Submitted:
05 January 2024
Posted:
05 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. A. oryzae Strains, Growth Conditions, and Transformation
2.2. Construction of KojR FL/BD Expression Vector
2.3. Expression of MBP-Fused KojR Using Cell-Free Protein Synthesis System
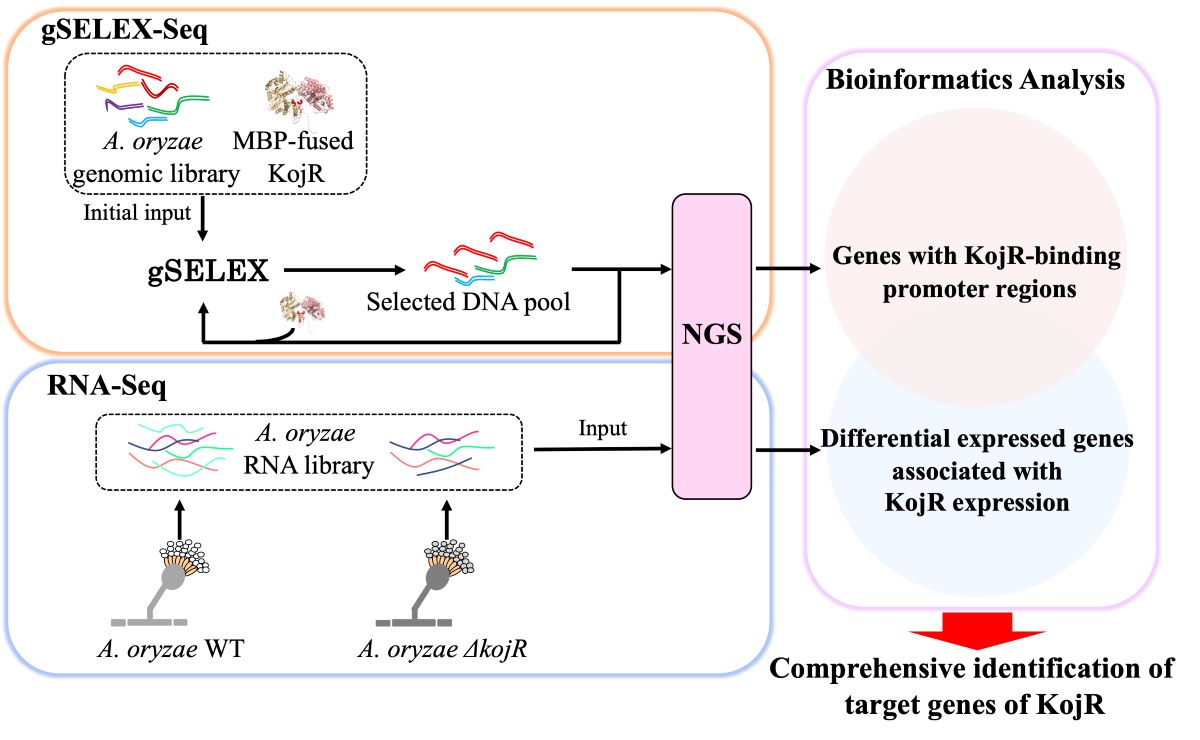
2.4. In Vitro Selection of KojR-Bound DNA Fragments from an A. oryzae Genomic Library Using gSELEX-Seq
2.5. DNA Sequencing and Data Analysis in gSELEX-Seq
2.6. Construction of an A. oryzae ΔkojR Strain Using CRISPR/Cas9
2.7. Identifications of DEGs Associated with KojR Expression Using RNA-Seq
2.8. KEGG Pathway Analysis of DEGs
3. Results
3.1. Preparation of MBP-Fused KojR for Use in gSELEX
3.2. Genome-Wide Identification of KojR Binding Sites Using gSELEX-Seq
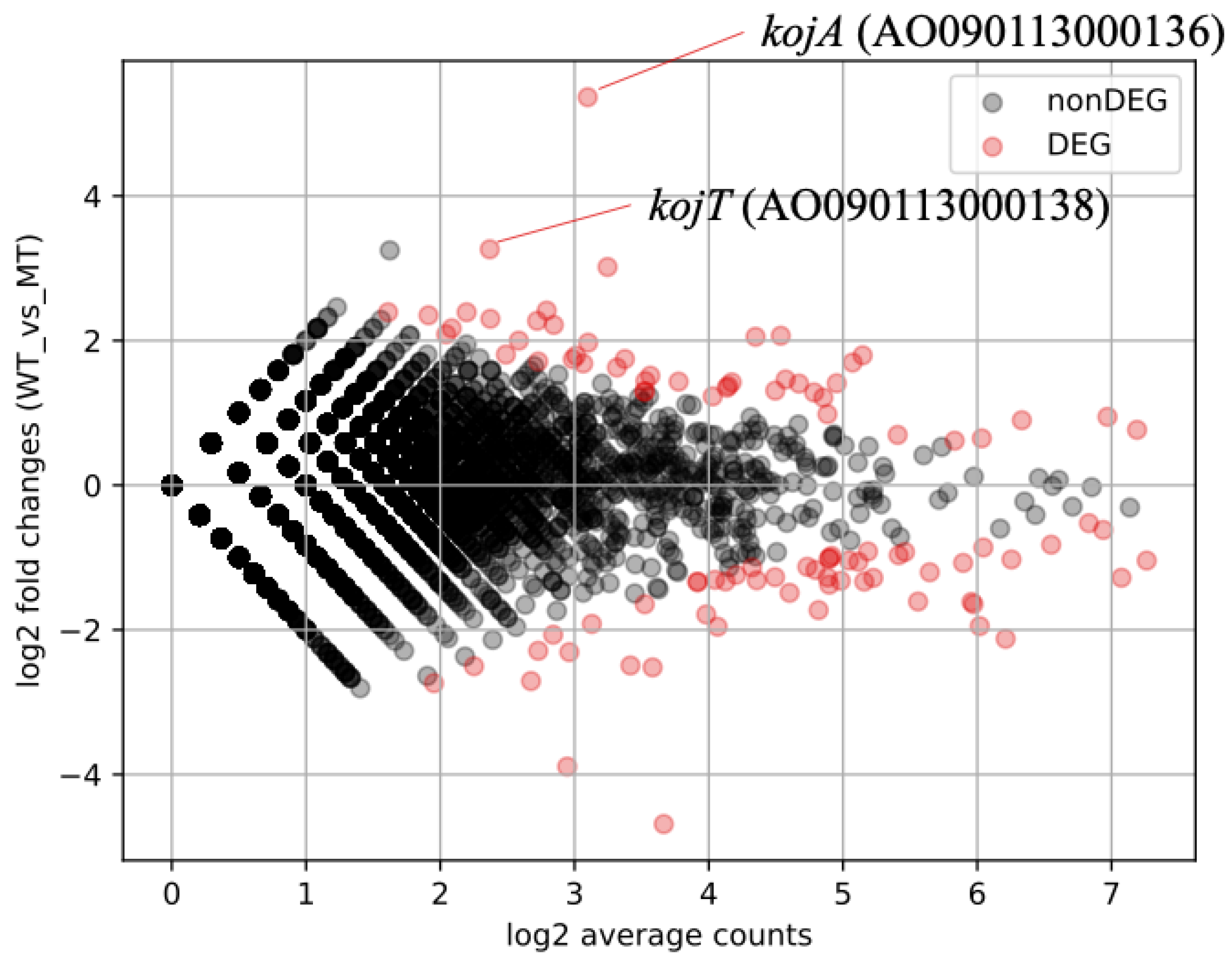
3.3. Identification of KojR Expression-Dependent DEGs Using RNA-Seq
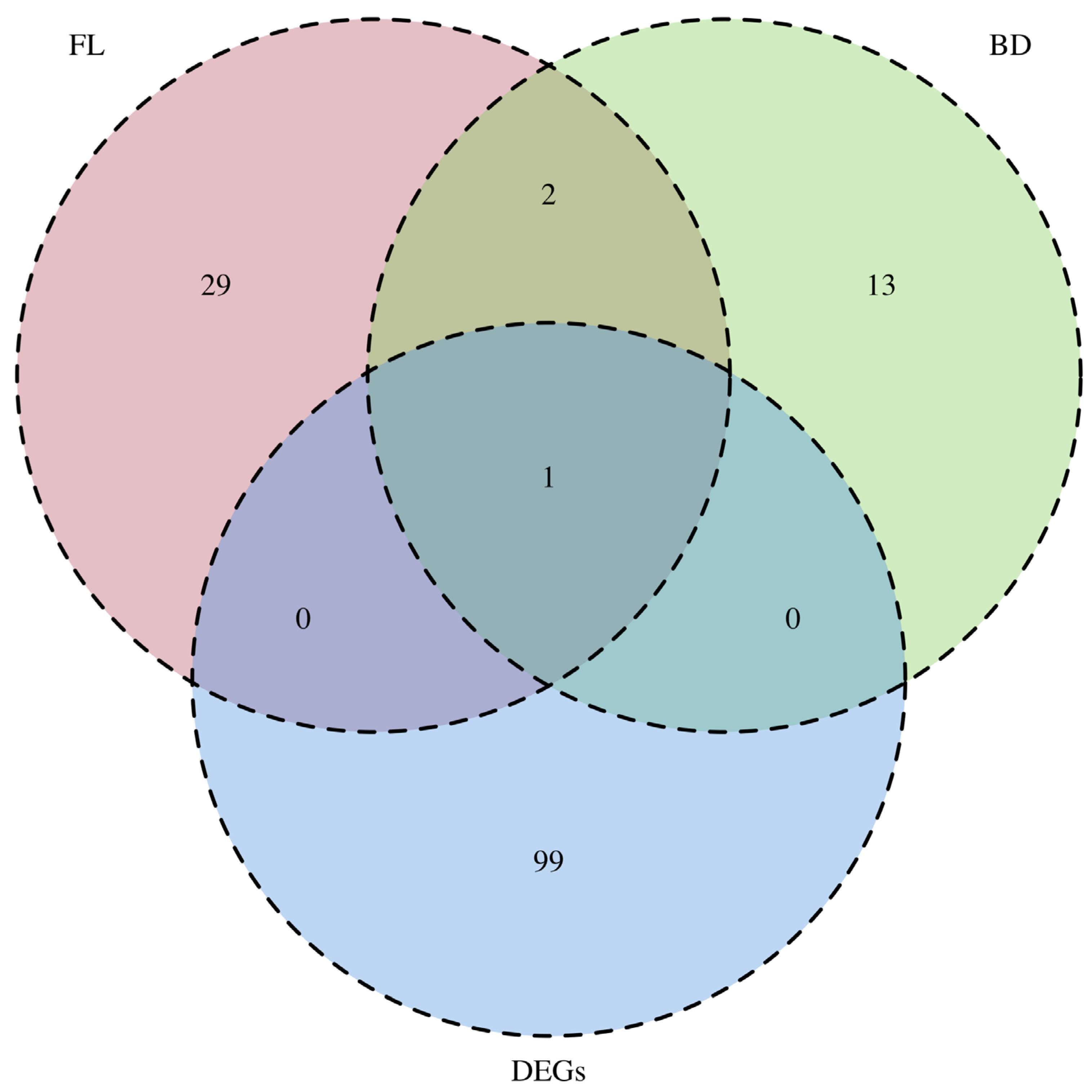
3.4. Comprehensive Identification of Target Genes of KojR Using Integrated Analysis-Based Binding Sites on A. oryzae Genome and DEGs Information
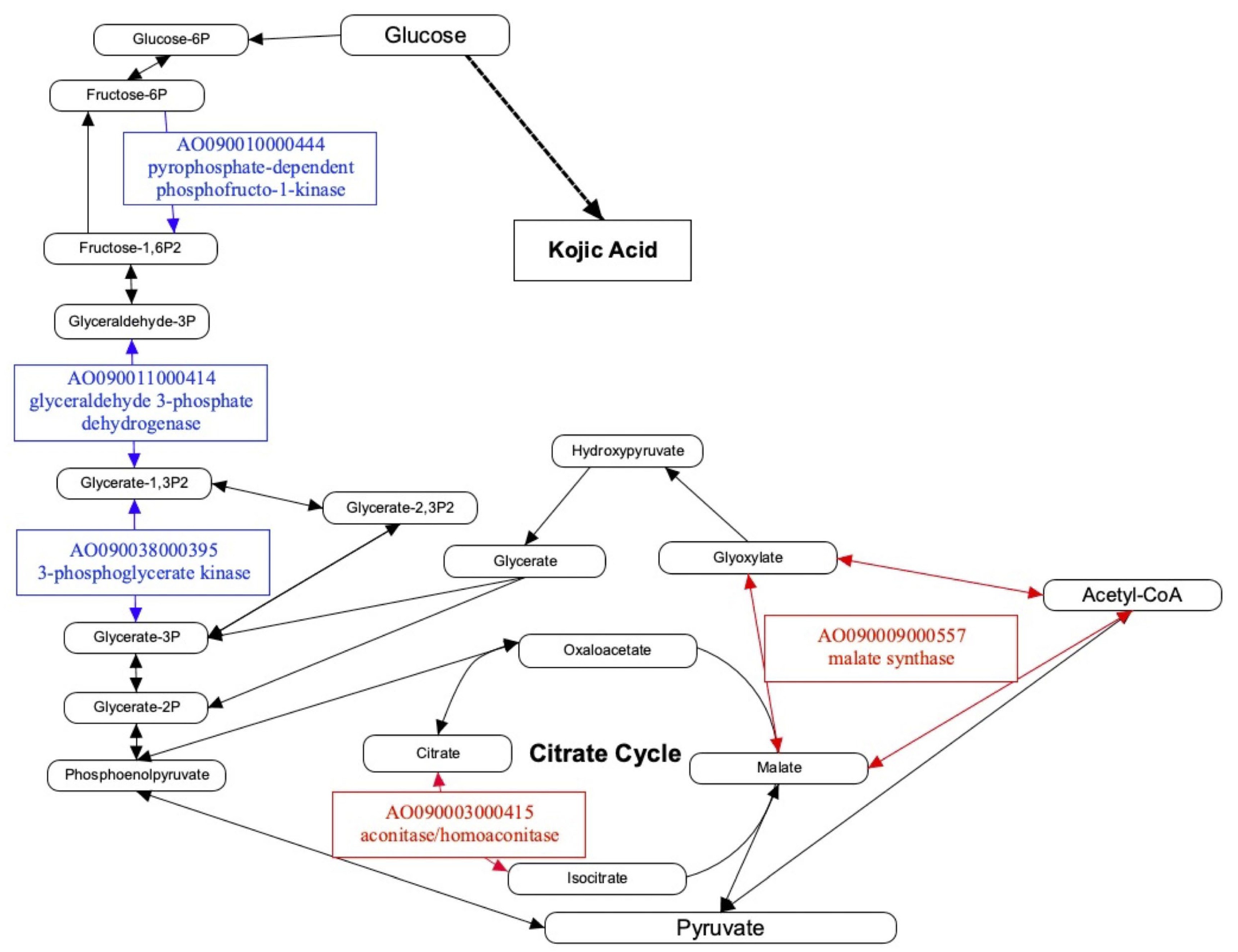
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saeedi:, M.; Eslamifar, M.; Khezri, K. Kojic Acid Applications in Cosmetic and Pharmaceutical Preparations. Biomedicine & Pharmacotherapy 2019, 110, 582–593. [Google Scholar] [CrossRef]
- Brtko, J. Biological Functions of Kojic Acid and Its Derivatives in Medicine, Cosmetics, and Food Industry: Insights into Health Aspects. Archiv der Pharmazie 2022, 355, 2200215. [Google Scholar] [CrossRef]
- Bentley, R. From Miso, Sake and Shoyu to Cosmetics: A Century of Science for Kojic Acid. ChemInform 2007, 38. [Google Scholar] [CrossRef]
- Chib, S.; Jamwal, V.L.; Kumar, V.; Gandhi, S.G.; Saran, S. Fungal Production of Kojic Acid and Its Industrial Applications. Appl Microbiol Biotechnol 2023, 107, 2111–2130. [Google Scholar] [CrossRef]
- Terabayashi, Y.; Sano, M.; Yamane, N.; Marui, J.; Tamano, K.; Sagara, J.; Dohmoto, M.; Oda, K.; Ohshima, E.; Tachibana, K.; et al. Identification and Characterization of Genes Responsible for Biosynthesis of Kojic Acid, an Industrially Important Compound from Aspergillus oryzae. Fungal Genetics and Biology 2010, 47, 953–961. [Google Scholar] [CrossRef]
- Marui, J.; Yamane, N.; Ohashi-Kunihiro, S.; Ando, T.; Terabayashi, Y.; Sano, M.; Ohashi, S.; Ohshima, E.; Tachibana, K.; Higa, Y.; et al. Kojic Acid Biosynthesis in Aspergillus oryzae Is Regulated by a Zn(II)2Cys6 Transcriptional Activator and Induced by Kojic Acid at the Transcriptional Level. Journal of Bioscience and Bioengineering 2011, 112, 40–43. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Chen, Z.; Fan, J.; Chen, T.; Zeng, B.; Zhang, Z. Construction of Single, Double, or Triple Mutants within Kojic Acid Synthesis Genes kojA, kojR, and kojT by the CRISPR/Cas9 Tool in Aspergillus oryzae. Folia Microbiol 2022, 67, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.-K.; Scharfenstein, L.L.; Mahoney, N.; Kong, Q. Kojic Acid Gene Clusters and the Transcriptional Activation Mechanism of Aspergillus flavus KojR on Expression of Clustered Genes. JoF 2023, 9, 259. [Google Scholar] [CrossRef]
- De Castro, P.A.; Colabardini, A.C.; Moraes, M.; Horta, M.A.C.; Knowles, S.L.; Raja, H.A.; Oberlies, N.H.; Koyama, Y.; Ogawa, M.; Gomi, K.; et al. Regulation of Gliotoxin Biosynthesis and Protection in Aspergillus Species. PLoS Genet 2022, 18, e1009965. [Google Scholar] [CrossRef] [PubMed]
- Kojima, T.; Kunitake, E.; Ihara, K.; Kobayashi, T.; Nakano, H. A Robust Analytical Pipeline for Genome-Wide Identification of the Genes Regulated by a Transcription Factor: Combinatorial Analysis Performed Using gSELEX-Seq and RNA-Seq. PLOS ONE 2016, 11, e0159011. [Google Scholar] [CrossRef] [PubMed]
- Oka, H.; Kojima, T.; Ihara, K.; Kobayashi, T.; Nakano, H. Comprehensive Investigation of the Gene Expression System Regulated by an Aspergillus oryzae Transcription Factor XlnR Using Integrated Mining of gSELEX-Seq and Microarray Data. BMC Genomics 2019, 20, 16. [Google Scholar] [CrossRef]
- Oka, H.; Kojima, T.; Kato, R.; Ihara, K.; Nakano, H. Construction of Transcript Regulation Mechanism Prediction Models Based on Binding Motif Environment of Transcription Factor AoXlnR in Aspergillus oryzae. bioRxiv 2021. [Google Scholar] [CrossRef]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.-I.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome Sequencing and Analysis of Aspergillus oryzae. Nature 2005, 438, 1157–1161. [Google Scholar] [CrossRef]
- Okegawa, Y.; Motohashi, K. A Simple and Ultra-Low Cost Homemade Seamless Ligation Cloning Extract (SLiCE) as an Alternative to a Commercially Available Seamless DNA Cloning Kit. Biochemistry and Biophysics Reports 2015, 4, 148–151. [Google Scholar] [CrossRef] [PubMed]
- Tani, S.; Itoh, T.; Kato, M.; Kobayashi, T.; Tsukagoshi, N. In vivo and in vitro analyses of the AmyR binding site of the Aspergillus nidulans agdA promoter; requirement of the CGG direct repeat for induction and high affinity binding of AmyR. Bioscience, Biotechnology, and Biochemistry 2001, 65, 1568–74. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet.journal 17, 10–12. [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nature Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Nakamura, H.; Zhang, Y.; Pascal, A.; Fujii, W.; Maruyama, J. Forced Recycling of an AMA1-Based Genome-Editing Plasmid Allows for Efficient Multiple Gene Deletion/Integration in the Industrial Filamentous Fungus Aspergillus oryzae. Appl Environ Microbiol 2019, 85, e01896–18. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y. KEGG Mapper for Inferring Cellular Functions from Protein Sequences. Protein Science 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Kutmon, M.; Van Iersel, M.P.; Bohler, A.; Kelder, T.; Nunes, N.; Pico, A.R.; Evelo, C.T. PathVisio 3: An Extendable Pathway Analysis Toolbox. PLoS Comput Biol 2015, 11, e1004085. [Google Scholar] [CrossRef]
- Tamano, K.; Kuninaga, M.; Kojima, N.; Umemura, M.; Machida, M.; Koike, H. Use of the kojA Promoter, Involved in Kojic Acid Biosynthesis, for Polyketide Production in Aspergillus oryzae: Implications for Long-Term Production. BMC Biotechnol 2019, 19, 70. [Google Scholar] [CrossRef]
- Arakawa, G.; Kudo, H.; Yanase, A.; Eguchi, Y.; Kodama, H.; Ogawa, M.; Koyama, Y.; Shindo, H.; Hosaka, M.; Tokuoka, M. A Unique Zn(II)2-Cys6-Type Protein, KpeA, Is Involved in Secondary Metabolism and Conidiation in Aspergillus oryzae. Fungal Genetics and Biology 2019, 127, 35–44. [Google Scholar] [CrossRef]
- Sano, M. Aspergillus oryzae nrtA Affects Kojic Acid Production. Bioscience, Biotechnology, and Biochemistry 2016, 80, 1776–1780. [Google Scholar] [CrossRef]
- Oda, K.; Kobayashi, A.; Ohashi, S.; Sano, M. Aspergillus oryzae laeA Regulates Kojic Acid Synthesis Genes. Bioscience, Biotechnology, and Biochemistry 2011, 75, 1832–1834. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, H.; Chen, Z.; Fan, J.; Chen, T.; Zeng, B.; Zhang, Z. Identification and Characterization of a Novel Gene Aokap1 Involved in Growth and Kojic Acid Synthesis in Aspergillus oryzae. Arch Microbiol 2022, 204, 67. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Chen, Z.; Fan, J.; Chen, T.; Xiao, Y.; Jie, J.; Zeng, B.; Zhang, Z. Overexpression of a Novel Gene Aokap2 Affects the Growth and Kojic Acid Production in Aspergillus oryzae. Mol Biol Rep 2022, 49, 2745–2754. [Google Scholar] [CrossRef]
- Chen, T.; Chen, Z.; Li, Y.; Zeng, B.; Zhang, Z. A Novel Major Facilitator Superfamily Transporter Gene Aokap4 near the Kojic Acid Gene Cluster Is Involved in Growth and Kojic Acid Production in Aspergillus oryzae. JoF 2022, 8, 885. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Z.; Zhang, F.; Chen, T.; Fan, J.; Deng, X.; Lei, X.; Zeng, B.; Zhang, Z. The C2H2-Type Zinc-Finger Regulator AoKap5 Is Required for the Growth and Kojic Acid Synthesis in Aspergillus oryzae. Fungal Genetics and Biology 2023, 167, 103813. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chen, T.; Wang, H.; Jiang, C.; Liu, Y.; Wu, X.; Li, Y.; Zeng, B.; Zhang, Z. Disruption of Aokap6 near the Kojic Acid Gene Cluster Affects the Growth and Kojic Acid Production in Aspergillus oryzae. World J Microbiol Biotechnol 2022, 38, 175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Fan, J.; Long, C.; He, B.; Hu, Z.; Jiang, C.; Li, Y.; Ma, L.; Wen, J.; Zou, X.; et al. Identification and Characterization of the ZRT, IRT-like Protein (ZIP) Family Genes Reveal Their Involvement in Growth and Kojic Acid Production in Aspergillus oryzae. Journal of Industrial Microbiology and Biotechnology 2019, 46, 1769–1780. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, T.; Zhang, H.; Li, Y.; Fan, J.; Yao, L.; Zeng, B.; Zhang, Z. Functional Role of a Novel Zinc Finger Protein, AoZFA, in Growth and Kojic Acid Synthesis in Aspergillus oryzae. Appl Environ Microbiol 2023, e00909–23. [Google Scholar] [CrossRef]

| Peak Region | Gene ID | Original Description |
|---|---|---|
| Chr5_A_oryzae_RIB40:4372480-4372692 | AO090113000138 | Putative transporter; present in the kojic acid biosynthetic gene cluster |
| Chr6_A_oryzae_RIB40:3584694-3584906 | AO090038000029 | Has domain(s) with predicted peroxiredoxin activity and role in oxidation-reduction process |
| Chr5_A_oryzae_RIB40:1165262-1165474 | AO090701000448 | Has domain(s) with predicted heme binding activity |
| Chr5_A_oryzae_RIB40:643370-643582 | AO090701000654 | Protein of unknown function |
| Chr5_A_oryzae_RIB40:1158260-1158472 | AO090701000450 | Has domain(s) with predicted catalytic activity, hydrolase activity, and role in metabolic process |
| Chr1_A_oryzae_RIB40:5569621-5569833 | AO090005000336 | Has domain(s) with predicted sequence-specific DNA binding RNA polymerase II transcription factor activity, zinc ion binding activity, and role in regulation of transcription, DNA-templated and nucleus localization |
| Chr7_A_oryzae_RIB40:229700-229912 | AO090011000078 | Ortholog of A. niger CBS 513.88 : An01g06430, A. versicolor : Aspve1_0082366, A. niger ATCC 1015 : 36036-mRNA, and A. zonatus : Aspzo1_0026572 |
| Chr1_A_oryzae_RIB40:4941497-4941709 | AO090005000571 | Has domain(s) with predicted NAD binding, oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor activity, and role in oxidation-reduction process |
| Chr2_A_oryzae_RIB40:4505321-4505533 | AO090003000890 | Protein of unknown function |
| Chr4_A_oryzae_RIB40:4149411-4149623 | AO090102000151 | Ortholog(s) have ubiquitin–protein transferase activity and role in protein import into peroxisome matrix and integral component of peroxisomal membrane localization |
| Chr1_A_oryzae_RIB40:6476509-6476721 | AO090308000007 | Protein of unknown function |
| Chr3_A_oryzae_RIB40:391752-391964 | AO090023000155 | Protein of unknown function |
| Chr6_A_oryzae_RIB40:3081726-3081938 | AO090038000221 | Protein of unknown function |
| Chr7_A_oryzae_RIB40:1923126-1923338 | AO090011000751 | Protein of unknown function |
| Chr2_A_oryzae_RIB40:5473553-5473765 | AO090003001242 | Has domain(s) with predicted ATP binding and inositol pentakisphosphate 2-kinase activity |
| Chr6_A_oryzae_RIB40:1409858-1410070 | AO090020000161 | Ortholog of A. nidulans FGSC A4 : AN6458, A. fumigatus Af293 : Afu3g07420, A. niger CBS 513.88 : An02g10960, An01g01420, An12g05390, and A. oryzae RIB40 : AO090005000921, AO090003001427 |
| Chr6_A_oryzae_RIB40:1409858-1410070 | AO090020000162 | Has domain(s) with predicted UDP-N-acetylmuramate dehydrogenase activity, flavin adenine dinucleotide binding, oxidoreductase activity, oxidoreductase activity, acting on CH-OH group of donors activity |
| Chr6_A_oryzae_RIB40:4172129-4172341 | AO090138000010 | Has domain(s) with predicted 2-dehydropantoate 2-reductase activity, NADP binding, coenzyme binding, oxidoreductase activity, oxidoreductase activity, acting on the CH-OH group of donors, and NAD or NADP as acceptor activity |
| Chr5_A_oryzae_RIB40:1114034-1114246 | AO090701000470 | Ortholog of A. fumigatus Af293 : Afu2g16985, A. wentii : Aspwe1_0171101, A. clavatus NRRL 1 : ACLA_075940, and A. zonatus : Aspzo1_0015705 |
| Chr3_A_oryzae_RIB40:1794701-1794913 | AO090023000683 | Protein of unknown function |
| Chr6_A_oryzae_RIB40:3558859-3559071 | AO090038000040 | Has domain(s) with predicted catalytic activity, catechol 1,2-dioxygenase activity, ferric iron binding, iron ion binding and oxidoreductase activity, more |
| Chr1_A_oryzae_RIB40:3825460-3825672 | AO090005000971 | Has domain(s) with predicted role in biosynthetic process |
| Chr5_A_oryzae_RIB40:4076380-4076592 | AO090113000012 | Ortholog of A. nidulans FGSC A4 : AN6909/BEST2, A. niger CBS 513.88 : An14g05100, A. wentii : Aspwe1_0153378, A. sydowii : Aspsy1_0054771, and A. terreus NIH2624 : ATET_06151 |
| Chr7_A_oryzae_RIB40:2355701-2355913 | AO090011000905 | Ortholog(s) have sequence-specific DNA binding transcription factor activity |
| Chr7_A_oryzae_RIB40:2498610-2498822 | AO090011000954 | Ortholog(s) have cytosol localization |
| Chr8_A_oryzae_RIB40:1654282-1654494 | AO090010000667 | Has domain(s) with predicted iron ion binding and oxidoreductase activity; role in fatty acid biosynthetic process and oxidation-reduction process |
| Chr3_A_oryzae_RIB40:244139-244351 | AO090023000096 | Has domain(s) with predicted catalytic activity, coenzyme binding activity, and role in cellular metabolic process |
| Chr8_A_oryzae_RIB40:1372821-1373033 | AO090010000775 | Ortholog(s) have UDP-N-acetylglucosamine transmembrane transporter activity and role in UDP-N-acetylglucosamine transport, UDP-glucose transport, fungal-type cell wall chitin biosynthetic process, and transmembrane transport |
| Chr1_A_oryzae_RIB40:3919266-3919478 | AO090005000937 | Ortholog of A. nidulans FGSC A4 : AN1323 and A. flavus NRRL 3357 : AFL2T_00913 |
| Chr6_A_oryzae_RIB40:442735-442947 | AO090020000539 | Ortholog(s) have Golgi apparatus, endoplasmic reticulum localization |
| Chr8_A_oryzae_RIB40:2804269-2804481 | AO090010000224 | Ortholog(s) have role in ethanol metabolic process and mitochondrial inner membrane localization |
| Chr8_A_oryzae_RIB40:2804269-2804481 | AO090010000223 | 40S ribosomal protein S2-like protein; predominantly expressed in the hyphal tip region |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





