [CrossRef]
- Barbitoff, Yury A., Darya N. Khmelkova, Ekaterina A. Pomerantseva, Aleksandr V. Slepchenkov, Nikita A. Zubashenko, Irina V. Mironova, Vladimir S. Kaimonov, Dmitrii E. Polev, Victoria V. Tsay, Andrey S. Glotov, Mikhail V. Aseev, Sergey G. Scherbak, Oleg S. Glotov, Arthur A. Isaev, and Alexander V. Predeus. Expanding the Russian Allele Frequency Reference Via Cross-Laboratory Data Integration: Insights from 7,452 Exome Samples. medRxiv (2022): 2021.11.02.21265801. [CrossRef]
- Richards, S., N. Aziz, S. Bale, D. Bick, S. Das, J. Gastier-Foster, W. W. Grody, M. Hegde, E. Lyon, E. Spector, K. Voelkerding, H. L. Rehm, and Acmg Laboratory Quality Assurance Committee. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17, no. 5 (2015): 405-24. [CrossRef]
- Simons, M., and M. Mlodzik. Planar Cell Polarity Signaling: From Fly Development to Human Disease. Annu Rev Genet 42 (2008): 517-40. [CrossRef]
- Hamann, J., G. Aust, D. Arac, F. B. Engel, C. Formstone, R. Fredriksson, R. A. Hall, B. L. Harty, C. Kirchhoff, B. Knapp, A. Krishnan, I. Liebscher, H. H. Lin, D. C. Martinelli, K. R. Monk, M. C. Peeters, X. Piao, S. Promel, T. Schoneberg, T. W. Schwartz, K. Singer, M. Stacey, Y. A. Ushkaryov, M. Vallon, U. Wolfrum, M. W. Wright, L. Xu, T. Langenhan, and H. B. Schioth. International Union of Basic and Clinical Pharmacology. Xciv. Adhesion G Protein-Coupled Receptors. Pharmacol Rev 67, no. 2 (2015): 338-67. [CrossRef]
- Formstone, C. J. 7tm-Cadherins: Developmental Roles and Future Challenges. Adv Exp Med Biol 706 (2010): 14-36. [CrossRef]
- Chen, W. S., D. Antic, M. Matis, C. Y. Logan, M. Povelones, G. A. Anderson, R. Nusse, and J. D. Axelrod. Asymmetric Homotypic Interactions of the Atypical Cadherin Flamingo Mediate Intercellular Polarity Signaling. Cell 133, no. 6 (2008): 1093-105. [CrossRef]
- Curtin, J. A., E. Quint, V. Tsipouri, R. M. Arkell, B. Cattanach, A. J. Copp, D. J. Henderson, N. Spurr, P. Stanier, E. M. Fisher, P. M. Nolan, K. P. Steel, S. D. Brown, I. C. Gray, and J. N. Murdoch. Mutation of Celsr1 Disrupts Planar Polarity of Inner Ear Hair Cells and Causes Severe Neural Tube Defects in the Mouse. Curr Biol 13, no. 13 (2003): 1129-33. [CrossRef]
- Formstone, C. J., C. Moxon, J. Murdoch, P. Little, and I. Mason. Basal Enrichment within Neuroepithelia Suggests Novel Function(S) for Celsr1 Protein. Mol Cell Neurosci 44, no. 3 (2010): 210-22. [CrossRef]
- Ghimire, S. R., E. M. Ratzan, and M. R. Deans. A Non-Autonomous Function of the Core Pcp Protein Vangl2 Directs Peripheral Axon Turning in the Developing Cochlea. Development 145, no. 12 (2018). [CrossRef]
- Simon, F., F. Tissir, V. Michel, G. Lahlou, M. Deans, and M. Beraneck. Implication of Vestibular Hair Cell Loss of Planar Polarity for the Canal and Otolith-Dependent Vestibulo-Ocular Reflexes in Celsr1(-/-) Mice. Front Neurosci 15 (2021): 750596. [CrossRef]
- Usami, F. M., M. Arata, D. Shi, S. Oka, Y. Higuchi, F. Tissir, M. Takeichi, and T. Fujimori. Intercellular and Intracellular Cilia Orientation Is Coordinated by Celsr1 and Camsap3 in Oviduct Multi-Ciliated Cells. J Cell Sci 134, no. 4 (2021). [CrossRef]
- Devenport, D., and E. Fuchs. Planar Polarization in Embryonic Epidermis Orchestrates Global Asymmetric Morphogenesis of Hair Follicles. Nat Cell Biol 10, no. 11 (2008): 1257-68. [CrossRef]
- Tatin, F., A. Taddei, A. Weston, E. Fuchs, D. Devenport, F. Tissir, and T. Makinen. Planar Cell Polarity Protein Celsr1 Regulates Endothelial Adherens Junctions and Directed Cell Rearrangements During Valve Morphogenesis. Dev Cell 26, no. 1 (2013): 31-44. [CrossRef]
- Krishnan, A., R. Samtani, P. Dhanantwari, E. Lee, S. Yamada, K. Shiota, M. T. Donofrio, L. Leatherbury, and C. W. Lo. A Detailed Comparison of Mouse and Human Cardiac Development. Pediatr Res 76, no. 6 (2014): 500-7. [CrossRef]
- Alitalo, K. The Lymphatic Vasculature in Disease. Nat Med 17, no. 11 (2011): 1371-80. [CrossRef]
- Qiao, X., Y. Liu, P. Li, Z. Chen, H. Li, X. Yang, R. H. Finnell, Z. Yang, T. Zhang, B. Qiao, Y. Zheng, and H. Wang. Genetic Analysis of Rare Coding Mutations of Celsr1-3 in Congenital Heart and Neural Tube Defects in Chinese People. Clin Sci (Lond) 130, no. 24 (2016): 2329-40. [CrossRef]
- Li, R., F. Fu, Q. Yu, D. Wang, X. Jing, Y. Zhang, F. Li, F. Li, J. Han, M. Pan, L. Zhen, D. Li, and C. Liao. Prenatal Exome Sequencing in Fetuses with Congenital Heart Defects. Clin Genet 98, no. 3 (2020): 215-30. [CrossRef]
- Narta, K., M. R. Teltumbade, M. Vishal, S. Sadaf, M. Faruq, H. Jama, N. Waseem, A. Rao, A. Sen, K. Ray, and A. Mukhopadhyay. Whole Exome Sequencing Reveals Novel Candidate Genes in Familial Forms of Glaucomatous Neurodegeneration. Genes (Basel) 14, no. 2 (2023). [CrossRef]
- Gonzalez-Garay, M. L., M. B. Aldrich, J. C. Rasmussen, R. Guilliod, P. E. Lapinski, P. D. King, and E. M. Sevick-Muraca. A Novel Mutation in Celsr1 Is Associated with Hereditary Lymphedema. Vasc Cell 8 (2016): 1. [CrossRef]
- Erickson, R. P., L. W. Lai, D. J. Mustacich, M. J. Bernas, P. H. Kuo, and M. H. Witte. Sex-Limited Penetrance of Lymphedema to Females with Celsr1 Haploinsufficiency: A Second Family. Clin Genet 96, no. 5 (2019): 478-82. [CrossRef]
- Maltese, P. E., S. Michelini, M. Ricci, S. Maitz, A. Fiorentino, R. Serrani, A. Lazzerotti, A. Bruson, S. Paolacci, S. Benedetti, and M. Bertelli. Increasing Evidence of Hereditary Lymphedema Caused by Celsr1 Loss-of-Function Variants. Am J Med Genet A 179, no. 9 (2019): 1718-24. [CrossRef]
- Seo, S. H., S. Lee, J. K. Park, E. J. Yang, B. Kim, J. S. Lee, M. J. Kim, S. S. Park, M. W. Seong, S. Y. Nam, C. Y. Heo, and Y. Myung. Clinical Staging and Genetic Profiling of Korean Patients with Primary Lymphedema Using Targeted Gene Sequencing. Sci Rep 12, no. 1 (2022): 13591. [CrossRef]
- Sudduth, Christopher L., Patrick J. Smits, Yu Sheng Cheng, Klaus Schmitz-Abe, Pankaj Agrawal, and Arin K. Greene. Primary Upper Extremity Lymphedema Caused by a Celsr1 Variant. Journal of Vascular Anomalies 3, no. 2 (2022): e041. [CrossRef]
- Brice, G, A H Child, A Evans, R Bell, S Mansour, K Burnand, M Sarfarazi, S Jeffery, and P Mortimer. Milroy Disease and the <Em>Vegfr-3</Em> Mutation Phenotype. Journal of Medical Genetics 42, no. 2 (2005): 98-102. [CrossRef]
- Connell, F. C., P. Ostergaard, C. Carver, G. Brice, N. Williams, S. Mansour, P. S. Mortimer, S. Jeffery, and Consortium Lymphoedema. Analysis of the Coding Regions of Vegfr3 and Vegfc in Milroy Disease and Other Primary Lymphoedemas. Hum Genet 124, no. 6 (2009): 625-31. [CrossRef]
- Mellor, R. H., C. E. Hubert, A. W. Stanton, N. Tate, V. Akhras, A. Smith, K. G. Burnand, S. Jeffery, T. Makinen, J. R. Levick, and P. S. Mortimer. Lymphatic Dysfunction, Not Aplasia, Underlies Milroy Disease. Microcirculation 17, no. 4 (2010): 281-96. [CrossRef]
- Klinner, J., M. Kruger, T. Brunet, C. Makowski, K. M. Riedhammer, A. Mollweide, M. Wagner, and J. Hoefele. Congenital Lymphedema as a Rare and First Symptom of Tuberous Sclerosis Complex. Gene 753 (2020): 144815. [CrossRef]
- Gordon, K., P. S. Mortimer, M. van Zanten, S. Jeffery, P. Ostergaard, and S. Mansour. The St George's Classification Algorithm of Primary Lymphatic Anomalies. Lymphat Res Biol 19, no. 1 (2021): 25-30. [CrossRef]
| Variant | Title 2 |
|---|---|
| Exon | 25 |
| Protein domain | Transmembrane |
| SIFT prediction | Deleterious (D) |
| Polyphen2 prediction | Probably damaging (D) |
| Mutation taster prediction | Disease causing (D) |
| 100 vertebrates conservation by PhastCons | 1.0 |
| DNA change | Protein change | Main features | Reference |
|---|---|---|---|
| c.5871G>A | p.Trp1957* | Inactivating variant of CELSR1 was identified in 5 family members with confirmed lower limb lymphedema and was absent in 6 healthy individuals. Proband had leg swelling appeared at ten years, a tortuous structure of vessels, extensive retrograde flow of lymph, and a unique "sheet flow" in both legs. |
[30] |
| c.5121dupC | p.Ile1708fs*44 | Inactivating mutation of CELSR1 in two patients with lymphatic disorders and their asymptomatic father. Lymphoscintigraphy of the lower limbs showed lymphatic abnormalities correlated with the disease burden. Sanger sequencing revealed carriers of this variant to be 5 women and 4 men, but clinical manifestations were only found in women. |
[31] |
| c.5526+2T>A | NA | 5 individuals with inactivating CELSR1 variants were identified in 95 probands with primary lymphedema. Four women with inactivating mutations had disease onset in childhood and adolescence, characterized by unilateral or bilateral lower limb edema. A man with the c.5702-1G>C mutation developed the disease at the age of 77, his mother and daughter had lymphedema. In probands’ families lymphedema was present in 5 out of 6 women with mutations (83%) and in 1 out of 4 men (25%). |
[32] |
| c.6739+1G>A | NA | ||
| c.868G>T | p.Glu290* | ||
| c.2042del | p.Asn681Metfs*16 | ||
| c.5702-1G>C | NA | ||
| c.8446C>T | p.Gln2816* | Among 60 patients with primary lymphedema, CELSR1 was found to be the most frequently mutated gene in 27 patients; c.8446C>T (p.Q2816X) was detected in a mother and daughter with lymphedema in the upper and lower limbs and classified as “likely pathogenic”; c.8871_8872del (p.C2957X) was found in the daughter, and may account for a more severe course of the disease |
[33] |
| c.8871_8872del | p.Cys2957* | ||
| c.4326_4332del | p.Thr1443Glyfs*14 | Non-syndromic lymphedema in a young male: right arm oedema appeared at 9 months and right leg oedema developed at 19 years. Patient and his mother with lower limb lymphedema, had a deletion in the CELSR1 gene resulting in a reading frame shift and premature stop codon formation. | [34] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





