Submitted:
10 January 2024
Posted:
11 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
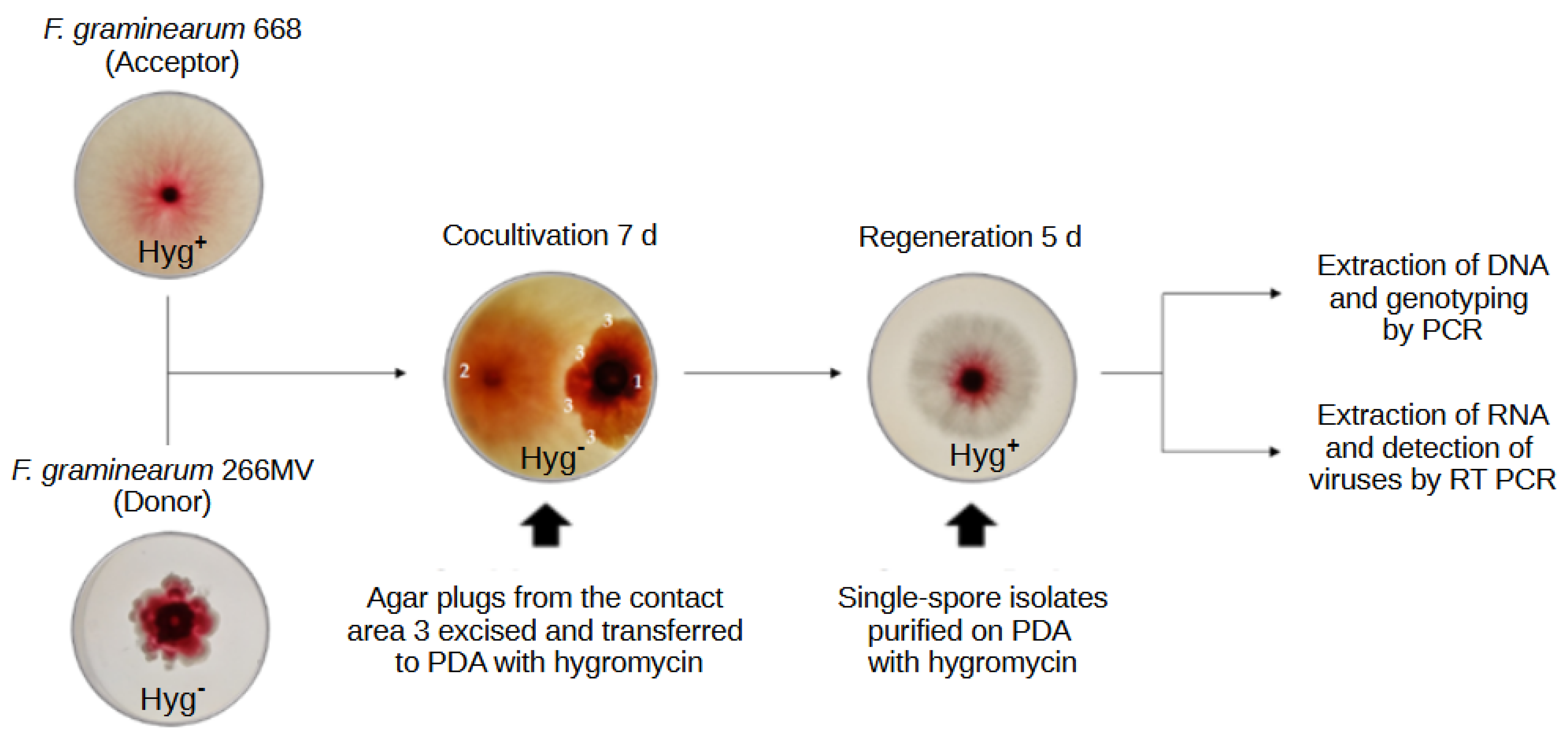
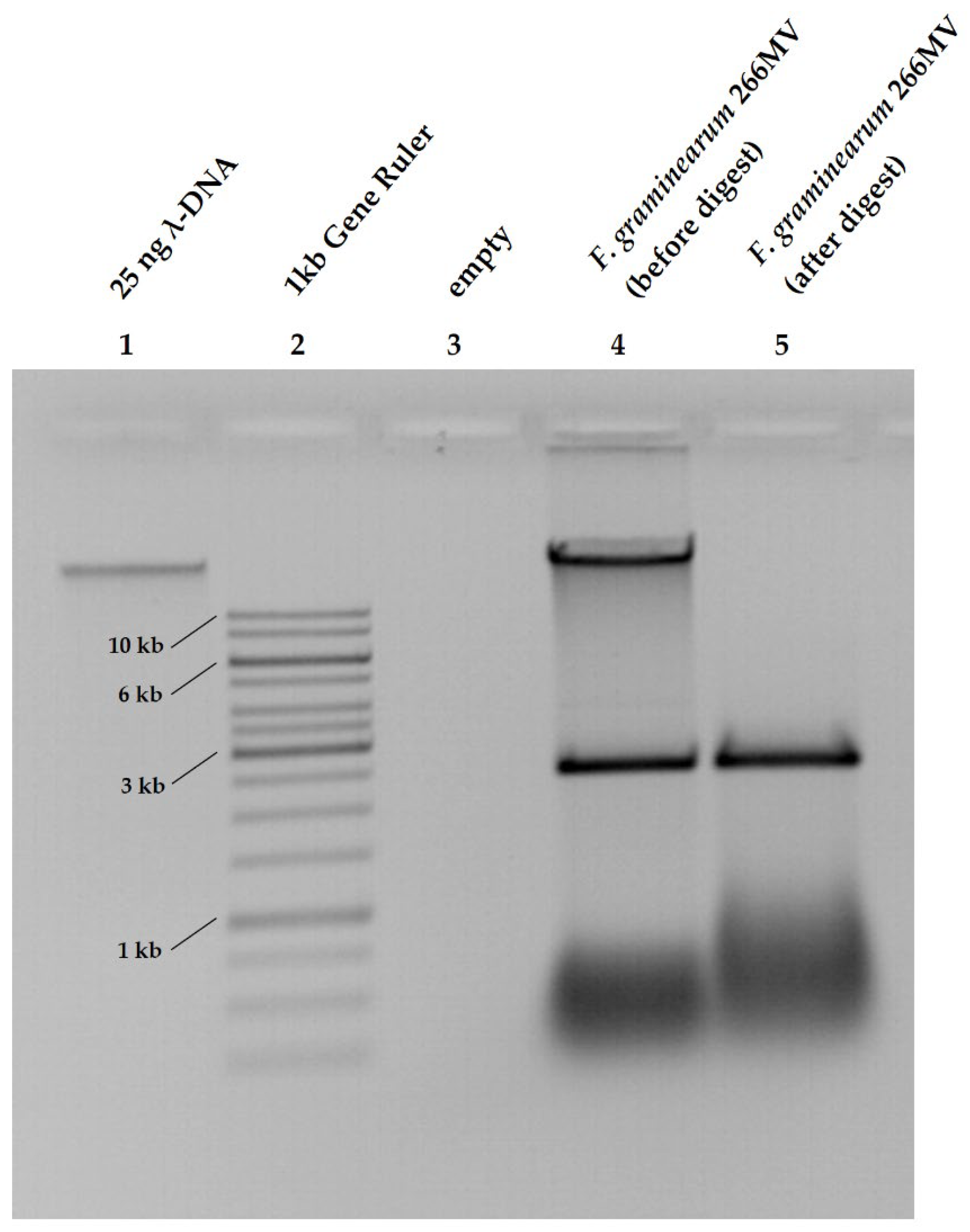
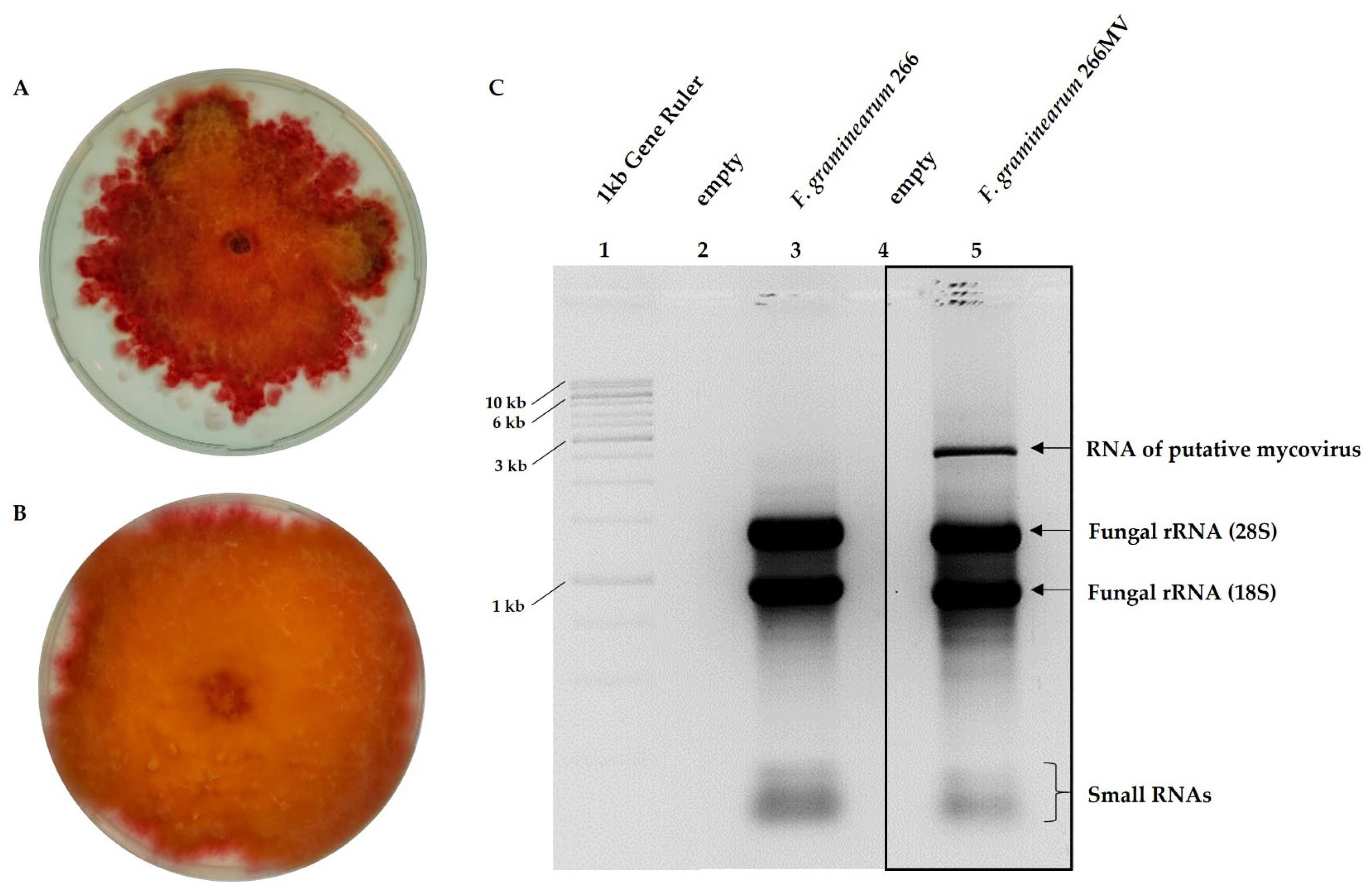
2.1. Characterization of F. graminearum 266MV carrying mycoviruses and its descendants that lost mycoviruses by mitotic segregation
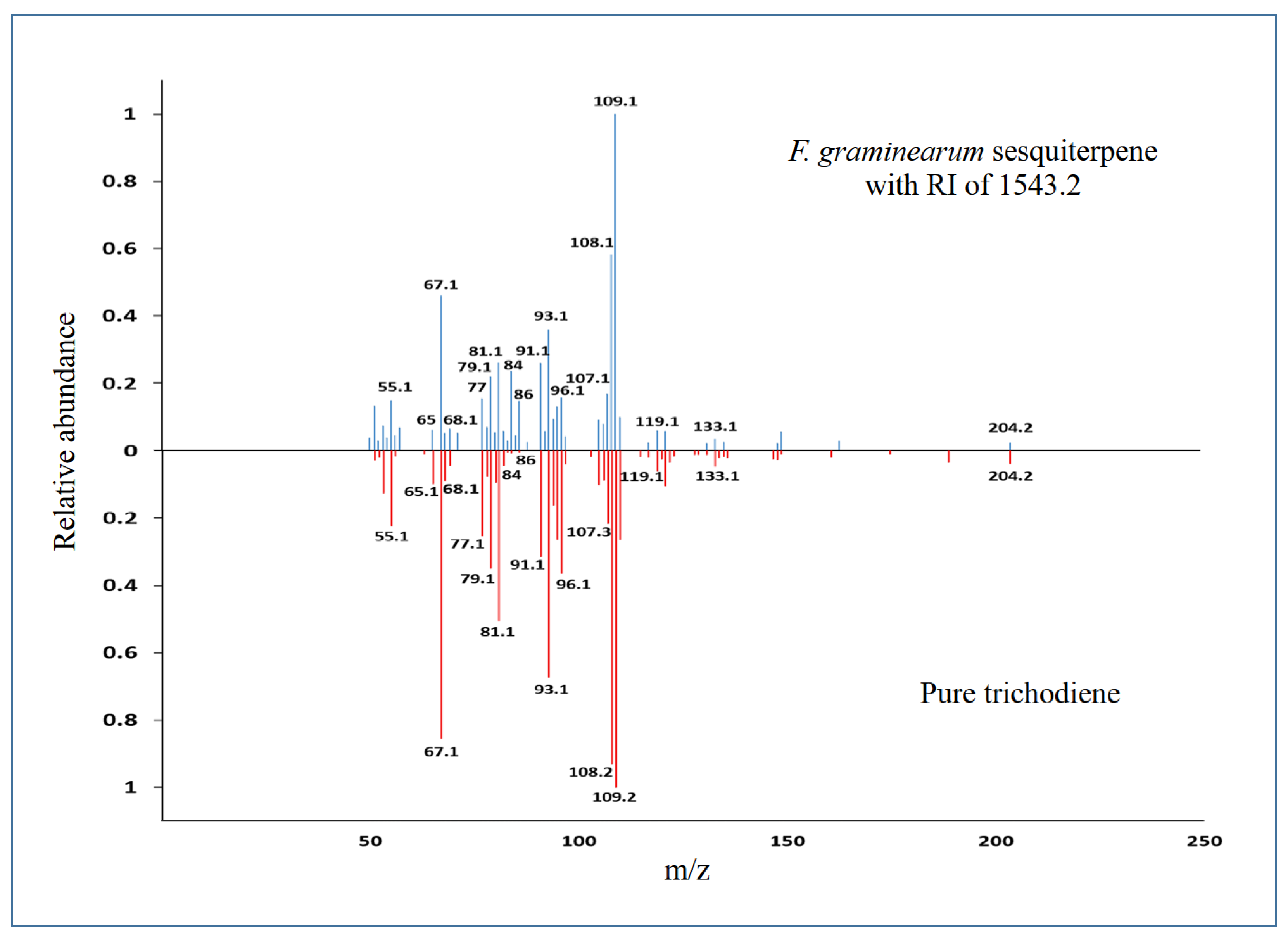
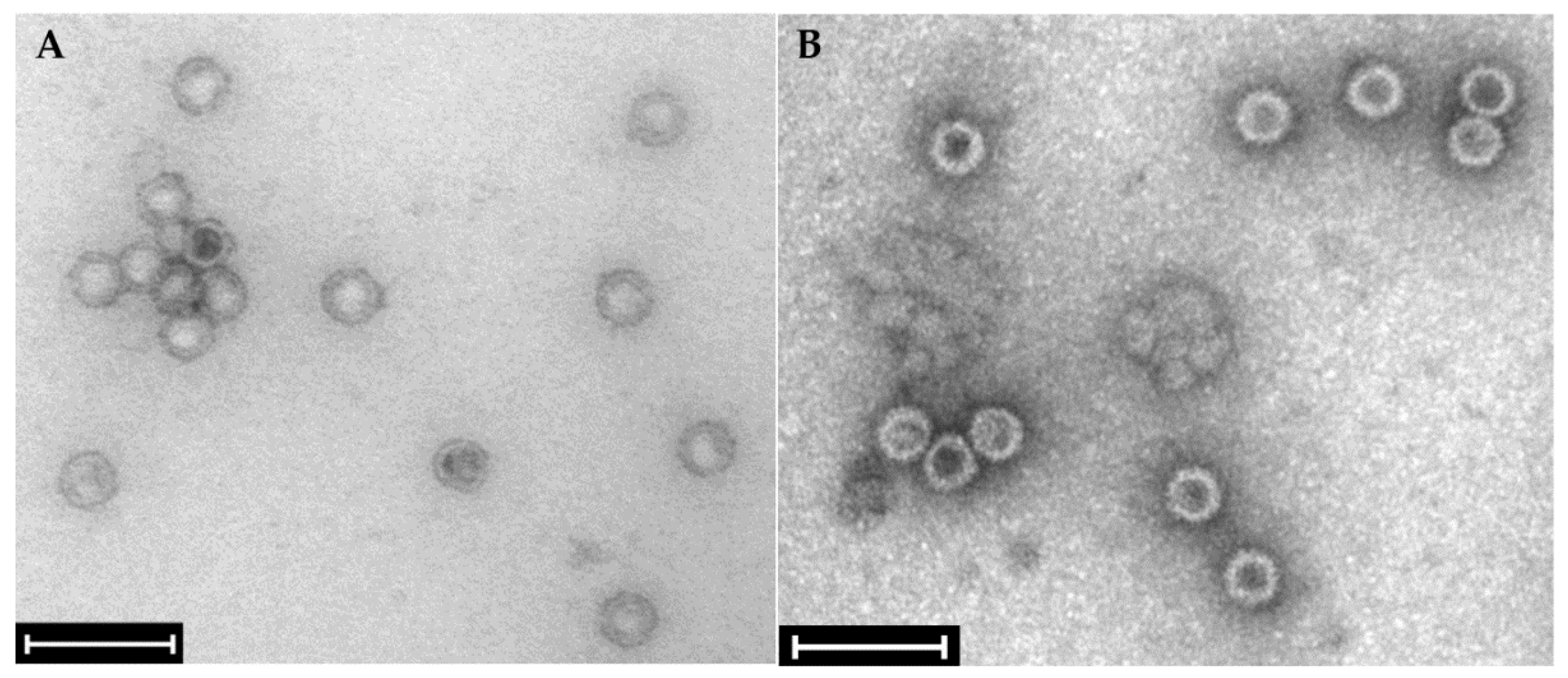
2.2. Confirmation of the presence of mycoviruses by RNA sequencing and electron microscopy
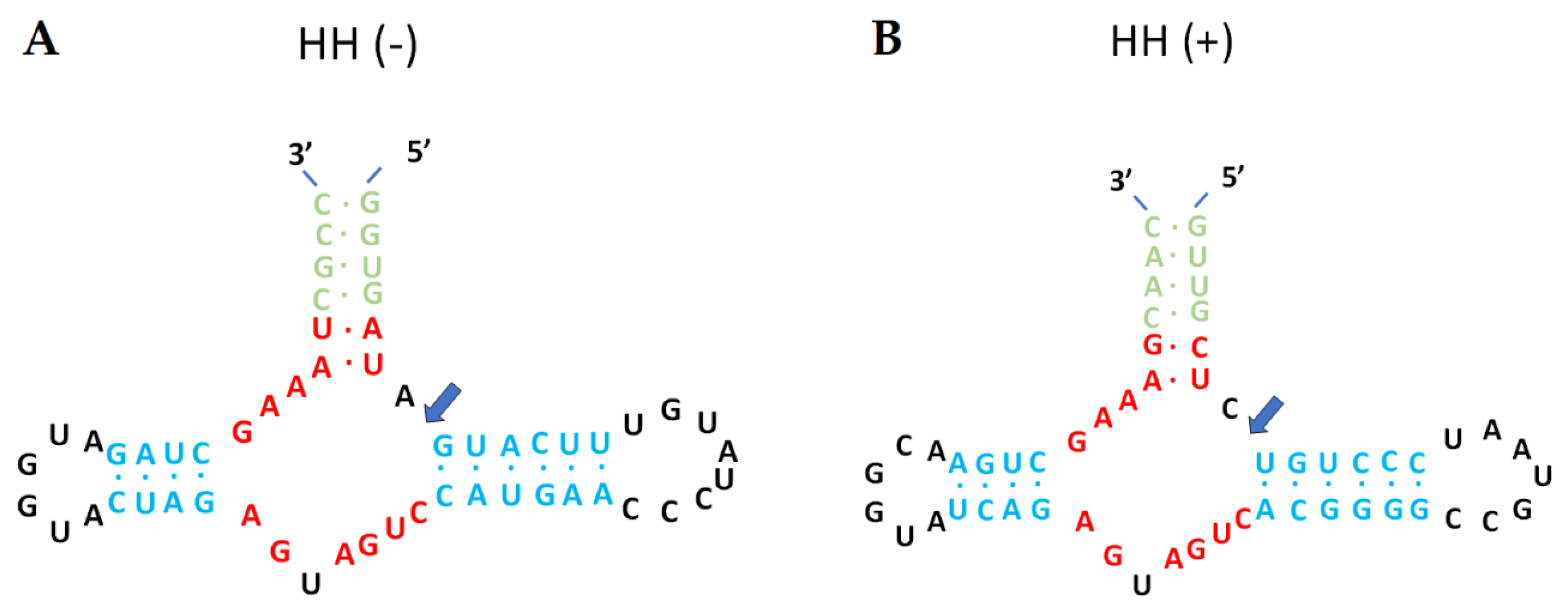
2.3. Characterization of mycoviral genomes
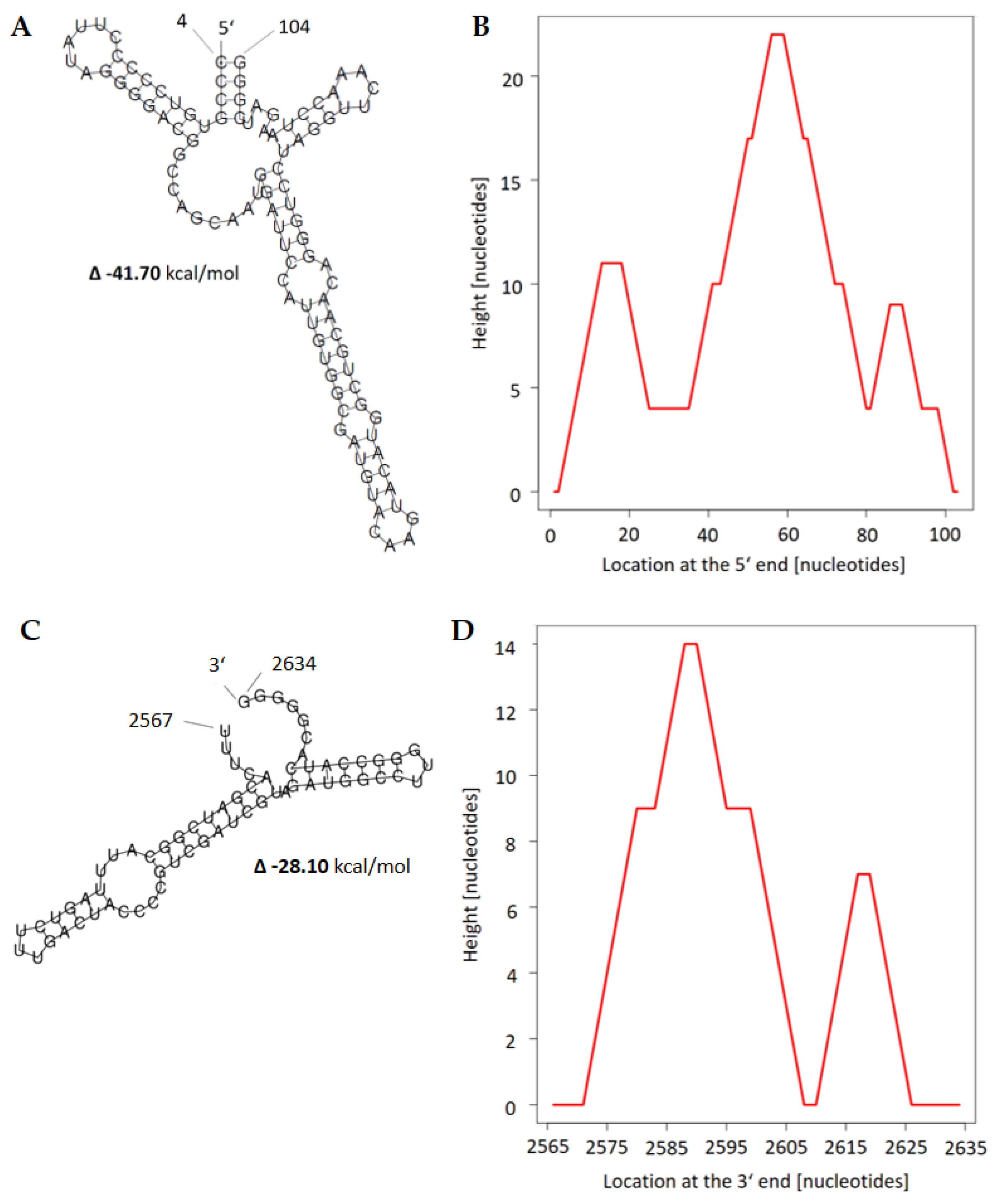
2.3.1. Fusarium graminearum ambivirus 1 (FgAV1)
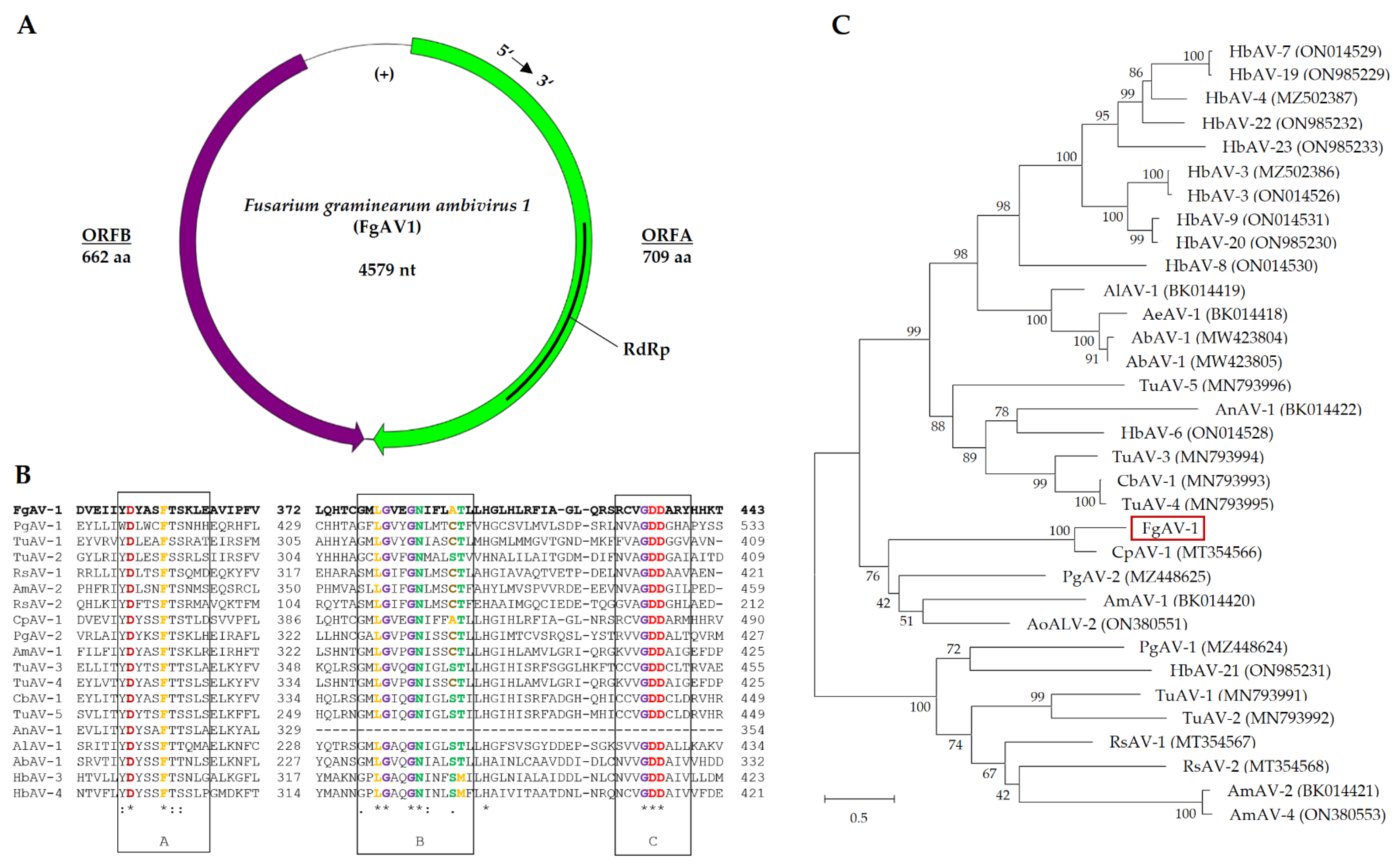
2.3.2. Fusarium graminearum negative-stranded RNA virus 2 (FgNSRV2)
2.3.3. Fusarium graminearum mitovirus 2 (FgMV2)
2.4. Transfection of mycoviruses from strain 266MV into mycovirus-free strain and the effect of mycoviruses on radial growth and sporulation
2.5. Effect of mycoviral infection on the production of secondary metabolites
2.5.1. Production of trichothecenes and aurofusarin
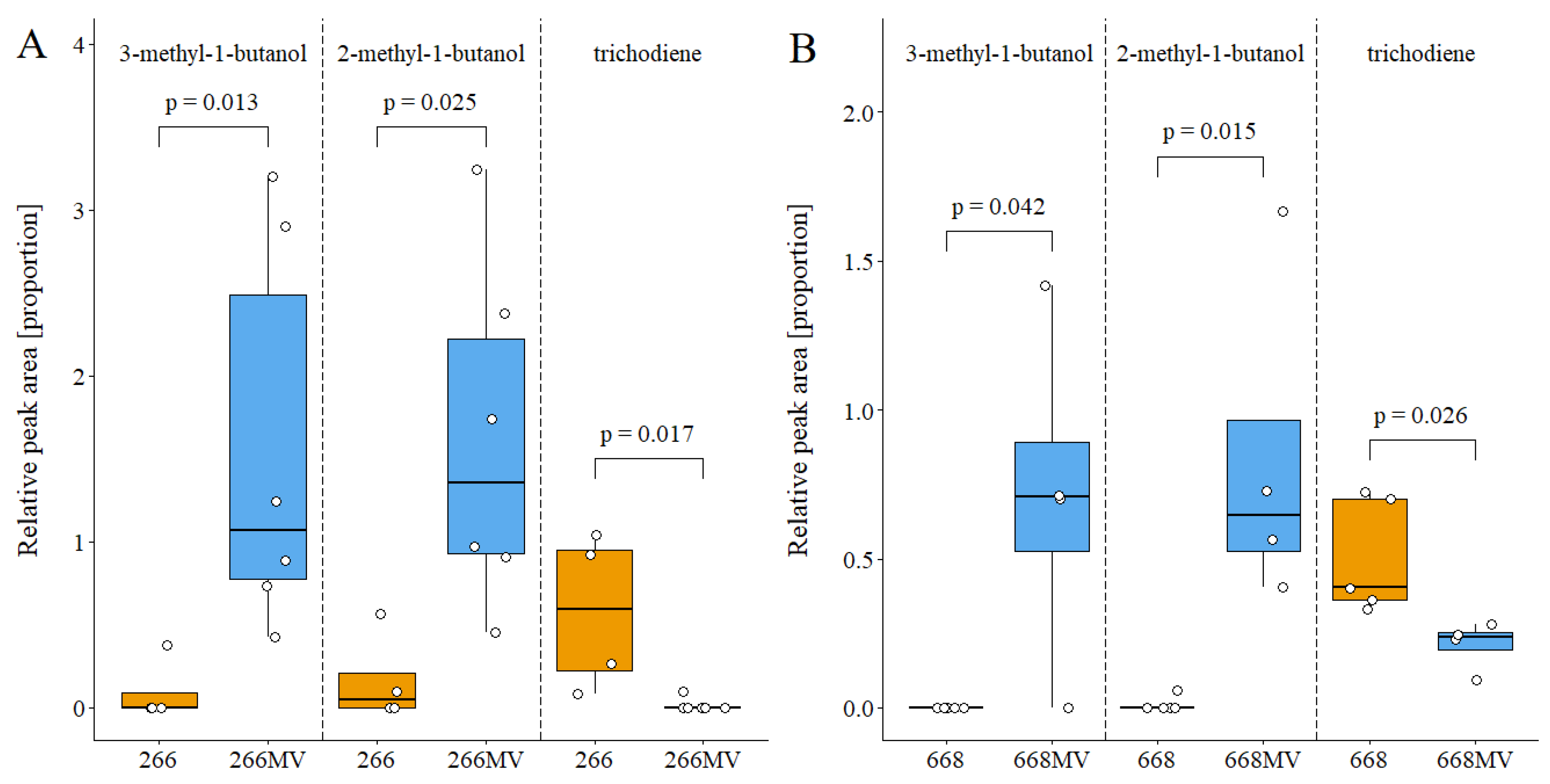
2.5.2. Effect of mycoviruses on the production of volatile compounds
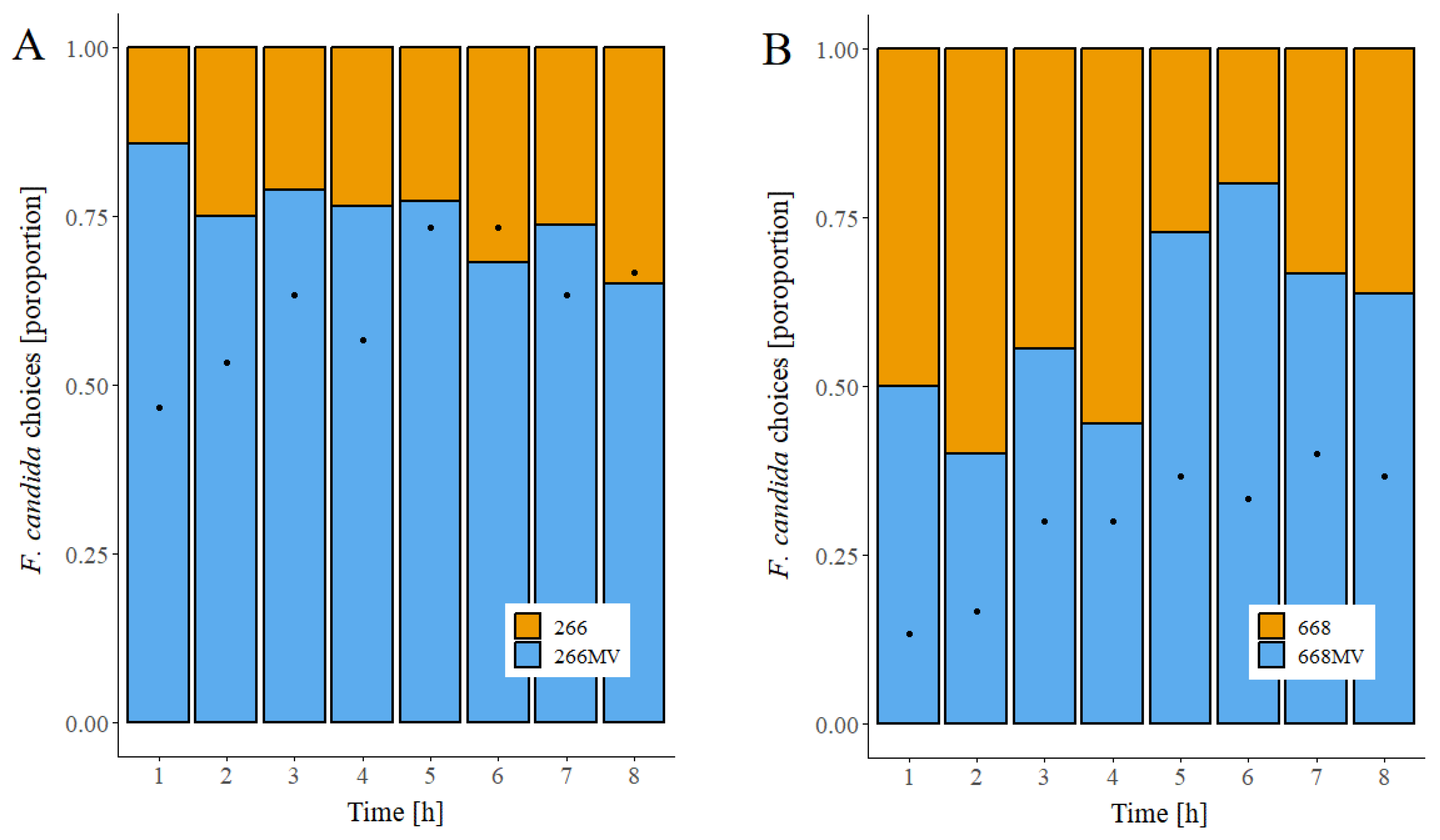
2.6. Effect of mycoviral infection on food preference in fungivorous arthropods
3. Discussion
3.1. Generation of isogenic strains differing in the presence of mycoviruses
3.2. Effect of mycoviral infection on fungal growth and sporulation
3.3. Effect of mycoviral infection on the accumulation of aurofusarin
3.4. Effect of mycoviral infection on the production of trichothecenes in F. graminearum
3.5. Effect of mycoviral infection on the emission of volatile metabolites
3.6. Effect of mycoviruses on the attractiveness of fungal mycelia for collembolans and its possible causes
4. Materials and Methods
4.1. Fungal strains and cultures
| Strain | Short label | Properties |
|---|---|---|
| F. graminearum DSM 116490* | 266MV | Natural isolate harboring mycoviruses |
| F. graminearum DSM 116489 | 266 | Descendant of 266MV that lost two mycoviruses |
| F. graminearum DSM 113709 | 668 | Hygromycin-resistant strain without mycoviruses |
| F. graminearum DSM 116488 | 668MV | Transfectant of 668 harboring two mycoviruses from 266MV |
4.2. RNA extraction, sequencing, RACE, RT-PCR, and phylogenetic analysis
4.3. DNA extraction, DNase treatment, and quantification of fungal biomass
4.4. Transmission of mycoviruses between F. graminearum strains
4.5. Determination of radial growth rate and sporulation
4.6. Quantification of trichothecenes and aurofusarin
4.7. Analysis of volatile compounds
4.8. Food preference assay
4.9. Transmission electron microscopy
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| name | sequence (5’-3’) | fragment length [bp] | target | source |
| FP-F1N | TATCGTCTCCACGAATCTCC | 786 | FpV RNA1 | [70] |
| FP-R1N | CTCGAATTGTTTCGAGACCG | |||
| FgSV1_F2 | CTCACACGTTTTTCGCAAGTT | 697 | FgSV1 | this study |
| FgSV1_R2 | GGCATCATTGGTAATCCTCCTA | |||
| Mito76/88uniF | GACARCCTATGGGAGCGTA | 670 | FgMV2 | this study |
| Mito76R20 | GCTTCAGTTTCTAGCACAGCT | |||
| FGPKS4f1 | CATGGTCCAAGTGCTACTGT | 800 | FGPKS4 | this study |
| FGPKS4r1 | CCACTAACGCTCATACGCTT | |||
| FgAV1_F1 | GCATCAAATGTAGCATAAAGATTGC | 783 | FgAV1 | this study |
| FgAV1_R1 | GCTCCTGGAGTATCTAAGGATGAG | |||
| EF1αF | ATGGGTAAGGARGACAAGAC | 690 | TEF-1α | [29] |
| EF1αR | GGARGTACCAGTRATCATGTT | |||
| Fg16NF | ACAGATGACAAGATTCAGGCACA | 280 | - | [71] |
| Fg16NR | TTCTTTGACATCTGTTCAACCCA | |||
| FpV_RNA1_cDNA_5endas | CGGATACTGTGGCGTTAGTGG | 560+X | FpV RNA1 | this study |
| FpV_RNA1_PCR_5endas | CGGGAGTAGCAGTGACGACG | 431+X | FpV RNA1 | this study |
| FpV_RNA1_cDNA_3ends | CAAACTCGTCGCACAGCTCTG | 422+X | FpV RNA1 | this study |
| FpV_RNA1_PCR_3ends | GTTTAACGATGATCGCGCCG | 284+X | FpV RNA1 | this study |
| FpV_RNA2_cDNA_5endas | CAGAGTGAGACAGTAGCCAGCG | 459+X | FpV RNA2 | this study |
| FpV_RNA2_PCR_5endas | CATAATCACCGGAGTCAAGTTCG | 319+X | FpV RNA2 | this study |
| FpV_RNA2_cDNA_3ends | CAATGATCGCTCCACTGCTGC | 574+X | FpV RNA2 | this study |
| FpV_RNA2_PCR_3ends | CTTACTCCCGAACTCCACCACG | 399+X | FpV RNA2 | this study |
| FgSV1_5end_cDNA_s | CTTTGTCCTGTTGCTTTTGAGC | 517+X | FgSV1 | this study |
| FgSV1_5end_PCR_s | CTTCCTTGACCGTGACATCCG | 334+X | FgSV1 | this study |
| FgSV1_3end_cDNA_as | CCGAGACAGAGATGCAGATGGTC | 516+X | FgSV1 | this study |
| FgSV1_3end_PCR_as | GAATTGATTATGCGAACCACCG | 328+X | FgSV1 | this study |
| Mito_5end_cDNA_as | GAATCAAAGGTTGAGTTAATAGGCG | 461+X | FgMV2 | this study |
| Mito_5end_PCR_as | GTTACCCACAAGACGACCATCAG | 293+X | FgMV2 | this study |
| Mito_3end_cDNA_s | CACACAATGGATGTTCTCATGCC | 743+X | FgMV2 | this study |
| Mito_3end_PCR1_s | GTACGCTAAGAATAGGAATGCCG | 544+X | FgMV2 | this study |
| Mito_3end_PCR2_s | GCCAATAAAACCGGCAATTTG | 498+X | FgMV2 | this study |
| Poly-A16 | CCTCGGGCAGTCCAAAAAAAAAAAAAAAA | - | respective tailed cDNA | this study |
| Poly-G15 | CTCAAACAGTCACGGGGGGGGGGGGGGG | - | respective tailed cDNA | this study |
| Poly-C14 | ATCCTGCAGGCGCGCCCCCCCCCCCCCC | - | respective tailed cDNA | this study |
| Poly-T18 | CCTCGGGCAGTCCTTTTTTTTTTTTTTTTTT | - | respective tailed cDNA | this study |
| Abbreviation | Full virus species name | GenBank accession number |
| RsAV1 | Rhizoctonia solani ambivirus 1 | QMP84024 |
| RsAV2 | Rhizoctonia solani ambivirus 2 | QMP84026 |
| AmAV2 | Armillaria mellea ambi-like virus 2 | DAD54839 |
| TuAV1 | Tulasnella ambivirus 1 | QPB44664 |
| HbAV1 | Heterobasidion ambi-like virus 1 | UHK02572 |
| PgAV1 | Phlebiopsis gigantea ambi-like virus 1 | UJT31805 |
| CpAV1 | Cryphonectria parasitica ambivirus 1 | QMP84022 |
| PgAV2 | Phlebiopsis gigantea ambi-like virus 2 | UJT31806 |
| AmAV1 | Armillaria mellea ambi-like virus 1 | DAD54837 |
| AbAV1 | Armillaria borealis ambi-like virus 1 | QUD20357 |
| AlAV1 | Armillaria luteobubalina ambi-like virus 1 | DAD54835 |
| HbAV3 | Heterobasidion ambi-like virus 3 | UHK02576 |
| TuAV5 | Tulasnella ambivirus 5 | QPB44674 |
| AnAV1 | Armillaria novae-zelandiae ambi-like virus 1 | DAD54841 |
| TuAV3 | Tulasnella ambivirus 3 | QPB44670 |
| TuAV4 | Tulasnella ambivirus 4 | QPB44672 |
| CbAV1 | Ceratobasidium ambivirus 1 | QPB44668 |
| CRV | Chimay rhabdovirus | AVM86063 |
| SsNSRV-1 | Sclerotinia sclerotiorum negative-stranded RNA virus 1 | YP009094317 |
| SsNSRV-3 | Sclerotinia sclerotiorum negative-stranded RNA virus 3 | YP009129259 |
| SsNSRV-9 | Sclerotinia sclerotiorum negative-stranded RNA virus 9 | QUE49142 |
| BcNSRV-3 | Botrytis cinerea negative-stranded RNA virus 3 | QJT73696 |
| CpSM1 | Cryphonectria parasitica sclerotimonavirus 1 | QMP84020 |
| SlNSRV-2 | Soybean leaf-associated negative-stranded RNA virus 2 | ALM62227 |
| AtNSRV1 | Alternaria tenuissima negative-stranded RNA virus 1 | QDB75013 |
| PvLAMNV2 | Plasmopara viticola lesion associated mononega virus 2 | QHD64785 |
| SlNSRV1 | Soybean leaf-associated negative-stranded RNA virus 1 | ALM62220 |
| FgNSRV1 | Fusarium graminearum negative-stranded RNA virus 1 | ATP75709 |
| BcNSRV4 | Botrytis cinerea negative-stranded RNA virus 4 | QJT73698 |
| Narnavirus sp. | Narnavirus sp. | QDH90729 |
| PvLAMV | Plasmopara viticola lesion associated mymonavirus 1 | QHD64779 |
| PcNSRV1 | Penicillium cairnsense negative-stranded RNA virus 1 | QDB75012 |
| SsNSRV7 | Sclerotinia sclerotiorum negative-stranded RNA virus 7 | AWY11040 |
| BcMV1 | Botrytis cinerea mymonavirus 1 | AXS76906 |
| CdNSRV1 | Coniothyrium diplodiella negative-stranded RNA virus 1 | QDB75015 |
| SlNSRV1 | Soybean leaf-associated negative-stranded RNA virus 1 | ALM62222 |
| FgNSRV1 | Fusarium graminearum negative-stranded RNA virus 1 | UNG44333 |
| FgNSRV1 | Fusarium graminearum negative-stranded RNA virus 1 | ATP75707 |
| FaNSRV1 | Fusarium asiaticum negative-stranded RNA virus 1 | UNG44330 |
| FmMV1 | Fusarium mangiferae mitovirus 1 | UBZ25880 |
| SlMV1 | Soybean leaf-associated mitovirus 1 | ALM62241 |
| BdMV1 | Botryosphaeria dothidea mitovirus 1 | QMU24933 |
| StMV1 | Setosphaeria turcica mitovirus 1 | AZT88625 |
| FsMV2 | Fusarium sambucinum mitovirus 2 | BCP96873 |
| CpMV1 | Cryphonectria parasitica mitovirus 1-NB631 | NP660174 |
| GaNV1 | Grapevine associated narnavirus-1 | CEZ26304 |
| SsMV7 | Sclerotinia sclerotiorum mitovirus 7-A | AWY10969 |
| BsMV1 | Buergenerula spartinae mitovirus 1 | AHY03257 |
| FaMV1 | Fusarium asiaticum mitovirus 1 | UNG44312 |
| PvMV13 | Plasmopara viticola lesion associated mitovirus 13 | QIR30236 |
| NpMV1 | Neofusicoccum parvum mitovirus 1 | QDB74992 |
| RsMV2 | Rhizoctonia solani mitovirus 2 | ALD89121 |
| FuMV1 | Fushun mitovirus 1 | UHM27612 |
| EnMV13 | Erysiphe necator associated mitovirus 13 | QKI79961 |
| BcMV1 | Botrytis cinerea mitovirus 1 | CEZ26300 |
| OpMV3 | Ophiostoma mitovirus 3b | CAJ32468 |
| MlSV | Mito-like spartanusvirus | QOW97240 |
| Toxin | Source | Polarity | Parent ion | Frag. [V] |
Collision [V] | Product ion | LOD [mg/kg] |
LOQ [mg/kg] |
| NIV | Romer Labs, Butzbach, Germany | Neg. | 357.1 | 95 | 10 10 |
311.1 281.1 |
0.02 | 0.08 |
| DON | Romer Labs, Butzbach, Germany | Pos. | 297.1 | 100 | 4 64 |
249.1 91.2 |
0.006 | 0.018 |
| 3-ADON | LGC Standards, Wesel, Germany | Pos. | 339.2 | 100 | 8 8 |
231.1 203.0 |
0.02 | 0.06 |
| 15-ADON | Sigma Aldrich, Steinheim am Albuch, Germany | Pos. | 339.2 | 90 | 10 10 10 |
321.1 261.0 137.0 |
0.04 | 0.13 |
| Aurofusarin | Bioviotica Naturstoffe GmbH, Dransfeld, Germany | Pos. | 571.0 | 195 | 21 21 |
556.0 541.0 |
7.8 | 26 |
References
- Munkvold, G.P. Epidemiology of Fusarium diseases and their mycotoxins in maize ears. Eur. J. Plant Pathol. 2003, 109, 705–713. [Google Scholar] [CrossRef]
- Desjardins, A.E. Fusarium Mycotoxins: Chemistry, Genetics and Biology; American Phytopathological Society, St. Paul. MN, USA, 2006; ISBN 0-89-54-335-6.
- Ferrigo, D.; Raiola, A.; Causin, R. Fusarium toxins in cereals: Occurrence, legislation, factors promoting the appearance and their management. Molecules 2016, 21, 627. [Google Scholar] [CrossRef]
- Foroud, N.A.; Baines, D.; Gagkaeva, T.Y.; Thakor, N.; Badea, A.; Steiner, B.; Bürstmayr, M.; Bürstmayr, H. Trichothecenes in cereal grains – an update. Toxins 2019, 11, 634. [Google Scholar] [CrossRef]
- Yoshizawa, T. Thirty-five years of research on deoxynivalenol, a trichothecene mycotoxin: with special reference to its discovery and co-occurrence with nivalenol in Japan. Food Safety 2013, 1, 2013002. [Google Scholar] [CrossRef]
- Tatsuno, T.; Saito, M.; Enomoto, M.; Tsunoda, H. Nivalenol, a toxic principle of Fusarium nivale. Chem. Pharm. Bull. 1968, 16, 2519–2520. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.H.; Hohn, T.M.; McCormick, S.P. reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol Plant Microbe Interact 1995, 8, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Jansen, C.; von Wettstein, D.; Schäfer, W.; Kogel, K.-H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 16892–16897. [Google Scholar] [CrossRef] [PubMed]
- Urry, W.H.; Wehrmeister, H.H.; Hodge, E.B.; Hidy, P.H. The structure of zearalenone. Tetrahedron Lett. 1966, 7, 3109–3114. [Google Scholar] [CrossRef]
- Utermark, J.; Karlovsky, P. Role of zearalenone lactonase in protection of Gliocladium roseum from fungitoxic effects of the mycotoxin zearalenone. Appl. Environ. Microbiol. 2007, 73, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Ashley, J.N.; Hobbs, B.C.; Raistrick, H. Studies in the biochemistry of micro-organisms: the crystalline colouring matters of Fusarium culmorum. Biochem. J. 1937, 31, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Vinas, M.; Alsarrag, A.; Su, L.; Pfohl, K.; Rohlfs, M.; Schäfer, W.; Chen, W.; Karlovsky, P. Bis-naphthopyrone pigments protect filamentous ascomycetes from a wide range of predators. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ghabrial, S.A.; Castón, J.R.; Jiang, D.; Nibert, M.L.; Suzuki, N. 50-plus years of fungal viruses. Virology 2015, 479–480, 356–368.
- Hough, B.; Steenkamp, E.; Wingfield, B.; Read, D. Fungal viruses unveiled: a comprehensive review of mycoviruses. Viruses 2023, 15, 1202. [Google Scholar] [CrossRef] [PubMed]
- te Velthuis, A. J. W.; Common and unique features of viral RNA-dependent polymerases. Cell. Mol. Life Sci., 2014, 71: 4403-20.
- Liu, S.; Xie, J.; Cheng, J.; Li, B.; Chen, T.; Fu, Y.; Li, G.; Wang, M.; Jin, H.; Wan, H.; et al. Fungal DNA virus infects a mycophagous insect and utilizes it as a transmission vector. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 12803–12808. [Google Scholar] [CrossRef] [PubMed]
- Nuss, D.L. Hypovirulence: Mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 2005, 3, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Hillman, B.I.; Annisa, A.; Suzuki, N. Viruses of plant-interacting fungi. Adv. Virus Res. 2018, 100, 99–116. [Google Scholar] [PubMed]
- Pearson, M.N.; Beever, R.E.; Boine, B.; Arthur, K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 2009, 10, 115–128. [Google Scholar] [CrossRef] [PubMed]
- García-Pedrajas, M.D.; Cañizares, M.C.; Sarmiento-Villamil, J.L.; Jacquat, A.G.; Dambolena, J.S. Mycoviruses in biological control: from basic research to field implementation. Phytopathology 2019, 109, 1828–1839. [Google Scholar] [CrossRef]
- Chu, Y.; Jeon, J.; Yea, S.; Kim, Y.; Yun, S.; Lee, Y.; Kim, K. Double-stranded RNA mycovirus from Fusarium graminearum. Appl. Environ. Microbiol. 2002, 68, 2529–2534. [Google Scholar] [CrossRef]
- Darissa, O.; Adam, G.; Schäfer, W. A dsRNA mycovirus causes hypovirulence of Fusarium graminearum to wheat and maize. Eur. J. Plant Path. 2012, 134, 181–189. [Google Scholar] [CrossRef]
- Li, P.; Zhang, H.; Chen, X.; Qiu, D.; Guo, L. Molecular characterization of a novel hypovirus from the plant pathogenic fungus Fusarium graminearum. Virology 2015, 481, 151–160. [Google Scholar] [CrossRef]
- Kotta-Loizou, I.; Coutts, R.H.A. Studies on the virome of the entomopathogenic fungus Beauveria bassiana reveal novel dsRNA elements and mild hypervirulence. PLoS Pathog. 2017, 13, 1–19. [Google Scholar] [CrossRef]
- Lee, K.-M.; Cho, W.K.; Yu, J.; Son, M.; Choi, H.; Min, K.; Lee, Y.-W.; Kim, K.-H. A comparison of transcriptional patterns and mycological phenotypes following infection of Fusarium graminearum by four mycoviruses. PLOS One 2014, 9, e100989. [Google Scholar] [CrossRef]
- Nerva, L.; Chitarra, W.; Siciliano, I.; Gaiotti, F.; Ciuffo, M.; Forgia, M.; Varese, G.C.; Turina, M. Mycoviruses mediate mycotoxin regulation in Aspergillus ochraceus. Environ. Microbiol. 2019, 21, 1957–1968. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, A.; Urayama, S.; Suo, R.; Itoi, S.; Fuji, S.; Moriyama, H.; Hagiwara, D. Mycovirus-induced tenuazonic acid production in a rice blast fungus Magnaporthe oryzae. Front. Microbiol. 2020, 11, 1641. [Google Scholar] [CrossRef] [PubMed]
- Schiwek, S.; Beule, L.; Vinas, M.; Pfordt, A.; von Tiedemann, A.; Karlovsky, P. High-resolution melting (HRM) curve assay for the identification of eight Fusarium species causing ear rot in maize. Pathogens 2020, 9, 270. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Forgia, M.; Navarro, B.; Daghino, S.; Cervera, A.; Gisel, A.; Perotto, S.; Aghayeva, D.N.; Akinyuwa, M.F.; Gobbi, E.; Zheludev, I.N.; et al. Hybrids of RNA viruses and viroid-like elements replicate in fungi. Nat. Commun. 2023, 14, 2591. [Google Scholar] [CrossRef]
- Sutela, S., Forgia, M., Vainio, E.J., Chiapello, M., Daghino, S., Vallino, M., Martino, E., Girlanda, M., Perotto, S., Turina, M., The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus Evolution 2020, 6, 76.
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Gonnet, G.H.; Cohen, M.A.; Benner, S.A. Exhaustive matching of the entire protein sequence database. Science 1992, 256, 1443–1445. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comp Appl Biosci 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Pringle, C.R.; Easton, A.J. Monopartite negative strand RNA genomes. Seminars in Virology 1997, 8, 49–57. [Google Scholar] [CrossRef]
- Liu, L.; Xie, J.; Cheng, J.; Fu, Y.; Li, G.; Yi, X.; Jiang, D. Fungal negative-stranded RNA virus that is related to Bornaviruses and Nyaviruses. Proc. Nat. Acad. Sci. U.S.A. 2014, 111, 12205–12210. [Google Scholar] [CrossRef]
- Wang, L.; He, H.; Wang, S.; Chen, X.; Qiu, D.; Kondo, H.; Guo, L. Evidence for a novel negative-stranded RNA mycovirus isolated from the plant pathogenic fungus Fusarium graminearum. Virology 2018, 518, 232–240. [Google Scholar] [CrossRef]
- Xu, Z.; Wu, S.; Liu, L.; Cheng, J.; Fu, Y.; Jiang, D.; Xie, J. A mitovirus related to plant mitochondrial gene confers hypovirulence on the phytopathogenic fungus Sclerotinia sclerotiorum. Virus Res. 2015, 197, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Research 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, M.E.; Pearson, M.N. Molecular characterization of three mitoviruses co-infecting a hypovirulent isolate of Sclerotinia sclerotiorum fungus. Virology 2013, 441, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.A.; Baig, D.I.; Bhatti, M.F. An overview of mycoviral curing strategies used in evaluating fungal host fitness. Mol. Biotechnol. 2023, 65, 1547–1564. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.M.; Lim, W.S.; Yea, S.J.; Cho, J.D.; Lee, Y.W.; Kim, K.H. Complexity of dsRNA mycovirus isolated from Fusarium graminearum. Virus Genes 2004, 28, 135–143. [Google Scholar] [CrossRef]
- Bormann, J.; Heinze, C.; Blum, C.; Mentges, M.; Brockmann, A.; Alder, A.; Landt, S.K.; Josephson, B.; Indenbirken, D.; Spohn, M.; et al. Expression of a structural protein of the mycovirus FgV-ch9 negatively affects the transcript level of a novel symptom alleviation factor and causes virus infection-like symptoms in Fusarium graminearum. J. Virol. 2018, 92, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Lee, K.M.; Son, M.; Kim, K.H. Effects of the deletion and over-expression of Fusarium graminearum gene FgHal2 on host response to mycovirus Fusarium graminearum virus 1. Mol. Plant Pathol. 2015, 16, 641–652. [Google Scholar] [CrossRef]
- Kim, J.-E.; Han, K.-H.; Jin, J.; Kim, H.; Kim, J.-C.; Yun, S.-H.; Lee, Y.-W. Putative polyketide synthase and laccase genes for biosynthesis of aurofusarin in Gibberella zeae. Appl. Env. Microbiol. 2005, 71, 1701–1708. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-M.; Lee, J.; Lee, Y.-W. Characterization of carotenoid biosynthetic genes in the ascomycete Gibberella zeae. FEMS Microbiol. Lett. 2010, 302, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Moonjely, S., Ebert, M., Paton-Glassbrook, D., Noel, Z. A., Roze, L., Shay, R., Watkins, T., Trail, F. Update on the state of research to manage Fusarium head blight. Fungal Genet. Biol. 2023, 169, 103829. [CrossRef]
- Son, M.; Yu, J.; Kim, K.-H. Five Questions about mycoviruses. PLOS Pathogens 2015, 11, e1005172. [Google Scholar] [CrossRef] [PubMed]
- Szabó, B.; Kocsis, R.; Mézes, M. Reproduction inhibiting effects of deoxynivalenol or T-2 toxin contaminated maize on Folsomia candida (Collembola). Acta Zoologica Academiae Scientiarum Hungaricae 2019, 65, 323–334.
- Szabó, B., Bálint, B., Mézes, M., Balogh, K. Agricultural trichothecene mycotoxin contamination affects the life-history and reduced glutathione content of Folsomia candida Willem (Collembola). Acta Zoologica Academiae Scientiarum Hungaricae 2020, 66, 379–392.
- Karlovsky, P. Biological detoxification of fungal toxins and its use in plant breeding, feed and food production. Natural Toxins 1999, 7, 1–23. [Google Scholar] [CrossRef]
- Staaden, S.; Milcu, A.; Rohlfs, M.; Scheu, S. Olfactory cues associated with fungal grazing intensity and secondary metabolite pathway modulate collembola foraging behaviour. Soil Biol. Biochem. 2011, 43, 1411–1416. [Google Scholar] [CrossRef]
- Hedlund, K.; Bengtsson, G.; Rundgren, S. Fungal odour discrimination in two sympatric species of fungivorous Collembolans. Funct. Ecol. 1995, 9, 869–875. [Google Scholar] [CrossRef]
- Bengtsson, G.; Hedlund, K.; Rundgren, S. selective odor perception in the soil Collembola Onychiurus armatus. J. Chem. Ecol. 1991, 17, 2113–2125. [Google Scholar] [CrossRef]
- Nebapure, S.M.; Shankarganesh, K.; Rajna, S.; Naga, K.C.; Pandey, D.; Gambhir, S.; Praveen, K.V.; Subramanian, S. Dynamic changes in virus-induced volatiles in cotton modulate the orientation and oviposition behavior of the whitefly Bemisia tabaci. Front. Physiol. 2022, 13, 1–16. [Google Scholar] [CrossRef]
- Werner, B.J.; Mowry, T.M.; Bosque-Pérez, N.A.; Ding, H.; Eigenbrode, S.D. Changes in green peach aphid responses to potato leafroll virus induced volatiles emitted during disease progression. Environ. Entomol. 2009, 38, 1429–1438. [Google Scholar] [CrossRef]
- Sherif, M.; Kirsch, N.; Splivallo, R.; Pfohl, K.; Karlovsky, P. The role of mycotoxins in interactions between Fusarium graminearum and F. verticillioides growing in saprophytic cultures and co-infecting maize plants. Toxins 2023, 15, 575. [CrossRef]
- Pfordt, A.; Ramos Romero, L.; Schiwek, S.; Karlovsky, P.; von Tiedemann, A. Impact of environmental conditions and agronomic practices on the prevalence of Fusarium species associated with ear- and stalk rot in maize. Pathogens 2020, 9, 236. [Google Scholar] [CrossRef]
- Lysøe, E.; Klemsdal, S.S.; Bone, K.R.; Frandsen, R.J.N.; Johansen, T.; Thrane, U.; Giese, H. The PKS4 Gene of Fusarium graminearum is essential for zearalenone production. Appl Environ. Microbiol. 2006, 72, 3924–3932. [Google Scholar] [CrossRef]
- Maniatis, T., Fritsch, E. F. & Sambrook, J. Molecular Cloning – a Laboratory Manual. Cold Spring Harbor Laboratory: New York, 1989, pp. 94-103.
- Pielhop, T.P.; Popp, C.; Knierim, D.; Margaria, P.; Maiß, E. Three new mycoviruses identified in the apple replant disease (ARD)-associated fungus Rugonectria rugulosa. Virus Genes, 2022,. [CrossRef]
- Koonin, E. V; Senkevich, T.G.; Dolja, V. V The ancient virus world and evolution of cells. Biol. Direct 2006, 1, 27. [Google Scholar]
- Nasir, A.; Caetano-Anollés, G. A phylogenomic data-driven exploration of viral origins and evolution. Sci. Adv. 2015, 1, e1500527. [Google Scholar] [CrossRef] [PubMed]
- Brandfass, C.; Karlovsky, P. Upscaled CTAB-Based DNA extraction and real-time PCR Assays for Fusarium culmorum and F. graminearum DNA in plant material with reduced sampling error. Int. J. Mol. Sci. 2008, 9, 2306–2321. [Google Scholar] [CrossRef] [PubMed]
- Nirenberg, H. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitt. Biol. Bundesanst. Land und Forstwirtsch. Berlin-Dahlem 1976, 169, 1–117.
- Schiwek, S.; Alhussein, M.; Rodemann, C.; Budragchaa, T.; Beule, L.; von Tiedemann, A.; Karlovsky, P. Fusarium culmorum produces NX-2 toxin simultaneously with deoxynivalenol and 3-acetyl-deoxynivalenol or nivalenol. Toxins 2022, 14, 456. [Google Scholar] [CrossRef] [PubMed]
- Osaki, H.; Sasaki, A.; Nomiyama, K.; Tomioka, K. Multiple virus infection in a single strain of Fusarium poae shown by deep sequencing. Virus Genes 2016, 52, 835–847. [Google Scholar] [CrossRef]
- Nicholson, P.; Simpson, D.R.; Weston, G.; Rezanoor, H.N.; Lees, A.K.; Parry, D.W.; Joyce, D. Detection and quantification of Fusarium culmorum and Fusarium graminearum in cereals using PCR assays. Physiol. Mol. Plant Path. 1998, 53, 17–37. [Google Scholar] [CrossRef]
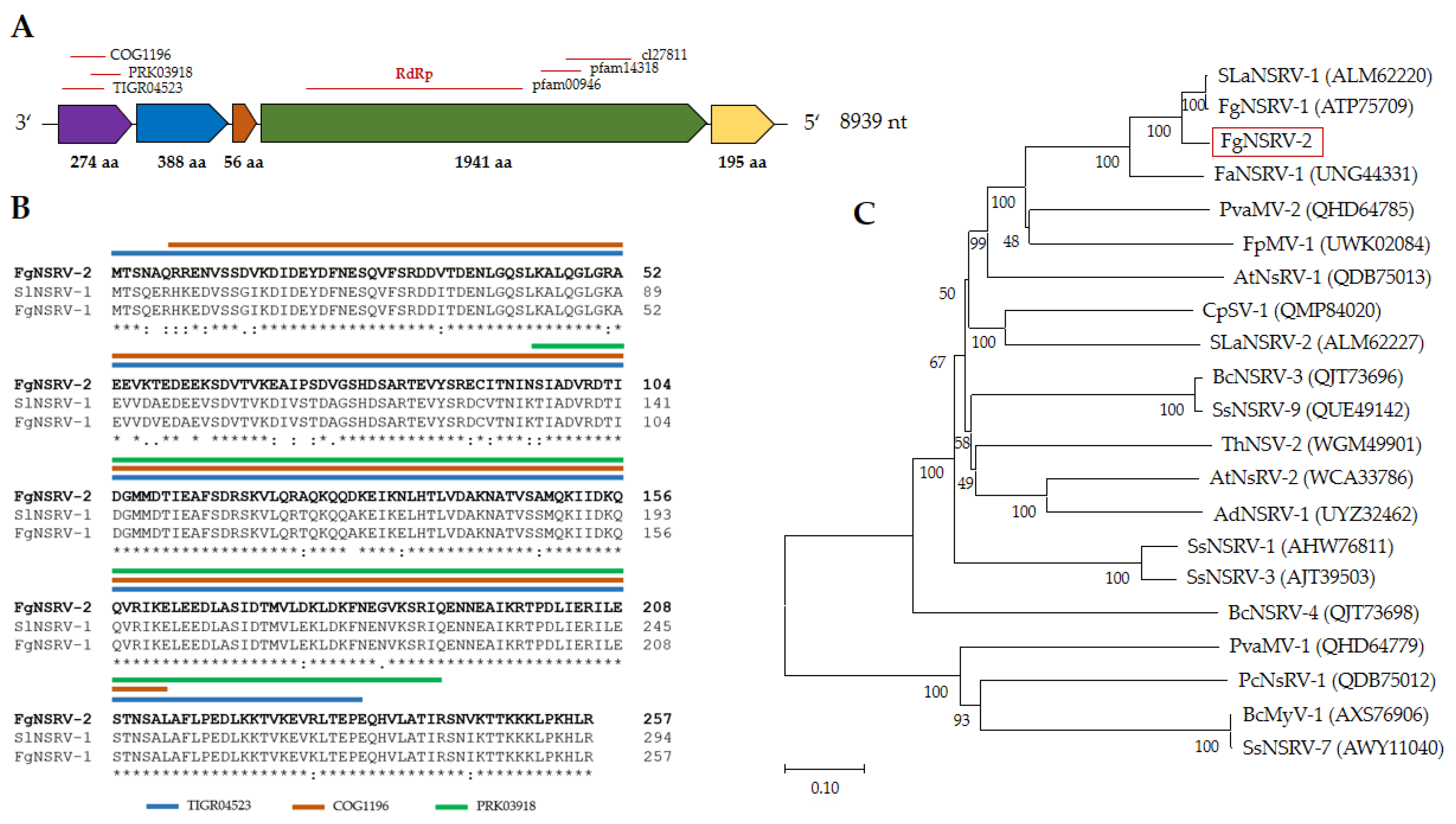

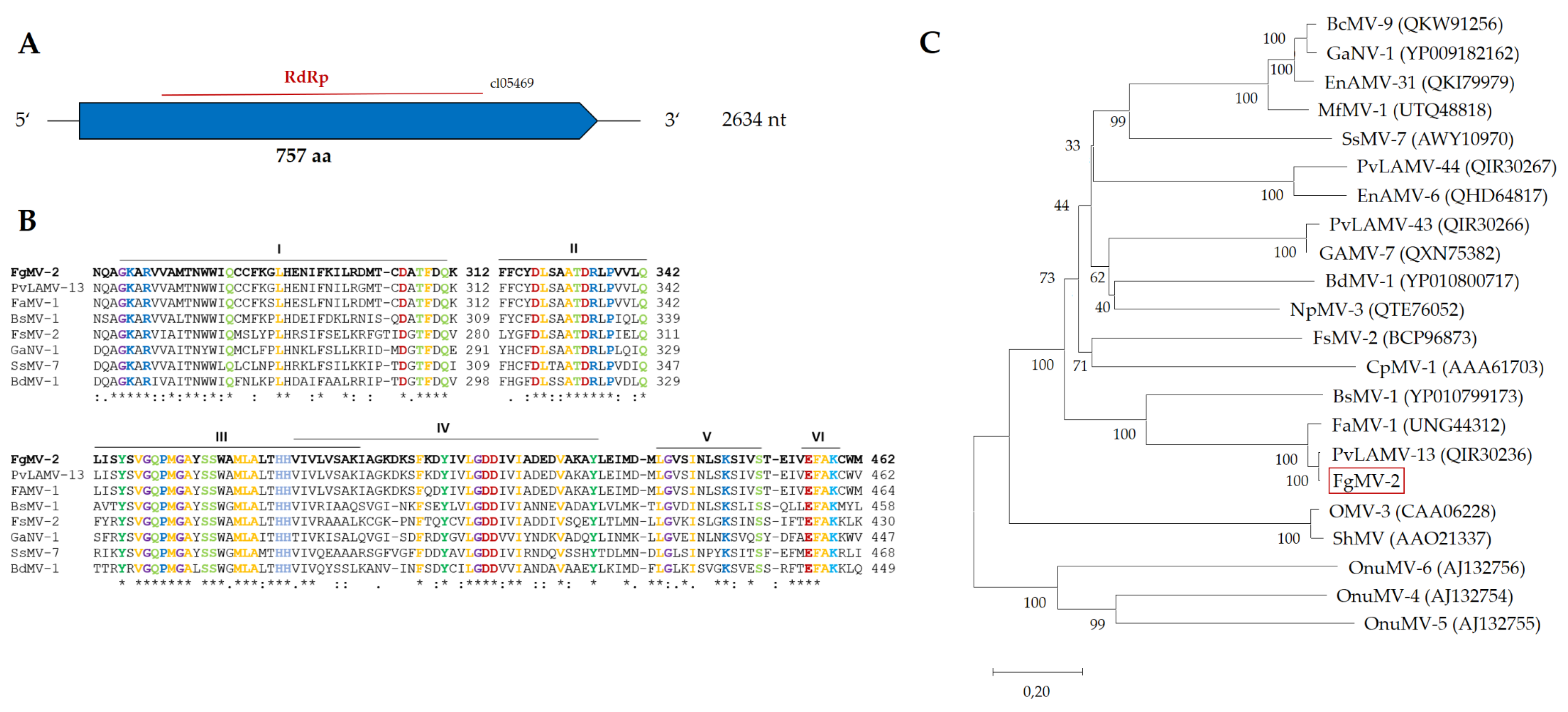
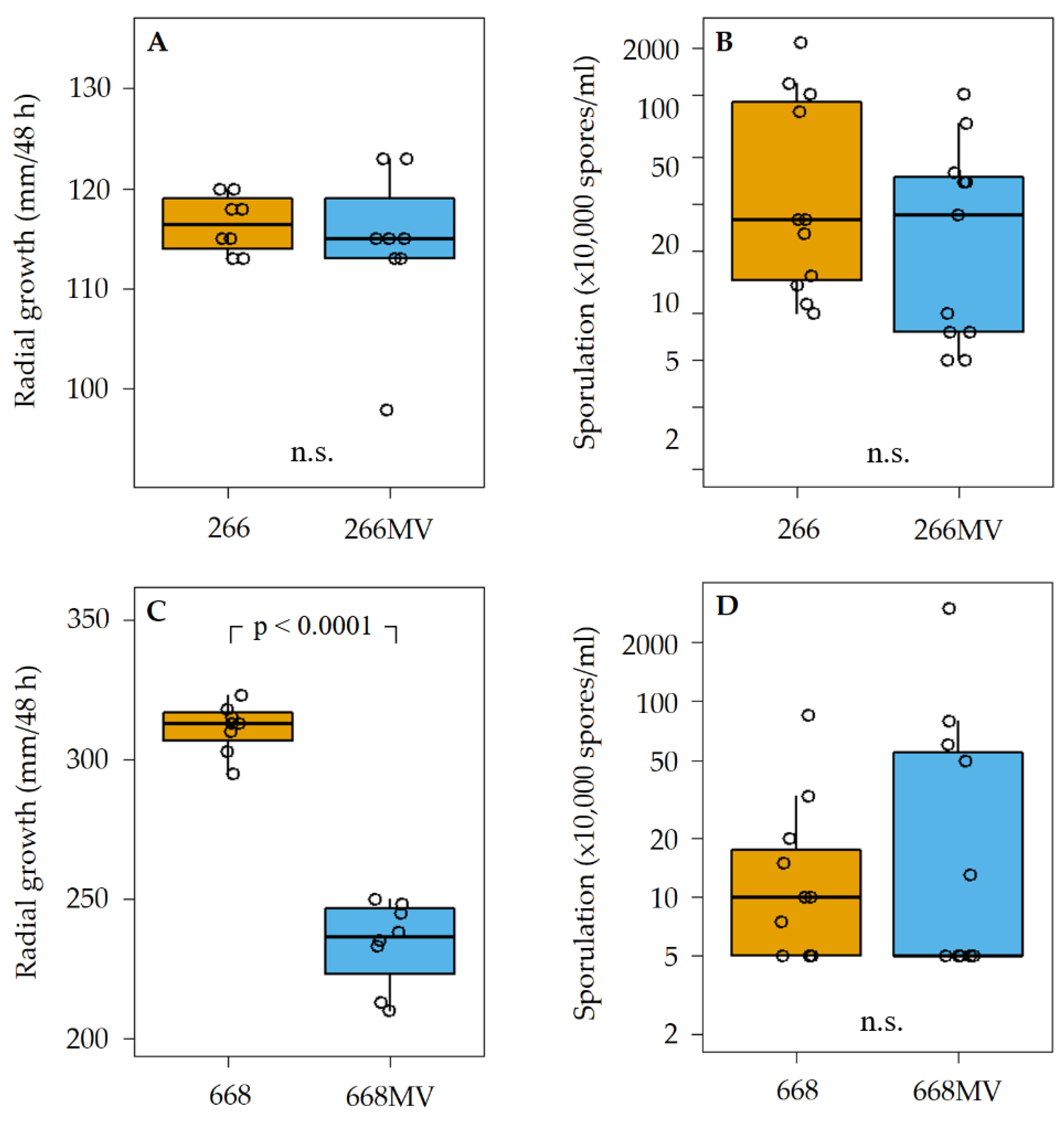
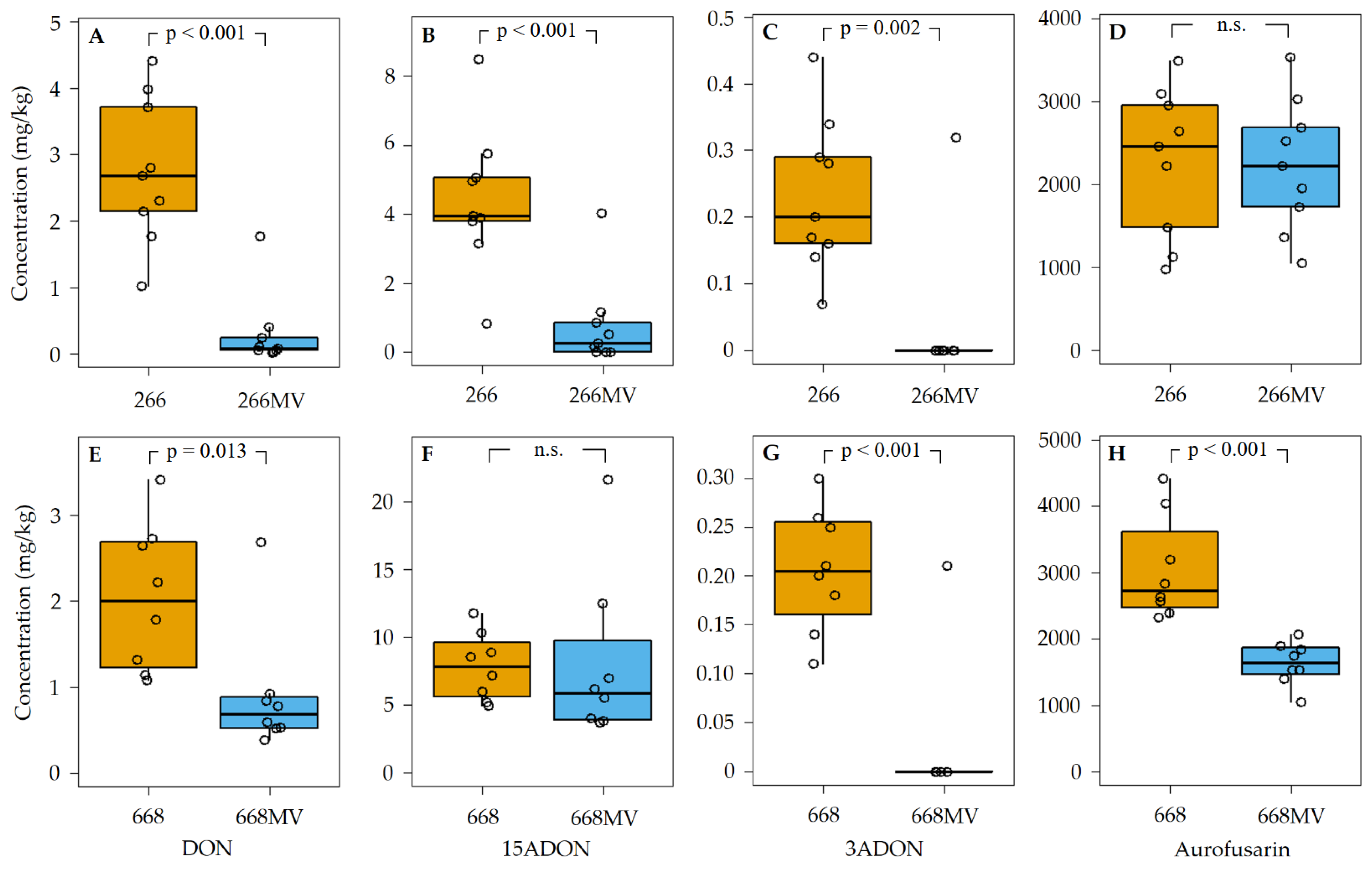
| Name | Size [nt] | Accession no. | Most similar genomes by NCBI BLASTn | |||
| Closest reference sequenceB | Accession no. | Genome [nt] | Similarity [%] | |||
| FpV1 | 2298 | ON969095 | Fusarium poae virus 1 RNA 1 | LC150606 | 2431 | 98.6 |
| 2205 | ON969096 | Fusarium poae virus 1 RNA 2 | OK524181 | 2212 | 98.8 | |
| FgNSRV2 | 8939 | ON969097 | Soybean leaf-associated negative-stranded RNA virus | NC075296 | 9041 | 98.0 |
| FgMV2 | 2634 | ON969098 | Plasmopara viticola lesion associated mitovirus 13 | MN539774 | 2607 | 96.3 |
| FgAV1 | 4579 | ON969099 | Cryphonectria parasitica ambivirus 1 | MT354566 | 4579 | 73.0 |
| Strain | FpV1 | FgNSRV1 | FgMV2 | FgAV1 |
| 266 | yes | no | yes | no |
| 266MV | yes | yes | yes | yes |
| 668 | no | no | no | no |
| 668MV | yes | no | no | yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





