Submitted:
28 December 2023
Posted:
29 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. GM approaches
2.1. Pathogen Derived Resistance
2.1.1. Protein-Mediated Resistance
2.1.2. Defective interfering virus mediated resistance
2.2. RNA silencing as a major antiviral weapon
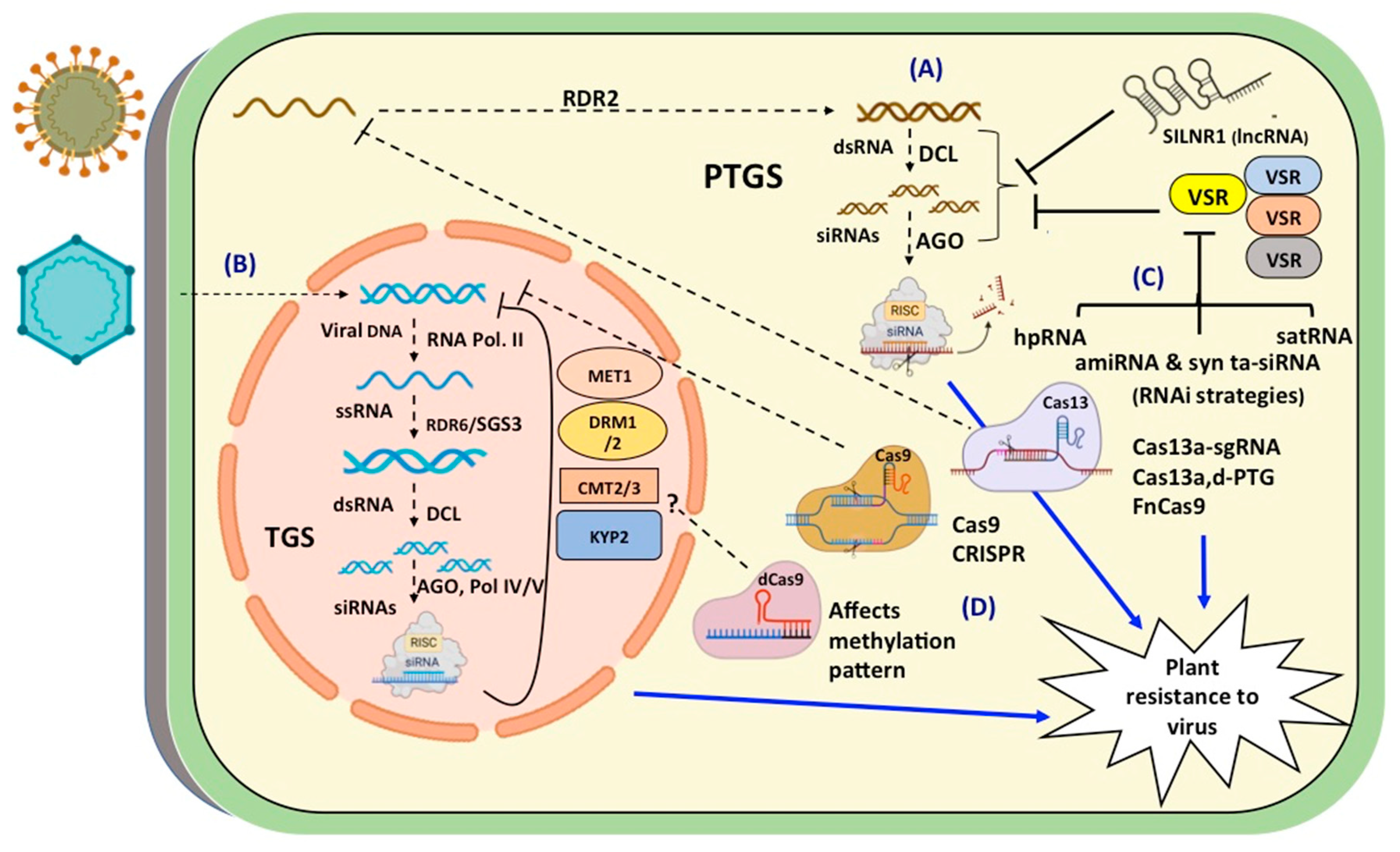
2.2.1. Role of TGS in virus infection
2.2.2. Role of PTGS in virus infection
2.2.3. small RNA shields in virus resistance
3. GE approaches
3.1. Different GE tools
3.2. Manipulation of genome through CRISPR/Cas system
3.2.1. Cas9 editing
3.2.2. Epigentic editors
3.2.3. RNA editors
3.2.4. Base editors
3.2.5. Prime editors
3.2.6. Multiplexing CRISPR/Cas editing
| Strategy | Virus | Plant | References |
|---|---|---|---|
| For DNA Viruses | |||
| ZFN | TYLCCNV, TbCSV BeYDV |
Tobacco |
[182] [237] |
| TALENS | TYLCCNV, TbCSV, TLCYV | [193] | |
| AZP | BSCTV, TYLCV RTBV |
[183], [238], [239], [240], [241], [242] | |
| CRISPR/Cas9 | TYLCV, CLcKV, BSCTV, BCTV, BeYDV, CLcV CaMV, BSCTV WDV BSV ACMV |
Tobacco Arabidopsis Barley Banana Cassava |
[243], [206] [244] [245] [246] [247] [248], [249] |
| For RNA Viruses | |||
| MNs | TRV | Tobacco | [250] |
| SpCas9 | BYSMV SYNV |
Tobacco | [251] [252] |
| FnCas9 | TMV, CMV CMV |
Tobacco Arabidopsis |
[217] |
| LshCas13a | TuMV TMV SRBSDV, RSMV PVY |
Tobacco Rice Potato |
[205] [221] |
| Cas13d/PTG | PVX or PLRV, PVY, PVS | Potato | [233] |
| Cas13a/PTG | PVY | Potato | [233] |
| Cas13a with multiplex gRNAs | PVY | Potato | [253] |
| LbCas12a | CLcMV TEV |
Cotton Tobacco |
[254] [255] |
| Cas12f | CLCuV | Cotton | [256] |
| Multiplex Cas9-gRNAs (9 duplex and 2 triplex) | ChiLCV | Tobacco | [245] |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends in ecology & evolution 2004, 19, 535–544. [Google Scholar]
- Jones, R.A. Global plant virus disease pandemics and epidemics. Plants 2021 10, 233. [CrossRef]
- Sastry, K.S.; A. Zitter, T.; Sastry, K.S.; Zitter, T.A. Management of virus and viroid diseases of crops in the tropics. Plant Virus and Viroid Diseases in the Tropics: Volume 2: Epidemiology and Management 2014, 149-480.
- Zhao, Y.; Yang, X.; Zhou, G.; Zhang, T. Engineering plant virus resistance: from RNA silencing to genome editing strategies. Plant Biotechnology Journal 2020, 18, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.A.; Gopal, D.S.; Sudhakar, C. GM Crops for Plant Virus Resistance: A Review. Genetically Modified Crops: Current Status, Prospects and Challenges 2021, 2, 257-337. [Google Scholar]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proceedings of the national academy of sciences 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed]
- Legg, J.P.; Shirima, R.; Tajebe, L.S.; Guastella, D.; Boniface, S.; Jeremiah, S.; Nsami, E.; Chikoti, P.; Rapisarda, C. Biology and management of Bemisia whitefly vectors of cassava virus pandemics in Africa. Pest management science 2014, 70, 1446–1453. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Stubbs, G.; Culver, J.N. Coat protein interactions involved in tobacco mosaic tobamovirus cross-protection. Virology 1998, 248, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Muller, G.W.; Rezende, J.A. Preimmunization: Applications and perspectives in virus disease control. In Diseases of Fruits and Vegetables Volume I: Diagnosis and Management, Dordrecht: Springer Netherlands 2004, (pp. 361-395).
- Sanford, J.C.; Johnston, S.A. The concept of parasite-derived resistance—deriving resistance genes from the parasite's own genome. Journal of Theoretical Biology 1985, 113, 395–405. [Google Scholar] [CrossRef]
- Nicaise, V. Crop immunity against viruses: outcomes and future challenges. Frontiers in plant science 2014, 5, 660. [Google Scholar] [CrossRef] [PubMed]
- Galvez, L.C.; Banerjee, J.; Pinar, H.; Mitra, A. Engineered plant virus resistance. Plant Science 2014, 228, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Csorba, T.; Pantaleo, V.; Burgyán, J. RNA silencing: an antiviral mechanism. Advances in virus research 2009, 75, 35–230. [Google Scholar] [PubMed]
- Sharma, K.K.; Bhatnagar-Mathur, P.; Thorpe, T.A. Genetic transformation technology: status and problems. In Vitro Cellular & Developmental Biology-Plant 2005, 41, 102–112. [Google Scholar]
- Fitchen, J.H.; Beachy, R.N. Genetically engineered protection against viruses in transgenic plants. Annual Review of Microbiology 1993, 47, 739–763. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D. Novel strategies for engineering virus resistance in plants. Current Opinion in Biotechnology 1994, 5, 117–124. [Google Scholar] [CrossRef]
- Abel, P.P.; Nelson, R.S.; De, B.; Hoffmann, N.; Rogers, S.G.; Fraley, R.T.; Beachy, R.N. Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science 1986, 232, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Beachy, R.N. Coat–protein–mediated resistance to tobacco mosaic virus: discovery mechanisms and exploitation. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 1999, 354, 659–664. [Google Scholar] [CrossRef]
- Dasgupta, I.; Malathi, V.G.; Mukherjee, S.K. Genetic engineering for virus resistance. Current science 2003, 84, 341–354. [Google Scholar]
- Nejidat, A.; Beachy, R.N. Transgenic tobacco plants expressing a coat protein gene. Molecular Plant-Microbe Interactions 1990, 3, 247–251. [Google Scholar] [CrossRef]
- Anderson, E.J.; Stark, D.M.; Nelson, R.S.; Powell, P.A.; Tumer, N.E.; Beachy, R.N. Transgenic plants that express the coat protein genes of tobacco mosaic virus or alfalfa mosaic virus interfere with disease development of some nonrelated viruses. Phytopathology 1989, 79, 1284–1290. [Google Scholar] [CrossRef]
- Lawson, C.; Kaniewski, W.; Haley, L.; Rozman, R.; Newell, C.; Sanders, P.; Tumer, N.E. Engineering resistance to mixed virus infection in a commercial potato cultivar: resistance to potato virus X and potato virus Y in transgenic Russet Burbank. Bio/technology 1990, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Chapman, S.; Kavanagh, T.; Baulcombe, D. Potato virus X as a vector for gene expression in plants. The Plant Journal 1992, 2, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Taschner, P.E.; Van Marle, G.; Brederode, F.T.; Tumer, N.E.; Bol, J.F. Plants transformed with a mutant alfalfa mosaic virus coat protein gene are resistant to the mutant but not to wild-type virus. Virology 1994, 203, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Reimann-Philipp, U.; Beachy, R.N. December. The mechanism (s) of coat protein-mediated resistance against tobacco mosaic virus. In Seminars in Virology, Academic Press 1993, (Vol. 4, No. 6, pp. 349-356).
- Yusibov, V.M.; Loesch-Fries, L.S. N-terminal basic amino acids of alfalfa mosaic virus coat protein involved in the initiation of infection. Virology 1995, 208, 405–407. [Google Scholar] [CrossRef] [PubMed]
- Clark, W.G.; Fitchen, J.; Nejidat, A.; Deom, C.M.; Beachy, R.N. Studies of coat protein-mediated resistance to tobacco mosaic virus (TMV). II. Challenge by a mutant with altered virion surface does not overcome resistance conferred by TMV coat protein. Journal of general virology 1995, 76, 2613–2617. [Google Scholar] [CrossRef] [PubMed]
- Asurmendi, S.; Berg, R.H.; Smith, T.J.; Bendahmane, M.; Beachy, R.N. Aggregation of TMV CP plays a role in CP functions and in coat-protein-mediated resistance. Virology 2007, 366, 98–106. [Google Scholar] [CrossRef]
- Lapidot, M.; Gafny, R.; Ding, B.; Wolf, S.; Lucas, W.J.; Beachy, R.N. A dysfunctional movement protein of tobacco mosaic virus that partially modifies the plasmodesmata and limits virus spread in transgenic plants. The Plant Journal 1993, 4, 959–970. [Google Scholar] [CrossRef]
- Cooper, B.; Lapidot, M.; Heick, J.A.; Dodds, J.A.; Beachy, R.N. A defective movement protein of TMV in transgenic plants confers resistance to multipleviruses whereas the functional analog increases susceptibility. Virology 1995, 206, 307–313. [Google Scholar] [CrossRef]
- Beck, D.L.; Van Dolleweerd, C.J.; Lough, T.J.; Balmori, E.; Voot, D.M.; Andersen, M.T.; O'Brien, I.E.; Forster, R.L. Disruption of virus movement confers broad-spectrum resistance against systemic infection by plant viruses with a triple gene block. Proceedings of the National Academy of Sciences 1994, 91, 10310–10314. [Google Scholar] [CrossRef] [PubMed]
- Golemboski, D.B.; Lomonossoff, G.P.; Zaitlin, M. Plants transformed with a tobacco mosaic virus nonstructural gene sequence are resistant to the virus. Proceedings of the National Academy of Sciences 1990, 87, 6311–6315. [Google Scholar] [CrossRef]
- Hanson, S.F.; Maxwell, D.P. trans-Dominant inhibition of geminiviral DNA replication by bean golden mosaic geminivirus rep gene mutants. Phytopathology 1999, 89, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Sangaré, A.; Deng, D.; Fauquet, C.M.; Beachy, R.N. Resistance to African cassava mosaic virus conferred by a mutant of the putative NTP-binding domain of the Rep gene (AC1) in Nicotiana benthamiana. Molecular Breeding 1999, 5, 95–102. [Google Scholar] [CrossRef]
- Donson, J.; Kearney, C.M.; Turpen, T.H.; Khan, I.A.; Kurath, G.; Turpen, A.M.; Jones, G.E.; Dawson, W.O.; Lewandowski, D.J. Broad resistance to tobamoviruses is mediated by a modified tobacco mosaic virus replicase transgene. Molecular plant-microbe interactions: MPMI 1993, 6, 635–642. [Google Scholar] [CrossRef]
- Tenllado, F.; García-Luque, I.; Serra, M.T.; Díaz-Ruíz, J.R. Resistance to pepper mild mottle tobamovirus conferred by the 54-kDa gene sequence in transgenic plants does not require expression of the wild-type 54-kDa protein. Virology 1996, 219, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Marano, M.R.; Baulcombe, D. Pathogen-derived resistance targeted against the negative-strand RNA of tobacco mosaic virus: RNA strand-specific gene silencing? The Plant Journal 1998, 13, 537–546. [Google Scholar] [CrossRef]
- Greene, A.E.; Allison, R.F. Recombination between viral RNA and transgenic plant transcripts. Science 1994, 263, 1423–1425. [Google Scholar] [CrossRef] [PubMed]
- Chávez-Calvillo, G.; Contreras-Paredes, C.A.; Mora-Macias, J.; Noa-Carrazana, J.C.; Serrano-Rubio, A.A.; Dinkova, T.D.; Carrillo-Tripp, M.; Silva-Rosales, L. Antagonism or synergism between papaya ringspot virus and papaya mosaic virus in Carica papaya is determined by their order of infection. Virology 2016, 489, 179–191. [Google Scholar] [CrossRef]
- Hu, C.C.; Hsu, Y.H.; Lin, N.S. Satellite RNAs and satellite viruses of plants. Viruses 2009, 1, 1325–1350. [Google Scholar] [CrossRef] [PubMed]
- Palukaitis, P.; Roossinck, M.J.; Dietzgen, R.G.; Francki, R.I. Cucumber mosaic virus. Advances in virus research 1992, 41, 281–348. [Google Scholar]
- Patil, B.L.; Dasgupta, I. Defective interfering DNAs of plant viruses. Critical reviews in plant sciences 2006, 25, 47–64. [Google Scholar] [CrossRef]
- Burgyan, J.; Grieco, F.; Russo, M. A defective interfering RNA molecule in cymbidium ringspot virus infections. Journal of General Virology 1989, 70, 235–239. [Google Scholar] [CrossRef]
- Stanley, J.; Frischmuth, T.; Ellwood, S. Defective viral DNA ameliorates symptoms of geminivirus infection in transgenic plants. Proceedings of the National Academy of Sciences 1990, 87, 6291–6295. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Wang, J.; Simon, A.E. Satellite RNA-mediated resistance to turnip crinkle virus in Arabidopsis involves a reduction in virus movement. The Plant Cell 1997, 9, 2051–2063. [Google Scholar] [PubMed]
- Budzyńska, D.; Zwart, M.P.; Hasiów-Jaroszewska, B. Defective RNA Particles of Plant Viruses—Origin, Structure and Role in Pathogenesis. Viruses 2022, 14, 2814. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.X.; Au, P.C.K.; Shi, B.J.; Smith, N.A.; Dennis, E.S.; Guo, H.S.; Zhou, C.Y.; Wang, M.B. Satellite RNAs interfere with the function of viral RNA silencing suppressors. Frontiers in Plant Science 2015, 6, 281. [Google Scholar] [CrossRef]
- Ibrahim, A.B.; Aragão, F.J. RNAi-mediated resistance to viruses in genetically engineered plants. Plant Gene Silencing: Methods and Protocols. 2015, pp.81-92.
- Mueller, E.; Gilbert, J.; Davenport, G.; Brigneti, G.; Baulcombe, D.C. Homology-dependent resistance: transgenic virus resistance in plants related to homology-dependent gene silencing. The Plant Journal 1995, 7, 1001–1013. [Google Scholar] [CrossRef]
- Schwind, N.; Zwiebel, M.; Itaya, A.; Ding, B.; WANG, M.B.; Krczal, G.; Wassenegger, M. RNAi-mediated resistance to Potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct. Molecular plant pathology 2009, 10, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Prins, M.; Goldbach, R. The emerging problem of tospovirus infection and nonconventional methods of control. Trends in microbiology 1998, 6, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, H.; Rajasubramaniam, S.; Rajam, M.V.; Dasgupta, I. RNA-interference in rice against Rice tungro bacilliform virus results in its decreased accumulation in inoculated rice plants. Transgenic research 2008, 17, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, D.N.; Martin, D.P.; Thomson, J.A. Transgenic strategies for developing crops resistant to geminiviruses. Plant Science 2009, 176, 1–11. [Google Scholar] [CrossRef]
- Sanan-Mishra, N.; Abdul Kader Jailani, A.; Mandal, B.; Mukherjee, S.K. Secondary siRNAs in plants: biosynthesis, various functions, and applications in virology. Frontiers in Plant Science 2021, 12, 610283. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, B.; Nasir, I.A.; Aslam, U.; Husnain, T. How RNA interference combat viruses in plants. Rijeka: InTech, Functional Genomics 2012, pp.113-130. [Google Scholar]
- Mette, M.F.; Aufsatz, W.; Van der Winden, J.; Matzke, M.A.; Matzke, A.J.M. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. The EMBO journal 2000, 19, 5194–5201. [Google Scholar] [CrossRef]
- Kooter, J.M.; Matzke, M.A.; Meyer, P. Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends in plant science 1999, 4, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, P.; Voinnet, O. The diversity of RNA silencing pathways in plants. TRENDS in Genetics 2006, 22, 268–280. [Google Scholar] [CrossRef]
- Palauqui, J.C.; Elmayan, T.; Pollien, J.M.; Vaucheret, H. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. The EMBO journal 1997, 16, 4738–4745. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O.; Vain, P.; Angell, S.; Baulcombe, D.C. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 1998, 95, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Kumakura, N.; Takeda, A.; Fujioka, Y.; Motose, H.; Takano, R.; Watanabe, Y. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies 2009, FEBS letters, 583, 1261-1266.
- Pélissier, T.; Thalmeir, S.; Kempe, D.; Sänger, H.L.; Wassenegger, M. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic acids research 1999, 27, 1625–1634. [Google Scholar] [CrossRef] [PubMed]
- Wassenegger, M. RNA-directed DNA methylation. Plant Gene Silencing 2000, 83–100. [Google Scholar]
- Melnyk, C.W.; Molnar, A.; Bassett, A.; Baulcombe, D.C. Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Current Biology 2011, 21, 1678–1683. [Google Scholar] [CrossRef] [PubMed]
- Brosnan, C.A.; Mitter, N.; Christie, M.; Smith, N.A.; Waterhouse, P.M.; Carroll, B.J. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proceedings of the National Academy of Sciences 2007, 104, 14741–14746. [Google Scholar] [CrossRef] [PubMed]
- Pikaard, C.S.; Haag, J.R.; Ream, T.; Wierzbicki, A.T. Roles of RNA polymerase IV in gene silencing. Trends in plant science 2008, 13, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Sigman, M.J.; Panda, K.; Kirchner, R.; McLain, L.L.; Payne, H.; Peasari, J.R.; Husbands, A.Y.; Slotkin, R.K.; McCue, A.D. An siRNA-guided ARGONAUTE protein directs RNA polymerase V to initiate DNA methylation. Nature Plants 2021, 7, 1461–1474. [Google Scholar] [CrossRef]
- Raja, P.; Sanville, B.C.; Buchmann, R.C.; Bisaro, D.M. Viral genome methylation as an epigenetic defense against geminiviruses. Journal of virology 2008, 82, 8997–9007. [Google Scholar] [CrossRef]
- Mirouze, M.; Paszkowski, J. Epigenetic contribution to stress adaptation in plants. Current opinion in plant biology 2011, 14, 267–274. [Google Scholar] [CrossRef]
- Wassenegger, M. The role of the RNAi machinery in heterochromatin formation. Cell 2005, 122, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Aufsatz, W.; Mette, M.; Matzke, A.; Matzke, M. The role of MET1 in RNA-directed de novo and maintenance methylation of CG dinucleotides. Plant molecular biology 2004, 54, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Aufsatz, W.; Zilberman, D.; Mette, M.F.; Huang, M.S.; Matzke, M.; Jacobsen, S.E. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Current biology 2003, 13, 2212–2217. [Google Scholar] [CrossRef] [PubMed]
- Pooggin, M.M. How can plant DNA viruses evade siRNA-directed DNA methylation and silencing? International journal of molecular sciences 2013, 14, 15233–15259. [Google Scholar] [CrossRef]
- Raja, P.; Wolf, J.N.; Bisaro, D.M. RNA silencing directed against geminiviruses: post-transcriptional and epigenetic components. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 2010, 1799, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, B.; Sanfaçon, H. Symptom recovery in virus-infected plants: revisiting the role of RNA silencing mechanisms. Virology 2015, 479, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Bischof, S.; Wang, H.; Feng, S.; Lee, T.F.; Teng, C.; Chen, X.; Park, S.Y.; Liu, L.; Gallego-Bartolome, J.; Liu, W. A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 2015, 163, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, K.I.; Eskelin, K.; Bašić, M.; De, S.; Lõhmus, A.; Varjosalo, M.; Mäkinen, K. Molecular insights into the function of the viral RNA silencing suppressor HC-Pro. The Plant Journal 2016, 85, 30–45. [Google Scholar] [CrossRef]
- Mäkinen, K.; De, S. The significance of methionine cycle enzymes in plant virus infections. Current opinion in plant biology 2019, 50, 67–75. [Google Scholar] [CrossRef]
- Vaucheret, H.; Béclin, C.; Fagard, M. Post-transcriptional gene silencing in plants. Journal of cell science 2001, 114, 3083–3091. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, K.B.G. Taking Some of the Mystery out of Host∶ Virus Interactions. PLoS Pathogens 2011, 7, e1002033. [Google Scholar] [CrossRef] [PubMed]
- Jaubert, M.; Bhattacharjee, S.; Mello, A.F.; Perry, K.L.; Moffett, P. ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant physiology 2011, 156, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, X.; Singh, J.; Li, D.; Qu, F. Temperature-dependent survival of Turnip crinkle virus-infected arabidopsis plants relies on an RNA silencing-based defense that requires dcl2, AGO2, and HEN1. Journal of virology 2012, 86, 6847–6854. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, P.; Obrępalska-Stęplowska, A. A single amino acid substitution in movement protein of tomato torrado virus influences ToTV infectivity in Solanum lycopersicum. Virus research 2016, 213, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.Y.; Weinmann, L.; Gaidatzis, D.; Pei, Y.; Zavolan, M.; Tuschl, T.; Meister, G. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. Rna 2008, 14, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, R.; Doudna, J.A. dsRNA with 5′ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. The EMBO journal 2009, 28, 545–555. [Google Scholar] [CrossRef]
- Glick, E.; Zrachya, A.; Levy, Y.; Mett, A.; Gidoni, D.; Belausov, E.; Citovsky, V.; Gafni, Y. Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proceedings of the National Academy of Sciences 2008, 105, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Induction and suppression of RNA silencing: insights from viral infections. Nature Reviews Genetics 2005, 6, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Fahim, M.; Millar, A.A.; Wood, C.C.; Larkin, P.J. Resistance to Wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant biotechnology journal 2012, 10, 150–163. [Google Scholar] [CrossRef] [PubMed]
- KUNG, Y.J.; LIN, S.S.; HUANG, Y.L.; CHEN, T.C.; Harish, S.S.; CHUA, N.H.; YEH, S.D. Multiple artificial microRNAs targeting conserved motifs of the replicase gene confer robust transgenic resistance to negative-sense single-stranded RNA plant virus. Molecular plant pathology 2012, 13, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Niu, Y.; Zhang, K.; Liu, Y.; Zhou, X. Virus-derived transgenes expressing hairpin RNA give immunity to Tobacco mosaic virus and Cucumber mosaic virus. Virology journal 2011, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nicola-Negri, E.D.; Brunetti, A.; Tavazza, M.; Ilardi, V. Hairpin RNA-mediated silencing of Plum pox virus P1 and HC-Pro genes for efficient and predictable resistance to the virus. Transgenic Research 2005, 14, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Kamachi, S.; Mochizuki, A.; Nishiguchi, M.; Tabei, Y. Transgenic Nicotiana benthamiana plants resistant to cucumber green mottle mosaic virus based on RNA silencing. Plant Cell Reports 2007, 26, 1283–1288. [Google Scholar] [CrossRef] [PubMed]
- Schwind, N.; Zwiebel, M.; Itaya, A.; Ding, B.; WANG, M.B.; Krczal, G.; Wassenegger, M. RNAi-mediated resistance to Potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct. Molecular plant pathology 2009, 10, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, A.; Ramos, P.L.; Fiallo, E.; Callard, D.; Sánchez, Y.; Peral, R.; Rodríguez, R.; Pujol, M. Intron–hairpin RNA derived from replication associated protein C1 gene confers immunity to Tomato yellow leaf curl virus infection in transgenic tomato plants. Transgenic Research 2006, 15, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Zrachya, A.; Kumar, P.P.; Ramakrishnan, U.; Levy, Y.; Loyter, A.; Arazi, T.; Lapidot, M.; Gafni, Y. Production of siRNA targeted against TYLCV coat protein transcripts leads to silencing of its expression and resistance to the virus. Transgenic research 2007, 16, 385–398. [Google Scholar] [CrossRef]
- Arif, M.; Azhar, U.; Arshad, M.; Zafar, Y.; Mansoor, S.; Asad, S. Engineering broad-spectrum resistance against RNA viruses in potato. Transgenic research 2012, 21, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Krubphachaya, P.; Juricek, M.; Kertbundit, S. Induction of RNA-mediated resistance to papaya ringspot virus type W. BMB Reports 2007, 40, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Tougou, M.; Furutani, N.; Yamagishi, N.; Shizukawa, Y.; Takahata, Y.; Hidaka, S. Development of resistant transgenic soybeans with inverted repeat-coat protein genes of soybean dwarf virus. Plant cell reports 2006, 25, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, M.J.; Pak, J.H.; Im, H.H.; Lee, D.H.; Kim, K.H.; Lee, J.H.; Kim, D.H.; Choi, H.K.; Jung, H.W.; Chung, Y.S. RNAi-mediated Soybean mosaic virus (SMV) resistance of a Korean Soybean cultivar. Plant Biotechnology Reports 2016, 10, 257–267. [Google Scholar] [CrossRef]
- Ludlow, E.J.; Mouradov, A.; Spangenberg, G.C. Post-transcriptional gene silencing as an efficient tool for engineering resistance to white clover mosaic virus in white clover (Trifolium repens). Journal of Plant Physiology 2009, 166, 1557–1567. [Google Scholar] [CrossRef] [PubMed]
- Vanderschuren, H.; Alder, A.; Zhang, P.; Gruissem, W. Dose-dependent RNAi-mediated geminivirus resistance in the tropical root crop cassava. Plant molecular biology 2009, 70, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, U.K.; Ganapathi, T.R.; Hadapad, A.B. Transgenic banana plants expressing small interfering RNAs targeted against viral replication initiation gene display high-level resistance to banana bunchy top virus infection. Journal of general virology 2012, 93, 1804–1813. [Google Scholar] [CrossRef] [PubMed]
- Soler, N.; Plomer, M.; Fagoaga, C.; Moreno, P.; Navarro, L.; Flores, R.; Pena, L. Transformation of Mexican lime with an intron-hairpin construct expressing untranslatable versions of the genes coding for the three silencing suppressors of Citrus tristeza virus confers complete resistance to the virus. Plant Biotechnology Journal 2012, 10, 597–608. [Google Scholar] [CrossRef]
- Winterhagen, P.; Dubois, C.; Sinn, M.; Wetzel, T.; Reustle, G.M. Gene silencing and virus resistance based on defective interfering constructs in transgenic Nicotiana benthamiana is not linked to accumulation of siRNA. Plant Physiology and Biochemistry 2009, 47, 739–742. [Google Scholar] [CrossRef] [PubMed]
- Patil, B.L.; Bagewadi, B.; Yadav, J.S.; Fauquet, C.M. Mapping and identification of cassava mosaic geminivirus DNA-A and DNA-B genome sequences for efficient siRNA expression and RNAi based virus resistance by transient agro-infiltration studies. Virus research 2016, 213, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Ogamino, T.; Hiraguri, A.; Nakazono-Nagaoka, E.; Uehara-Ichiki, T.; Nakajima, M.; Akutsu, K.; Omura, T.; Sasaya, T. Strong resistance against Rice grassy stunt virus is induced in transgenic rice plants expressing double-stranded RNA of the viral genes for nucleocapsid or movement proteins as targets for RNA interference. Phytopathology 2013, 103, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Liu, H.; Hao, J.; Li, J.; Luo, L. Expression profiling and regulatory network of cucumber microRNAs and their putative target genes in response to cucumber green mottle mosaic virus infection. Archives of virology 2019, 164, 1121–1134. [Google Scholar] [CrossRef] [PubMed]
- Ai, T.; Zhang, L.; Gao, Z.; Zhu, C.X.; Guo, X. Highly efficient virus resistance mediated by artificial microRNAs that target the suppressor of PVX and PVY in plants. Plant Biology 2011, 13, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Lafforgue, G.; Martínez, F.; Niu, Q.W.; Chua, N.H.; Daròs, J.A.; Elena, S.F. Improving the effectiveness of artificial microRNA (amiR)-mediated resistance against Turnip mosaic virus by combining two amiRs or by targeting highly conserved viral genomic regions. Journal of Virology 2013, 87, 8254–8256. [Google Scholar] [CrossRef] [PubMed]
- Gago-Zachert, S.; Schuck, J.; Weinholdt, C.; Knoblich, M.; Pantaleo, V.; Grosse, I.; Gursinsky, T.; Behrens, S.E. Highly efficacious antiviral protection of plants by small interfering RNAs identified in vitro. Nucleic acids research 2019, 47, 9343–9357. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cheng, X.; Cai, J.; Zhan, L.; Wu, X.; Liu, Q.; Wu, X. Multiple virus resistance using artificial trans-acting siRNAs. Journal of virological methods 2016, 228, 16–20. [Google Scholar] [CrossRef]
- López-Dolz, L.; Spada, M.; Daròs, J.A.; Carbonell, A. Fine-tune control of targeted RNAi efficacy by plant artificial small RNAs. Nucleic Acids Research 2020, 48, 6234–6250. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J. Classification and comparison of small RNAs from plants. Annual review of plant biology 2013, 64, 137–159. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Bazzini, A.A.; Hopp, H.E.; Beachy, R.N.; Asurmendi, S. Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proceedings of the National Academy of Sciences 2007, 104, 12157–12162. [Google Scholar] [CrossRef] [PubMed]
- Tagami, Y.; Inaba, N.; Kutsuna, N.; Kurihara, Y.; Watanabe, Y. Specific enrichment of miRNAs in Arabidopsis thaliana infected with Tobacco mosaic virus. DNA research 2007, 14, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, Z.; Wang, Y.; Zheng, L.; Ye, R.; Ji, Y.; Zhao, S.; Ji, S.; Liu, R.; Xu, L.; Zheng, H. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. Elife 2015, 4, e05733. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jiao, X.; Kong, X.; Hamera, S.; Wu, Y.; Chen, X.; Fang, R.; Yan, Y. A signaling cascade from miR444 to RDR1 in rice antiviral RNA silencing pathway. Plant Physiology 2016, 170, 2365–2377. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Zhai, Y.; Zhou, L.; Ai, X.; Chen, J.; Yan, F. A Novel miRNA in Rice Associated with the Low Seed Setting Rate Symptom of Rice Stripe Virus. International Journal of Molecular Sciences 2023, 24, 3675. [Google Scholar] [CrossRef]
- Niu, Q.W.; Lin, S.S.; Reyes, J.L.; Chen, K.C.; Wu, H.W.; Yeh, S.D.; Chua, N.H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nature biotechnology 2006, 24, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Vaucheret, H.; Vazquez, F.; Crété, P.; Bartel, D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & development 2004, 18, 1187–1197. [Google Scholar]
- Lafforgue, G.; Martínez, F.; Sardanyés, J.; De La Iglesia, F.; Niu, Q.W.; Lin, S.S.; Solé, R.V.; Chua, N.H.; Daròs, J.A.; Elena, S.F. Tempo and mode of plant RNA virus escape from RNA interference-mediated resistance. Journal of virology 2011, 85, 9686–9695. [Google Scholar] [CrossRef] [PubMed]
- Cisneros, A.E.; Carbonell, A. Artificial small RNA-based silencing tools for antiviral resistance in plants. Plants 2020, 9, 669. [Google Scholar] [CrossRef]
- Schwab, R.; Ossowski, S.; Riester, M.; Warthmann, N.; Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. The Plant Cell 2006, 18, 1121–1133. [Google Scholar] [CrossRef]
- Ramesh, S.V.; Ratnaparkhe, M.B.; Kumawat, G.; Gupta, G.K.; Husain, S.M. Plant miRNAome and antiviral resistance: a retrospective view and prospective challenges. Virus Genes 2014, 48, 1–14. [Google Scholar] [CrossRef]
- Reynolds, A.; Leake, D.; Boese, Q.; Scaringe, S.; Marshall, W.S.; Khvorova, A. Rational siRNA design for RNA interference. Nature biotechnology 2004, 22, 326–330. [Google Scholar] [CrossRef]
- Duan, C.G.; Wang, C.H.; Fang, R.X.; Guo, H.S. Artificial microRNAs highly accessible to targets confer efficient virus resistance in plants. Journal of virology 2008, 82, 11084–11095. [Google Scholar] [CrossRef]
- Van Vu, T.; Choudhury, N.R.; Mukherjee, S.K. Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, Tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Research 2013, 172, 35–45. [Google Scholar]
- Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bartel, B. MicroRNAs directing siRNA biogenesis. Nature structural & molecular biology 2005, 12, 569–571. [Google Scholar]
- Wang, M.B.; Masuta, C.; Smith, N.A.; and Shimura, H. RNA Silencing and Plant Viral Diseases. 1275 MPMI 2012, 25, 1275–1285. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nature Reviews Microbiology 2013, 11, 745–760. [Google Scholar] [CrossRef]
- Revers, F.; Nicaise, V. Plant resistance to infection by viruses. eLS 2014. [Google Scholar]
- Khalid, A.; Zhang, Q.; Yasir, M.; Li, F. Small RNA based genetic engineering for plant viral resistance: application in crop protection. Frontiers in microbiology 2017, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.D.; Gan, Q.H.; Chi, X.Y.; Qin, S. Roles of microRNA in plant defense and virus offense interaction. Plant cell reports 2008, 27, 1571–1579. [Google Scholar] [CrossRef]
- Waterhouse, P.M.; Graham, M.W.; Wang, M.B. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proceedings of the National Academy of Sciences 1998, 95, 13959–13964. [Google Scholar] [CrossRef]
- Rosa, C.; Kuo, Y.W.; Wuriyanghan, H.; Falk, B.W. RNA interference mechanisms and applications in plant pathology. Annual review of phytopathology 2018, 56, 581–610. [Google Scholar] [CrossRef] [PubMed]
- Gaffar, F.Y.; Koch, A. Catch me if you can! RNA silencing-based improvement of antiviral plant immunity. Viruses 2019, 11, 673. [Google Scholar] [CrossRef] [PubMed]
- Voloudakis, A.E.; Kaldis, A.; Patil, B.L. RNA-based vaccination of plants for control of viruses. Annual Review of Virology 2022, 9, 521–548. [Google Scholar] [CrossRef] [PubMed]
- Tenllado, F.; Dıaz-Ruız, J.R. Double-stranded RNA-mediated interference with plant virus infection. Journal of virology 2001, 75, 12288–12297. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, A.; Carlos, N.; Ruiz, Y.; Callard, D.; Sánchez, Y.; Ochagavía, M.E.; Seguin, J.; Malpica-López, N.; Hohn, T.; Lecca, M.R.; Pérez, R. Field trial and molecular characterization of RNAi-transgenic tomato plants that exhibit resistance to tomato yellow leaf curl geminivirus. Molecular Plant-Microbe Interactions 2016, 29, 197–209. [Google Scholar] [CrossRef]
- Walsh, H.A.; Vanderschuren, H.; Taylor, S.; Rey, M.E.C. RNA silencing of South African cassava mosaic virus in transgenic cassava expressing AC1/AC4 hp-RNA induces tolerance. Biotechnology Reports 2019, 24, e00383. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tanti, B.; Patil, B.L.; Mukherjee, S.K.; Sahoo, L. RNAi-derived transgenic resistance to Mungbean yellow mosaic India virus in cowpea. PLoS One 2017, 12, e0186786. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Mao, L.; Qi, Y. Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant physiology 2012, 160, 990–999. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Peragine, A.; Park, M.Y.; Poethig, R.S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes & development 2005, 19, 2164–2175. [Google Scholar]
- Vazquez, F.; Gasciolli, V.; Crété, P.; Vaucheret, H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Current Biology 2004, 14, 346–351. [Google Scholar] [CrossRef]
- Gasciolli, V.; Mallory, A.C.; Bartel, D.P.; Vaucheret, H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Current Biology 2005, 15, 1494–1500. [Google Scholar] [CrossRef]
- Carbonell, A.; Takeda, A.; Fahlgren, N.; Johnson, S.C.; Cuperus, J.T.; Carrington, J.C. New generation of artificial MicroRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant physiology 2014, 165, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A. Secondary small interfering RNA-based silencing tools in plants: an update. Frontiers in Plant Science 2019, 10, 687. [Google Scholar] [CrossRef] [PubMed]
- Wierzbicki, A.T.; Blevins, T.; Swiezewski, S. Long noncoding RNAs in plants. Annual Review of Plant Biology 2021, 72, 245–271. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.I.; Alam, M.; Lightfoot, D.A.; Gurha, P.; Afzal, A.J. Classification and experimental identification of plant long non-coding RNAs. Genomics 2019, 111, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Kaya, S.B.; Cagirici, H.B. Long non-coding RNA in plants in the era of reference sequences. Frontiers in Plant Science 2020, 11, 276. [Google Scholar] [CrossRef] [PubMed]
- Taliansky, M.; Samarskaya, V.; Zavriev, S.K.; Fesenko, I.; Kalinina, N.O.; Love, A.J. RNA-based technologies for engineering plant virus resistance. Plants 2021, 10, 82. [Google Scholar] [CrossRef]
- Gelaw, T.A.; Sanan-Mishra, N. Non-coding RNAs in response to drought stress. International journal of molecular sciences 2021, 22, 12519. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S. The state of long non-coding RNA biology. Non-coding RNA 2018, 4, 17. [Google Scholar] [CrossRef]
- Prasad, A.; Prasad, M. Host-virus interactions mediated by long non-coding RNAs. Virus Research 2021, 298, 198402. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, W.; Yang, Y.; Li, X.; Chen, T.; Liu, T.; Ma, N.; Yang, X.; Liu, R.; Zhang, B. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Scientific reports 2015, 5, 16946. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.H.; Sun, Y.D.; Atallah, O.O.; Huguet-Tapia, J.C.; Noble, J.D.; Folimonova, S.Y. A long non-coding RNA of Citrus tristeza virus: Role in the virus interplay with the host immunity. Viruses 2019, 11, 436. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, N.; Bujarski, J.J. Long noncoding RNAs in plant viroids and viruses: a review. Pathogens 2020, 9, 765. [Google Scholar] [CrossRef]
- Navarro, B.; Gisel, A.; Rodio, M.E.; Delgado, S.; Flores, R.; Di Serio, F. Viroids: how to infect a host and cause disease without encoding proteins. Biochimie 2012, 94, 1474–1480. [Google Scholar] [CrossRef] [PubMed]
- Dadami, E.; Boutla, A.; Vrettos, N.; Tzortzakaki, S.; Karakasilioti, I.; Kalantidis, K. DICER-LIKE 4 but not DICER-LIKE 2 may have a positive effect on potato spindle tuber viroid accumulation in Nicotiana benthamiana. Molecular plant 2013, 6, 232–234. [Google Scholar] [CrossRef]
- Karlik, E.; Ari, S.; Gozukirmizi, N. LncRNAs: genetic and epigenetic effects in plants. Biotechnology & Biotechnological Equipment 2019, 33, 429–439. [Google Scholar]
- Yang, Y.; Liu, T.; Shen, D.; Wang, J.; Ling, X.; Hu, Z.; Chen, T.; Hu, J.; Huang, J.; Yu, W.; Dou, D. Tomato yellow leaf curl virus intergenic siRNAs target a host long noncoding RNA to modulate disease symptoms. PLoS pathogens 2019, 15, e1007534. [Google Scholar] [CrossRef] [PubMed]
- Bak, R.O.; Gomez-Ospina, N.; Porteus, M.H. Gene editing on center stage. Trends in Genetics 2018, 34, 600–611. [Google Scholar] [CrossRef]
- Saurabh, S. Genome editing: revolutionizing the crop improvement. Plant Molecular Biology Reporter 2021, 39, 752–772. [Google Scholar] [CrossRef]
- Mushtaq, M.; Mukhtar, S.; Sakina, A.; Dar, A.A.; Bhat, R.; Deshmukh, R.; Molla, K.; Kundoo, A.A.; Dar, M.S. Tweaking genome-editing approaches for virus interference in crop plants. Plant Physiology and Biochemistry 2020, 147, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Jurica, M.S.; Stoddard, B.L. Homing endonucleases: structure, function and evolution. Cellular and Molecular Life Sciences CMLS 1999, 55, 1304–1326. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Joshi, R.K.; Zhao, K. Genome editing in rice: recent advances, challenges, and future implications. Frontiers in Plant Science 2018, 9, 1361. [Google Scholar] [CrossRef] [PubMed]
- Townsend, J.A.; Wright, D.A.; Winfrey, R.J.; Fu, F.; Maeder, M.L.; Joung, J.K.; Voytas, D.F. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 2009, 459, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Puchta, H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. Journal of experimental botany 2005, 56, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, K.; Osakabe, Y.; Toki, S. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proceedings of the National Academy of Sciences 2010, 107, 12034–12039. [Google Scholar] [CrossRef]
- Zhang, F.; Maeder, M.L.; Unger-Wallace, E.; Hoshaw, J.P.; Reyon, D.; Christian, M.; Li, X.; Pierick, C.J.; Dobbs, D.; Peterson, T.; Joung, J.K. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proceedings of the National Academy of Sciences 2010, 107, 12028–12033. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D. Genome engineering with zinc-finger nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Curtin, S.J.; Zhang, F.; Sander, J.D.; Haun, W.J.; Starker, C.; Baltes, N.J.; Reyon, D.; Dahlborg, E.J.; Goodwin, M.J.; Coffman, A.P.; Dobbs, D. Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant physiology 2011, 156, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in biotechnology 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Curtin, S.J.; Voytas, D.F.; Stupar, R.M. Genome engineering of crops with designer nucleases. The Plant Genome 2012, 5. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ren, S.; Yu, S.; Pan, H.; Li, T.; Ge, S.; Zhang, J.; Xia, N. Methods favoring homology-directed repair choice in response to CRISPR/Cas9 induced-double strand breaks. International Journal of Molecular Sciences 2020, 21, 6461. [Google Scholar] [CrossRef]
- Chen, W.; Qian, Y.; Wu, X.; Sun, Y.; Wu, X.; Cheng, X. Inhibiting replication of begomoviruses using artificial zinc finger nucleases that target viral-conserved nucleotide motif. Virus Genes 2014, 48, 494–501. [Google Scholar] [CrossRef]
- Sera, T. ; Inhibition of virus DNA replication by artificial zinc finger proteins. Journal of virology 2005, 79, 2614–2619. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Khan, S.H.; Ahmed, A.; Iqbal, M.U.; Mubarik, M.S.; Ghouri, M.Z.; Ahmad, F.; Yaseen, S.; Ali, Z.; Khan, A.A.; Azhar, M.T. Genome editing in cotton: challenges and opportunities. Journal of Cotton Research 2023, 6, 1–21. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: masters at redirecting and reprogramming plant processes. Nature Reviews Microbiology 2013, 11, 777–788. [Google Scholar] [CrossRef]
- Lloyd, A.; Plaisier, C.L.; Carroll, D.; Drews, G.N. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proceedings of the National Academy of Sciences 2005, 102, 2232–2237. [Google Scholar] [CrossRef]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; DeKelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X.; Choi, V.M. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 2009, 459, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.K.; Sander, J.D. TALENs: a widely applicable technology for targeted genome editing. Nature reviews Molecular cell biology 2013, 14, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Bonas, U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annual review of phytopathology 2010, 48, 419–436. [Google Scholar] [CrossRef]
- Kay, S.; Bonas, U. How Xanthomonas type III effectors manipulate the host plant. Current opinion in microbiology 2009, 12, 37–43. [Google Scholar] [CrossRef]
- Miller, J.C.; Tan, S.; Qiao, G.; Barlow, K.A.; Wang, J.; Xia, D.F.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; Dulay, G.P. A TALE nuclease architecture for efficient genome editing. Nature biotechnology 2011, 29, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Sugio, A.; White, F.F. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proceedings of the National Academy of Sciences 2006, 103, 10503–10508. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Li, F.; Cai, J.; Chen, W.; Zhao, N.; Sun, Y.; Guo, Y.; Yang, X.; Wu, X. Artificial TALE as a convenient protein platform for engineering broad-spectrum resistance to begomoviruses. Viruses 2015, 7, 4772–4782. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of bacteriology 1987, 169, 5429–5433. [Google Scholar] [CrossRef]
- Jansen, R.; Embden, J.D.V.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Molecular microbiology 2002, 43, 1565–1575. [Google Scholar] [CrossRef]
- Wright, A.V.; Nuñez, J.K.; Doudna, J.A. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 2016, 164, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S. The heroes of CRISPR. Cell 2016, 164, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 structures and mechanisms. Annual review of biophysics 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Konermann, S.; Shehata, S.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O.; Nishimasu, H.; Ran, F.; Hsu, P. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156. [Google Scholar]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef]
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 as a versatile tool for engineering biology. Nature methods 2013, 10, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, S.H.; Redding, S.; Jinek, M.; Greene, E.C.; Doudna, J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Biophysical Journal 2014, 106, 695a. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; Severinov, K. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome biology 2018, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Ali, S.; Tashkandi, M.; Zaidi, S.S.E.A.; Mahfouz, M.M. CRISPR/Cas9-mediated immunity to geminiviruses: differential interference and evasion. Scientific reports 2016, 6, 26912. [Google Scholar] [CrossRef]
- Ali, Z.; Abul-Faraj, A.; Li, L.; Ghosh, N.; Piatek, M.; Mahjoub, A.; Aouida, M.; Piatek, A.; Baltes, N.J.; Voytas, D.F.; Dinesh-Kumar, S. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Molecular plant 2015, 8, 1288–1291. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.A.; Kumar, R.; Dasgupta, I. CRISPR/Cas-mediated resistance against viruses in plants. International Journal of Molecular Sciences 2022, 23, 2303. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; Horvath, P. An updated evolutionary classification of CRISPR–Cas systems. Nature Reviews Microbiology 2015, 13, 722–736. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Van Der Oost, J.; Regev, A.; Koonin, E.V. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Fonfara, I.; Richter, H.; Bratovič, M.; Le Rhun, A.; Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 2016, 532, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Selma, S.; Orzáez, D. Perspectives for epigenetic editing in crops. Transgenic Research 2021, 30, 381–400. [Google Scholar] [CrossRef] [PubMed]
- Uranga, M.; Daròs, J.A. Tools and targets: The dual role of plant viruses in CRISPR–Cas genome editing. The Plant Genome 2023, 16, e20220. [Google Scholar] [CrossRef] [PubMed]
- Dubois, A.; Roudier, F. Deciphering plant chromatin regulation via CRISPR/dCas9-based epigenome engineering. Epigenomes 2021, 5, 17. [Google Scholar] [CrossRef]
- Price, A.A.; Sampson, T.R.; Ratner, H.K.; Grakoui, A.; Weiss, D.S. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proceedings of the National Academy of Sciences 2015, 112, 6164–6169. [Google Scholar] [CrossRef]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; Zhang, F. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Molecular cell 2015, 60, 385–397. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, Q.; Yi, X.; An, H.; Zhao, Y.; Ma, S.; Zhou, G. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant biotechnology journal 2018, 16, 1415–1423. [Google Scholar] [CrossRef]
- Marqués, M.C.; Sánchez-Vicente, J.; Ruiz, R.; Montagud-Martínez, R.; Márquez-Costa, R.; Gómez, G.; Carbonell, A.; Daròs, J.A.; Rodrigo, G. Diagnostics of infections produced by the plant viruses TMV, TEV, and PVX with CRISPR-Cas12 and CRISPR-Cas13. ACS Synthetic Biology 2022, 11, 2384–2393. [Google Scholar] [CrossRef]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; Severinov, K. Diversity and evolution of class 2 CRISPR–Cas systems. Nature reviews microbiology 2017, 15, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Kavuri, N.R.; Ramasamy, M.; Qi, Y.; Mandadi, K. Applications of CRISPR/Cas13-based RNA editing in plants. Cells 2022, 11, 2665. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, Y.; Ye, J.; Cao, X.; Xu, C.; Chen, B.; An, H.; Jiao, Y.; Zhang, F.; Yang, X.; Zhou, G. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant biotechnology journal 2019, 17, 1185. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Ghosh, A.; Chakravarti, R.; Singh, R.; Ravichandiran, V.; Swarnakar, S.; Ghosh, D. Cas13d: a new molecular scissor for transcriptome engineering. Frontiers in Cell and Developmental Biology 2022, 10, 866800. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Molla, K.A.; Yang, Y. CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends in biotechnology 2019, 37, 1121–1142. [Google Scholar] [CrossRef] [PubMed]
- Bastet, A.; Zafirov, D.; Giovinazzo, N.; Guyon-Debast, A.; Nogué, F.; Robaglia, C.; Gallois, J.L. Mimicking natural polymorphism in eIF 4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnology Journal 2019, 17, 1736–1750. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Li, C.; Zhang, R.; Meng, X.; Chen, S.; Zong, Y.; Lu, C.; Qiu, J.L.; Chen, Y.H.; Li, J.; Gao, C. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nature biotechnology 2020, 38, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nature biotechnology 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Hong, X.; Zhang, S.; Yao, R.; Xiao, Y.I. CRISPR base editing and prime editing: DSB and template-free editing systems for bacteria and plants. Synthetic and Systems Biotechnology 2020, 5, 277–292. [Google Scholar]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; Liu, D.R. Prime genome editing in rice and wheat. Nature biotechnology 2020, 38, 582–585. [Google Scholar] [CrossRef]
- Binyameen, B.; Khan, Z.; Khan, S.H.; Ahmad, A.; Munawar, N.; Mubarik, M.S.; Riaz, H.; Ali, Z.; Khan, A.A.; Qusmani, A.T.; Abd-Elsalam, K.A. Using multiplexed CRISPR/Cas9 for suppression of cotton leaf curl virus. International journal of molecular sciences 2021, 22, 12543. [Google Scholar] [CrossRef]
- Cao, J.; Xiao, Q.; Yan, Q. The multiplexed CRISPR targeting platforms. Drug Discovery Today: Technologies 2018, 28, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Liu, W.; Nie, B.; Zhang, F.; Zhang, J. Cas13d-mediated multiplex RNA targeting confers a broad-spectrum resistance against RNA viruses in potato. Communications Biology 2023, 6, 855. [Google Scholar] [CrossRef] [PubMed]
- Eid, A.; Alshareef, S.; Mahfouz, M.M. CRISPR base editors: genome editing without double-stranded breaks. Biochemical Journal 2018, 475, 1955–1964. [Google Scholar] [CrossRef]
- Mubarik, M.S.; Wang, X.; Khan, S.H.; Ahmad, A.; Khan, Z.; Amjid, M.W.; Razzaq, M.K.; Ali, Z.; Azhar, M.T. Engineering broad-spectrum resistance to cotton leaf curl disease by CRISPR-Cas9 based multiplex editing in plants. GM Crops & Food 2021, 12, 647–658. [Google Scholar]
- Ji, X.; Si, X.; Zhang, Y.; Zhang, H.; Zhang, F.; Gao, C. Conferring DNA virus resistance with high specificity in plants using virus-inducible genome-editing system. Genome biology 2018, 19, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA replicons for plant genome engineering. The Plant Cell 2014, 26, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, K.; Koshino-Kimura, Y.; Aoyama, Y.; Sera, T. Inhibition of tomato yellow leaf curl virus replication by artificial zinc-finger proteins. In Nucleic Acids Symposium Series, Oxford University Press 2007, Vol. 51, No. 1, pp. 429-430.
- Koshino-Kimura, Y.; Takenaka, K.; Domoto, F.; Ohashi, M.; Miyazaki, T.; Aoyama, Y.; Sera, T. Construction of plants resistant to TYLCV by using artificial zinc-finger proteins. In Nucleic Acids Symposium Series, Oxford University Press 2009, Vol. 53, No. 1, pp. 281-282. Oxford University Press.
- Mori, T.; Takenaka, K.; Domoto, F.; Aoyama, Y.; Sera, T. Inhibition of binding of tomato yellow leaf curl virus rep to its replication origin by artificial zinc-finger protein. Molecular biotechnology 2013, 54, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Romay, G.; Bragard, C. Antiviral defenses in plants through genome editing. Frontiers in microbiology 2017, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Ordiz, M.I.; Magnenat, L.; Barbas, C.F.; Beachy, R.N. Negative regulation of the RTBV promoter by designed zinc finger proteins. Plant molecular biology 2010, 72, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nature Plants 2015, 1, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Baltes, N.J.; Hummel, A.W.; Konecna, E.; Cegan, R.; Bruns, A.N.; Bisaro, D.M.; Voytas, D.F. Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nature Plants 2015, 1, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Zhai, Y.; Ortiz, J.; Neff, M.; Mandal, B.; Mukherjee, S.K.; Pappu, H.R. Multiplexed editing of a begomovirus genome restricts escape mutant formation and disease development. PloS one 2019, 14, e0223765. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Soyars, C.L.; Li, J.; Fei, Q.; He, G.; Peterson, B.A.; Meyers, B.C.; Nimchuk, Z.L.; Wang, X. CRISPR/Cas9-mediated resistance to cauliflower mosaic virus. Plant direct 2018, 2, e00047. [Google Scholar] [CrossRef]
- Kis, A.; Hamar, É.; Tholt, G.; Bán, R.; Havelda, Z. Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant biotechnology journal 2019, 17, 1004. [Google Scholar] [CrossRef]
- Tripathi, J.N.; Ntui, V.O.; Ron, M.; Muiruri, S.K.; Britt, A.; Tripathi, L. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Communications Biology 2019, 2, 46. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Stürchler, A.; Anjanappa, R.B.; Zaidi, S.S.E.A.; Hirsch-Hoffmann, M.; Gruissem, W.; Vanderschuren, H. Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome biology 2019, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Honig, A.; Marton, I.; Rosenthal, M.; Smith, J.J.; Nicholson, M.G.; Jantz, D.; Zuker, A.; Vainstein, A. Transient expression of virally delivered meganuclease in planta generates inherited genomic deletions. Molecular plant 2015, 8, 1292–1294. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Xu, W.Y.; Yan, T.; Fang, X.D.; Cao, Q.; Zhang, Z.J.; Ding, Z.H.; Wang, Y.; Wang, X.B. Rescue of a plant cytorhabdovirus as versatile expression platforms for planthopper and cereal genomic studies. New Phytologist 2019, 223, 2120–2133. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, X.; Liu, H.; Li, Z. Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nature Plants 2020, 6, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Noureen, A.; Zuhaib Khan, M.; Amin, I.; Zainab, T.; Ahmad, N.; Haider, S.; Mansoor, S. Broad-spectrum resistance against multiple PVY-strains by CRSIPR/Cas13 system in Solanum tuberosum crop. GM Crops & Food 2022, 13, 97–111. [Google Scholar]
- Ashraf, S.; Ahmad, A.; Khan, S.H.; Jamil, A.; Sadia, B.; Brown, J.K. LbCas12a mediated suppression of Cotton leaf curl Multan virus. Frontiers in Plant Science 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Uranga, M.; Vazquez-Vilar, M.; Orzáez, D.; Daròs, J.A. CRISPR-Cas12a genome editing at the whole-plant level using two compatible RNA virus vectors. The CRISPR Journal 2021, 4, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.; Faiq, A.; Khan, M.Z.; Mansoor, S.; Amin, I. Fully Transient CRISPR/Cas12f system in plants capable of broad-spectrum resistance against Begomovirus. bioRxiv 2022, 2022–06. [Google Scholar]
- Senthil-Kumar, M.; Mysore, K.S. New dimensions for VIGS in plant functional genomics. Trends in plant science 2011, 16, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Dalakouras, A.; Ganopoulos, I. Induction of promoter DNA methylation upon high-pressure spraying of double-stranded RNA in plants. Agronomy 2021, 11, 789. [Google Scholar] [CrossRef]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nature plants 2017, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, Y.; Xu, K.; Li, D.; Hu, H.; Zhou, F.; Song, P.; Yu, Y.; Wei, Q.; Liu, Q.; Wang, W. Clay nanosheet-mediated delivery of recombinant plasmids expressing artificial miRNAs via leaf spray to prevent infection by plant DNA viruses. Horticulture Research 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Martín, J.; Ruiz, L.; Janssen, D.; Velasco, L. Exogenous application of dsRNA for the control of viruses in cucurbits. Frontiers in Plant Science 2022, 13, 895953. [Google Scholar] [CrossRef] [PubMed]
- Rego-Machado, C.M.; Nakasu, E.Y.; Silva, J.M.; Lucinda, N.; Nagata, T.; Inoue-Nagata, A.K. siRNA biogenesis and advances in topically applied dsRNA for controlling virus infections in tomato plants. Scientific Reports 2020, 10, 22277. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.E.; Mazumdar, P.; Hee, T.W.; Song, A.L.A.; Othman, R.Y.; Harikrishna, J.A. Crude extracts of bacterially-expressed dsRNA protect orchid plants against Cymbidium mosaic virus during transplantation from in vitro culture. The Journal of Horticultural Science and Biotechnology 2014, 89, 569–576. [Google Scholar] [CrossRef]
- Holeva, M.C.; Sklavounos, A.; Rajeswaran, R.; Pooggin, M.M.; Voloudakis, A.E. Topical application of double-stranded RNA targeting 2b and CP genes of Cucumber mosaic virus protects plants against local and systemic viral infection. Plants 2021, 10, 963. [Google Scholar] [CrossRef]
- Xu, X.; Yu, T.; Zhang, D.; Song, H.; Huang, K.; Wang, Y.; Shen, L.; Li, Y.; Wang, F.; Zhang, S.; Jiao, Y. Evaluation of the anti-viral efficacy of three different dsRNA nanoparticles against potato virus Y using various delivery methods. Ecotoxicology and Environmental Safety 2023, 255, 114775. [Google Scholar] [CrossRef] [PubMed]
| Strategy | Virus | Target | Plant | References |
|---|---|---|---|---|
|
hpRNA (IR) hpRNA |
TMV CMV PPV CGMMV PSTVd TYLCV PVX PVY PLRV PRSV SbDV SMV WCMV ACMV BBTV CTV |
Movement protein Replicase P1 and HC-Pro Coat protein Viroid sequence Replicase (C1) Coat protein ORF2 HC-Pro CP CP CP HC-Pro (VSR) Replicase Rep (AC1) Replicase P20, p23, p25 (VSRs) |
Tobacco Tomato Potato Melon Soyabean White clover Cassava Banana Mexican lime |
[91] [92] [93] [94] [95] [96] [97] [98] [99] [100] [101] [102] [103] [104] |
| DI Virus-derived RNA | PVY TBSV GLFV |
Coat protein (DI virus) Movement protein (DI virus) |
Tobacco | [105] |
| dsRNA | ACMV RGSV RTBV |
DNA-A and DNA-B pC5, pC6 ORF IV |
Tobacco Rice |
[106] [107] [53] |
|
amiRNA (precursor miR395) amiRNA (precursor miR319) amiRNA/single monocistronic AthMIR156 AthMIR159a AthMIR167b AthMIR169a amiRNA/single polycistronic OsaMIR395 amiRNA/single monocistronic in tandem repeats AthMIR159a amiRNA/multiple monocistronic in trans AthMIR159a AthMIR390a |
WSMV GFLV CGMMV WSMV PVX, PVY TuMV, TYMV RBSDV, RSV TuMV TBSV |
Virus genome (conserved region) CP CP 5’UTR+P1+HC-Pro+ P3 P25 (PVX) + HC-Pro (PVY) HC- Pro (TuMV) + P69 (TYMV) CP + HC-Pro 5’ terminal TBSV (+) RNA |
Wheat Grape Tobacco Wheat N. tabacum Arabidopsis Tobacco |
[89] [103] [108] [89] [109] [110] [111] |
| ta-siRNAs AthTAS3a | TuMV, CMV | Multiple genomic positions | Arabidopsis |
[112] |
|
syn-tasiRNA/ single polycistronic AthTAS1c |
TSWV |
RdRP |
Tobacco |
[113] |
| dsRNA coating and inoculation | Plant | Virus | References |
|---|---|---|---|
| LDH nanosheets | Cowpea Tomato |
CMV TYLCV |
[250] [260] |
| Agroinfiltration and Direct spray | Cucumber | CGMMV ToLCNDV |
[261] |
| High pressure spraying of free siRNAs | Tobacco | CaMV 35S promoter | [258] |
| Mechanical inoculation of free dsRNAs in vivo and in vitro synthesized dsRNAs |
Tomato Orchid Tobacco Chenopodium Quinoa |
ToMV CymMV CMV |
[262] [263] [264] |
| CQAS-dsRNA nanoparticles and Carbon Quantum Dots (CQDs) through Root soaking, infiltration | Tobacco Tomato Pepper |
PVY | [265] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





