Submitted:
23 December 2023
Posted:
25 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant material
2.2. Methods
2.2.1. Field experiment
2.2.2. Weather conditions
2.2.3. DNA isolation
2.2.4. Genotyping
2.2.5. Association mapping using GWAS analysis
2.2.6. Statistical Analysis and Association Mapping
2.2.7. Physical mapping
2.2.8. Functional analysis of gene sequences
2.2.9. Design of primers for identified SilicoDArT and SNP polymorphisms related to yield and its characteristics
3. Results
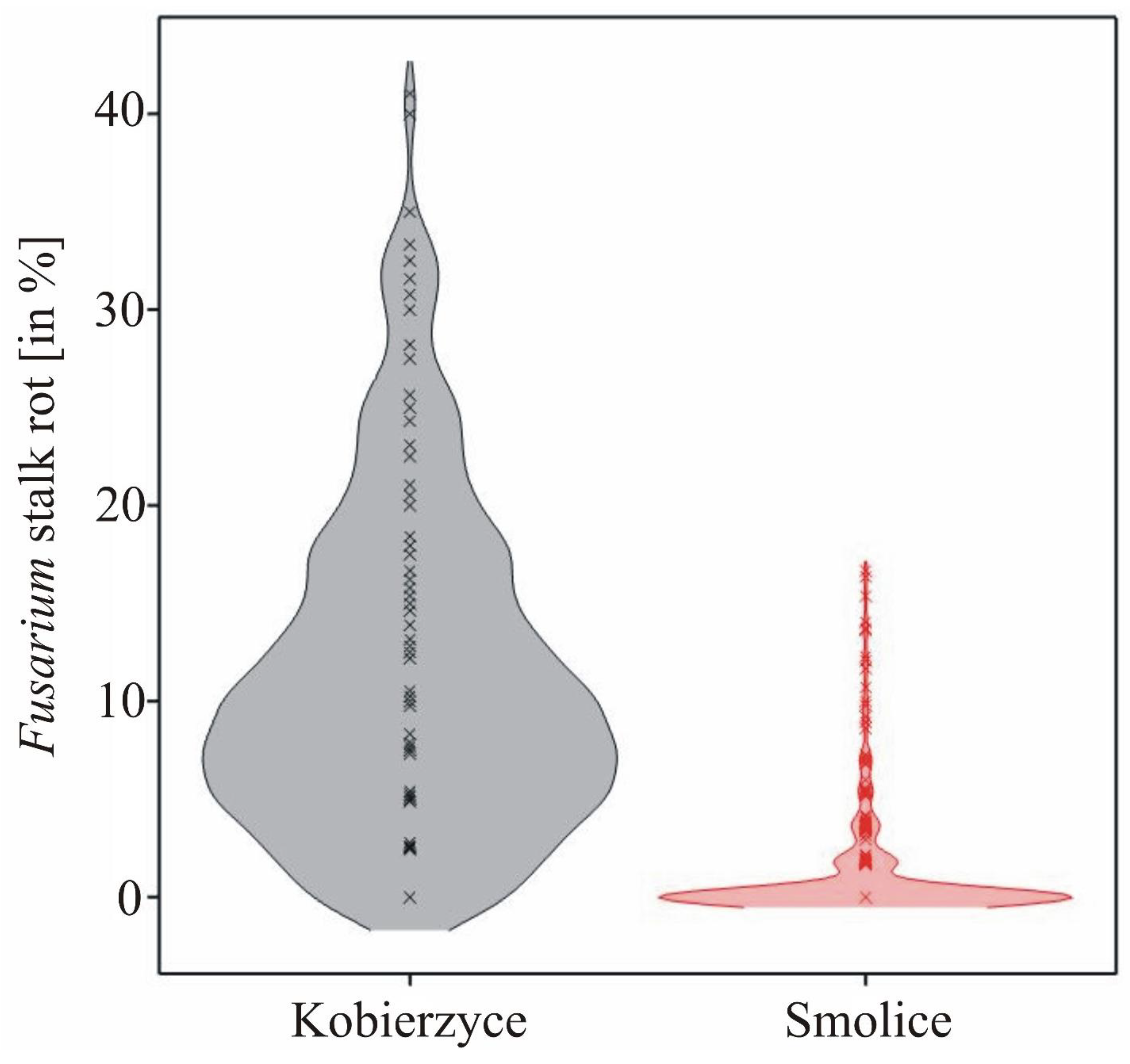
3.1. Phenotyping
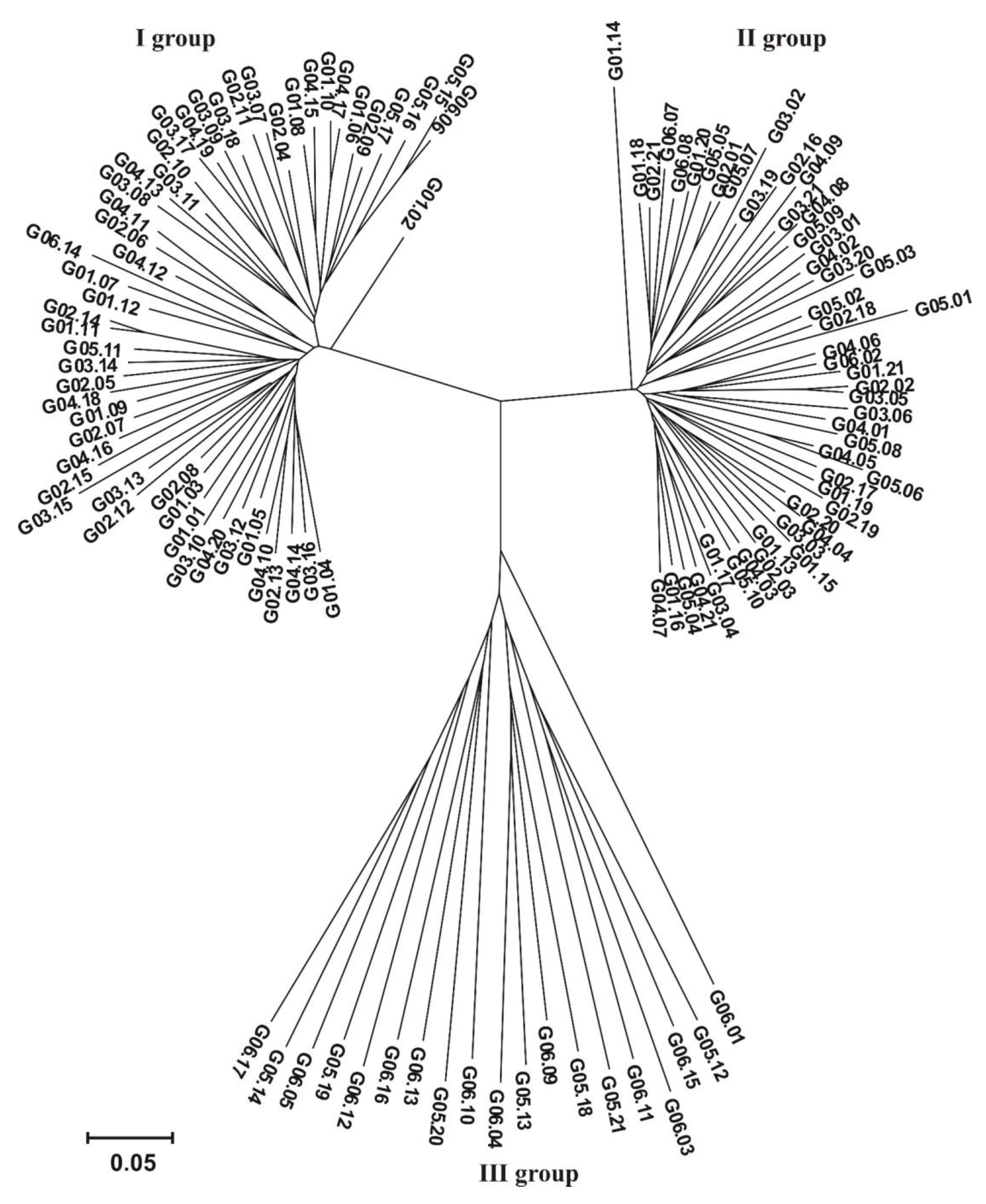
3.2. Genotyping
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zijlstra, C.; Lund, I.; Justesen, A.; Nicolaisen, M.; Bianciotto, V.; Posta, K.; Balestrini, R.; Przetakiewicz, A.; Czembor, E.; van de Zande, J. Combining novel monitoring tools and precision application technologies for integrated high-tech crop protection in the future (a discussion document). Pest Manag. Sci. 2011, 67, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Galiano-Carneiro, A.; Miedaner, T. Genetics of resistance and pathogenicity in the maize/Setosphaeria turcica pathosystem and implications for breeding. Front. Plant Sci. 2017, 8, 1490. [Google Scholar] [CrossRef]
- Vigier, B.; Reid, L.M.; Dwyer, L.M.; Stewart, D.W.; Sinha, R.C.; Arnason, J.T.; Butler, G. Maize resistance to gibberella ear rot: symptoms, deoxynivalenol, and yield. Can. J. Plant Pathol. 2001, 23, 99–105. [Google Scholar] [CrossRef]
- Sun, Y.; Ruan, X.; Wang, Q.; Zhou, Y.; Wang, F.; Ma, L.; Wang, Z.; Gao, X. Integrated gene co-expression analysis and metabolites profiling highlight the important role of ZmHIR3 in maize resistance to Gibberella stalk rot. Front. Plant Sci. 2021, 12, 664–733. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ceron, D.; Lowe, R.G.T.; McKenna, J.A.; Brain, L.M.; Dawson, C.S.; Clark, B.; Berkowitz, O.P.; Whelan, J.; Bleackley, M.R.; Anderson, M.A. Extracellular vesicles from Fusarium graminearum contain protein effectors expressed during infection of corn. J. Fungi 2021, 7, 977. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, R.A.; Jebur, A.; Kang, W.; El-Demerdash, F. An overview on the major mycotoxins in food products: characteristics, toxicity, and analysis. J. Future Foods 2022, 2, 91–102. [Google Scholar] [CrossRef]
- Hoppe, K.; Chełkowski, J.; Błaszczyk, L.; Bocianowski, J. Observation of changes in Fusarium mycotoxin profiles in maize grain over the last decade in Poland. Food Control 2024, 158, 110248. [Google Scholar] [CrossRef]
- Deepthi, B.V.; Deepa, N.; Vanitha, P.R.; Sreenivasa, M.Y. Stress responses on the growth and mycotoxin biosynthesis of Fusarium proliferatum associated with stored poultry feeds. Appl. Food Res. 2022, 2, 100091. [Google Scholar] [CrossRef]
- Doohan, F.M.; Brennan, J.; Cooke, B.M. Influence of climatic factors on Fusarium species pathogenic to cereals. Europ. J. Plant Pathol. 2003, 109, 755–768. [Google Scholar] [CrossRef]
- Alsamir, M.; Al-Samir, E.; Kareem, T.A.; Abass, M.; Trethowan, R. The application of zinc fertilizer reduces Fusarium infection and development in wheat. Aust. J. Crop Sci. 2020, 14, 1088–1094. [Google Scholar] [CrossRef]
- Nowembabaz, A.; Taulya, G.; Tinzaara, W.; Karamura, E. Effect of integrated potassium nutrition on Fusarium wilt tolerance in apple bananas. Afr. J. Plant Sci. 2021, 15, 257–265. [Google Scholar] [CrossRef]
- Gilardi, G.; Vasileiadou, A.; Garibaldi, A.; Gullino, M.L. The effects of biological control agents, potassium phosphite and calcium oxide on race 1 of Fusarium oxysporum f. sp. lactucae of lettuce in closed soilless cultivation systems. J. Phytopathol. 2022, 170, 626–634. [Google Scholar] [CrossRef]
- Szóke, C.; Zsubori, Z.; Pók, I.; Rácz, F.; Illés, O.; Szegedi, I. Significance of the European corn borer (Ostrinia nubilalis Hóbn.) in maize production. Acta Agron. Hungarica 2002, 50, 447–461. [Google Scholar] [CrossRef]
- Blandino, M.; Reyneri, A.; Vanara, F.; Pascale, M.; Haidukowski, M.; Campagna, C. Management of fumonisin contamination in maize kernels through the timing of insecticide application against European corn borer Ostrinia nubilalis Hübner. Food Addit. Contam. A 2009, 26, 1501–1514. [Google Scholar] [CrossRef] [PubMed]
- Champeil, A.; Doré, T.; Fourbet, J.F. Fusarium head blight: epidemiological origin of the effects of cultural practices on head blight attacks and the production of mycotoxins by Fusarium in wheat grains. Plant Sci. 2004, 166, 1389–1415. [Google Scholar] [CrossRef]
- Unterseer, S.; Bauer, E.; Haberer, G.; Seidel, M.; Knaak, C.; Ouzunova, M.; Meitinger, T.; Strom, T.M.; Fries, R.; Pausch, H.; Bertani, C.; Davassi, A.; Mayer, K.F.; Schön, C.C. A powerful tool for genome analysis in maize: development and evaluation of the high density 600 k SNP genotyping array. BMC Genomics 2014, 15, 823. [Google Scholar] [CrossRef] [PubMed]
- Shah, T.R.; Prasad, K.; Kumar, P. Maize – A potential source of human nutrition and health. Cogent Food Agriculture 2016, 2, 1. [Google Scholar] [CrossRef]
- Zila, C.T.; Ogut, F.; Romay, M.C.; Gardner, C.A.; Buckler, E.S. Genome wide association study of Fusarium ear rot disease in the U.S.A. maize inbred line collection. BMC Plant Biol. 2014, 14, 372. [Google Scholar] [CrossRef] [PubMed]
- Zila, C.T.; Samayoa, L.F.; Santiago, R.; Butrón, A.; Holland, J.B. A genome-wide association study reveals genes associated with fusarium ear rot resistance in a maize core diversity panel. G3-Genes Genom. Genetic 2013, 3, 2095–2104. [Google Scholar] [CrossRef]
- Eathington, S.R.; Crosbie, T.M.; Edwards, M.D.; Reiter, R.S.; Bull, J.K. Molecular markers in a commercial breeding program. Crop Sci. 2007, 47, S-154–S-163. [Google Scholar] [CrossRef]
- Xu, C.; Ren, Y.; Jian, Y.; Guo, Z.; Zhang, Y.; Xie, C.; Fu, J.; Wang, H.; Wang, G.; Xu, Y.; Li, P.; Zou, C. Development of a maize 55 K SNP array with improved genome coverage for molecular breeding. Mol. Breed. 2017, 37, 20. [Google Scholar] [CrossRef] [PubMed]
- Cerrudo, D.; Cao, S.; Yuan, Y.; Martinez, C.; Suarez, E.A.; Babu, R.; Zhang, X.; Trachsel, S. Genomic selection outperforms marker assisted selection for grain yield and physiological traits in a maize doubled haploid population across water treatments. Front. Plant Sci. 2018, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; Dreisigacker, S.; Singh, R.; Zhang, X.; Gowda, M.; Roorkiwal, M.; Rutkoski, J.; Varshney, R.K. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Heslot, N.; Jannink, J.-L.; Sorrells, M.E. Perspectives for genomic selection applications and research in plants. Crop Sci. 2015, 55, 1–12. [Google Scholar] [CrossRef]
- Shikha, M.; Kanika, A.; Rao, A.R.; Mallikarjuna, M.G.; Gupta, H.S.; Nepolean, T. Genomic selection for drought tolerance using genome-wide SNPs in maize. Front. Plant Sci. 2017, 8, 550. [Google Scholar] [CrossRef] [PubMed]
- Dhliwayo, T.; Pixley, K.; Menkir, A.; Warburton, M. Combining ability, genetic distances, and heterosis among elite CIMMYT and IITA tropical maize inbred lines. Crop Sci. 2009, 49, 1201–1210. [Google Scholar] [CrossRef]
- Chander, S.; Guo, Y.Q.; Yang, X.H.; Zhang, Z.; Lu, X.Q.; Yan, J.B.; Song, T.M.; Rocheford, T.R.; Li, J.S. Using molecular markers to identify two major loci controlling carotenoid contents in maize grain. Theor. Appl. Genet. 2008, 116, 223–233. [Google Scholar] [CrossRef] [PubMed]
- VSN International Genstat for Windows, 23rd ed.; VSN International: Hemel Hempstead, UK, 2023.
- Chung, U.; Gbegbelegbe, S.; Shiferaw, B.; Robertson, R.; Yun, J.I.; Tesfaye, K.; Hoogenboom, G.; Sonder, K. Modeling the effect of a heat wave on maize production in the USA and its implications on food security in the developing world. Weather Clim. Extremes 2014, 5–6, 67–77. [Google Scholar] [CrossRef]
- Byerlee, D. The globalization of hybrid maize, 1921–70. J. Global History 2020, 15, 101–122. [Google Scholar] [CrossRef]
- Cassidy, E.S.; West, P.C.; Gerber, J.S.; Foley, J.A. Redefining agricultural yields: From tonnes to people nourished per hectare. Environ. Res. Lett. 2013, 8, 034015. [Google Scholar] [CrossRef]
- Burdon, J.J.; Zhan, J. Climate change and disease in plant communities. PLoS Biol. 2020, 18, e3000949. [Google Scholar] [CrossRef] [PubMed]
- Bocianowski, J.; Szulc, P.; Nowosad, K. Soil tillage methods by years interaction for dry matter of plant yield of maize (Zea mays L.) using additive main effects and multiplicative interaction model. J. Integr. Agric. 2018, 17, 2836–2839. [Google Scholar] [CrossRef]
- Bocianowski, J.; Nowosad, K.; Szulc, P. Soil tillage methods by years interaction for harvest index of maize (Zea mays L.) using additive main effects and multiplicative interaction model. Acta Agric. Scand. Sect. B 2019, 69, 75–81. [Google Scholar] [CrossRef]
- Picot, A.; Barreau, C.; Pinson-Gadais, L.; Caron, D.; Lannou, C.; Richard-Forget, F. Factors of the Fusarium verticillioides-maize environment modulating fumonisin production. Crit. Rev. Microbiol. 2010, 36, 221–231. [Google Scholar] [CrossRef]
- Goswami, R.S.; Kistler, H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Wenda-Piesik, A.; Lemańczyk, G.; Twarużek, M.; Błajet-Kosicka, A.; Kazek, M.; Grajewski, J. Fusarium head blight incidence and detection of Fusarium toxins in wheat in relation to agronomic factors. Eur. J. Plant Pathol. 2017, 149, 515–531. [Google Scholar] [CrossRef]
- Ogliari, J.B.; Guimarães, M.A.; Geraldi, I.O.; Camargo, L.E.A. New resistance genes in the Zea mays – Exserohilum turcicum pathosystem. Genet. Mol. Biol. 2005, 28, 435–439. [Google Scholar] [CrossRef]
- Tagele, S.B.; Kim, S.W.; Lee, H.G.; Lee, Y.S. Potential of novel sequence type of Burkholderia cenocepacia for biological control of root rot of maize (Zea mays L.) caused by Fusarium temperatum. Int. J. Mol. Sci. 2019, 20, 1–18. [Google Scholar] [CrossRef]
- Bocianowski, J.; Szulc, P.; Waśkiewicz, A.; Nowosad, K.; Kobus-Cisowska, J. Ergosterol and Fusarium mycotoxins content in two maize cultivars under different forms of nitrogen fertilizers. J. Phytopathol. 2019, 167, 516–526. [Google Scholar] [CrossRef]
- Bocianowski, J.; Szulc, P.; Waśkiewicz, A.; Cyplik, A. The Effect of Agrotechnical Factors on Fusarium Mycotoxins Level in Maize. Agriculture 2020, 10, 528. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, S.; Li, W.X.; Liu, J.; Guo, W.; Xu, M.; Xu, Y. QTL mapping and genomic selection for Fusarium ear rot resistance using two F2:3 populations in maize. Euphytica 2022, 218, 131. [Google Scholar] [CrossRef]
- Maschietto, V.; Cinzia, C.; Pirona, R.; Pea, G.; Strozzi, F.; Marocco Adriano-Rossini, L.; Lanubile, A. QTL Mapping and candidate genes for resistance to Fusarium Ear Rot and fumonisin contamination in Maize. BMC Plant Biol. 2017, 17, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Brachi, B.; Faure, N.; Horton, M.; Flahauw, E.; Vazquez, A.; Nordborg, M.; Bergelson, J.; Cuguen, J.; Roux, F. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 2010, 6, e1000940. [Google Scholar] [CrossRef] [PubMed]
- Bocianowski, J.; Nowosad, K.; Wróbel, B.; Szulc, P. Identification of Associations between SSR Markers and Quantitative Traits of Maize (Zea mays L.). Agronomy 2021, 11, 182. [Google Scholar] [CrossRef]
- Sobiech, A.; Tomkowiak, A.; Bocianowski, J.; Szymańska, G.; Nowak, B.; Lenort, M. Identification and analysis of candidate genes associated with maize fusarium cob resistance using next-generation sequencing technology. Int. J. Mol. Sci. 2023, 24, 16712. [Google Scholar] [CrossRef] [PubMed]
- Bocianowski, J.; Tomkowiak, A.; Bocianowska, M.; Sobiech, A. The use of DArTseq technology to identify markers related to the heterosis effects in selected traits in maize. Curr. Issues Mol. Biol. 2023, 45, 2644–2660. [Google Scholar] [CrossRef] [PubMed]
- Tomkowiak, A.; Nowak, B.; Sobiech, A.; Bocianowski, J.; Wolko, Ł.; Spychała, J. The use of DArTseq technology to identify new SNP and SilicoDArT markers related to the yield-related traits components in maize. Genes 2022, 13, 848. [Google Scholar] [CrossRef] [PubMed]
- Nowak, B.; Tomkowiak, A.; Bocianowski, J.; Sobiech, A.; Bobrowska, R.; Kowalczewski, P.; Bocianowska, M. The Use of DArTseq Technology to Identify Markers Linked to Genes Responsible for Seed Germination and Seed Vigor in Maize. Int. J. Mol. Sci. 2022, 23, 14865. [Google Scholar] [CrossRef] [PubMed]
- Muñiz, L.M.; Royo, J.; Gómez, E.; Baudot, G.; Paul, W.; Hueros, G. Atypical response regulators expressed in the maize endosperm transfer cells link canonical two component systems and seed biology. BMC Plant Biol. 2010, 10, 84. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Dong, H.; Mu, J.; Ren, B.; Zheng, B.; Ji, Z.; Yang, W.-C.; Liang, Y.; Zuo, J. Arabidopsis histidine kinase CKI1 acts upstream of HISTIDINE PHOSPHOTRANSFER PROTEINS to regulate female gametophyte development and vegetative growth. Plant Cell 2010, 22, 1232–1248. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shen, J.; Liang, J. Genome-wide identification, expression profile, and alternative splicing analysis of the brassinosteroid-signaling kinase (BSK) family genes in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 1138. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Willige, B.C.; Jaillais, Y.; Geng, S.; Park, M.Y.; Gray, W.M.; Chory, J. BRASSINOSTEROIDSIGNALING KINASE 3, a plasma membrane-associated scaffold protein involved in early brassinosteroid signaling. PLoS Genet. 2019, 15, e1007904. [Google Scholar] [CrossRef] [PubMed]
| Source of variation | The number of degrees of freedom | Sum of square | Mean square | F statistics |
|---|---|---|---|---|
| Genotype | 121 | 8468.68 | 69.99 | 2.45 *** |
| Location | 1 | 18988.97 | 18988.97 | 664.68 *** |
| Genotype × Location | 121 | 5765.31 | 47.65 | 1.67 *** |
| Residual | 488 | 13941.44 | 28.57 | |
| Total | 731 | 47164.40 |
| Location | Kobierzyce | Smolice | Average | Location | Kobierzyce | Smolice | Average | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Mean | s.d. | Mean | s.d. | Mean | s.d. | Genotype | Mean | s.d. | Mean | s.d. | Mean | s.d. |
| G01.01 | 5.83 | 2.89 | 0.00 | 0.00 | 2.92 | 3.68 | G03.20 | 5.83 | 8.04 | 0.61 | 1.05 | 3.22 | 5.87 |
| G01.02 | 6.84 | 3.92 | 0.00 | 0.00 | 3.42 | 4.49 | G03.21 | 25.98 | 7.39 | 5.45 | 9.45 | 15.72 | 13.56 |
| G01.03 | 6.80 | 1.57 | 1.82 | 3.15 | 4.31 | 3.52 | G04.01 | 12.68 | 2.24 | 0.00 | 0.00 | 6.34 | 7.09 |
| G01.04 | 9.11 | 10.12 | 0.00 | 0.00 | 4.55 | 8.11 | G04.02 | 11.67 | 1.44 | 5.85 | 1.92 | 8.76 | 3.53 |
| G01.05 | 7.50 | 2.50 | 0.57 | 0.99 | 4.04 | 4.16 | G04.03 | 8.33 | 8.04 | 0.62 | 1.07 | 4.48 | 6.65 |
| G01.06 | 5.83 | 1.44 | 0.00 | 0.00 | 2.92 | 3.32 | G04.04 | 4.19 | 2.87 | 1.16 | 1.00 | 2.67 | 2.54 |
| G01.07 | 15.11 | 8.97 | 1.25 | 1.09 | 8.18 | 9.50 | G04.05 | 1.67 | 2.89 | 0.61 | 1.05 | 1.14 | 2.03 |
| G01.08 | 4.17 | 1.45 | 0.00 | 0.00 | 2.09 | 2.46 | G04.06 | 11.86 | 6.43 | 2.44 | 2.13 | 7.15 | 6.71 |
| G01.09 | 10.22 | 6.75 | 0.00 | 0.00 | 5.11 | 7.04 | G04.07 | 3.33 | 3.82 | 0.00 | 0.00 | 1.67 | 3.03 |
| G01.10 | 1.65 | 1.43 | 0.00 | 0.00 | 0.82 | 1.28 | G04.08 | 14.46 | 7.48 | 5.18 | 7.46 | 9.82 | 8.39 |
| G01.11 | 14.53 | 7.84 | 0.58 | 1.01 | 7.56 | 9.13 | G04.09 | 5.00 | 4.33 | 0.00 | 0.00 | 2.50 | 3.87 |
| G01.12 | 8.55 | 4.18 | 0.00 | 0.00 | 4.28 | 5.38 | G04.10 | 12.52 | 11.43 | 3.95 | 4.36 | 8. 23 | 9.05 |
| G01.13 | 7.61 | 8.56 | 0.00 | 0.00 | 3.81 | 6.84 | G04.11 | 7.65 | 8.92 | 0.58 | 1.01 | 4.12 | 6.87 |
| G01.14 | 4.27 | 3.92 | 0.00 | 0.00 | 2.14 | 3.41 | G04.12 | 5.09 | 5.00 | 0.00 | 0.00 | 2.54 | 4.22 |
| G01.15 | 23.72 | 2.80 | 0.00 | 0.00 | 11.86 | 13.11 | G04.13 | 3.33 | 2.89 | 0.00 | 0.00 | 1.67 | 2.58 |
| G01.16 | 11.97 | 7.83 | 0.00 | 0.00 | 5.98 | 8.22 | G04.14 | 16.05 | 7.59 | 0.57 | 0.99 | 8.31 | 9.76 |
| G01.17 | 15.83 | 3.82 | 3.33 | 5.77 | 9.58 | 8.13 | G04.15 | 13.33 | 16.65 | 0.00 | 0.00 | 6.67 | 12.81 |
| G01.18 | 8.38 | 6.23 | 0.72 | 1.25 | 4.55 | 5.81 | G04.16 | 23.61 | 9.22 | 1.85 | 3.21 | 12.73 | 13.42 |
| G01.19 | 16.89 | 7.03 | 0.00 | 0.00 | 8.44 | 10.26 | G04.17 | 10.83 | 8.04 | 0.00 | 0.00 | 5.42 | 7.81 |
| G01.20 | 16.09 | 12.78 | 0.00 | 0.00 | 8.05 | 11.96 | G04.18 | 19.38 | 8.27 | 3.57 | 6.18 | 11.48 | 10.85 |
| G01.21 | 16.77 | 9.40 | 0.00 | 0.00 | 8.39 | 10.94 | G04.19 | 10.00 | 2.50 | 0.71 | 1.23 | 5.36 | 5.39 |
| G02.01 | 1.67 | 1.44 | 0.00 | 0.00 | 0.83 | 1.29 | G04.20 | 21.03 | 3.07 | 1.23 | 2.14 | 11.13 | 11.10 |
| G02.02 | 10.00 | 2.50 | 3.61 | 3.64 | 6.81 | 4.48 | G04.21 | 9.40 | 10.36 | 1.36 | 2.36 | 5.38 | 8.04 |
| G02.03 | 2.50 | 0.00 | 0.00 | 0.00 | 1.25 | 1.37 | G05.01 | 5.83 | 1.44 | 1.96 | 2.10 | 3.90 | 2.66 |
| G02.04 | 15.06 | 8.94 | 0.00 | 0.00 | 7.53 | 10.00 | G05.02 | 11.84 | 5.49 | 0.00 | 0.00 | 5.92 | 7.36 |
| G02.05 | 13.42 | 7.50 | 0.00 | 0.00 | 6.71 | 8.75 | G05.03 | 15.11 | 4.24 | 6.36 | 6.06 | 10.74 | 6.70 |
| G02.06 | 15.83 | 1.44 | 0.58 | 1.01 | 8.21 | 8.43 | G05.04 | 9.07 | 3.69 | 0.00 | 0.00 | 4.53 | 5.49 |
| G02.07 | 11.10 | 6.84 | 0.00 | 0.00 | 5.55 | 7.46 | G05.05 | 15.17 | 5.26 | 4.85 | 4.32 | 10.01 | 7.10 |
| G02.08 | 21.90 | 5.60 | 5.64 | 5.84 | 13.77 | 10.27 | G05.06 | 4.23 | 3.90 | 1.26 | 2.18 | 2.74 | 3.26 |
| G02.09 | 4.25 | 3.85 | 0.00 | 0.00 | 2.13 | 3.37 | G05.07 | 9.65 | 8.46 | 2.30 | 3.98 | 5.98 | 7.15 |
| G02.10 | 10.96 | 7.33 | 4.68 | 8.11 | 7.82 | 7.72 | G05.08 | 6.67 | 2.89 | 1.75 | 3.04 | 4.21 | 3.78 |
| G02.11 | 7.91 | 7.31 | 0.62 | 1.07 | 4.26 | 6.15 | G05.09 | 15.83 | 10.10 | 3.07 | 2.76 | 9.45 | 9.63 |
| G02.12 | 16.71 | 13.70 | 1.89 | 1.96 | 9.30 | 11.94 | G05.10 | 5.88 | 3.81 | 0.62 | 1.07 | 3.25 | 3.81 |
| G02.13 | 8.33 | 2.89 | 0.00 | 0.00 | 4.17 | 4.92 | G05.11 | 16.67 | 11.82 | 2.90 | 3.50 | 9.78 | 10.84 |
| G02.14 | 17.27 | 6.23 | 1.19 | 2.06 | 9.23 | 9.74 | G05.12 | 13.14 | 6.84 | 0.57 | 0.99 | 6.86 | 8.16 |
| G02.15 | 4.30 | 5.33 | 0.00 | 0.00 | 2.15 | 4.11 | G05.13 | 21.67 | 7.22 | 0.67 | 1.16 | 11.17 | 12.40 |
| G02.16 | 8.51 | 3.05 | 0.00 | 0.00 | 4.26 | 5.05 | G05.14 | 14.17 | 9.47 | 0.00 | 0.00 | 7.08 | 9.80 |
| G02.17 | 6.67 | 3.82 | 0.00 | 0.00 | 3.33 | 4.38 | G05.15 | 21.94 | 10.55 | 1.19 | 2.06 | 11.57 | 13.25 |
| G02.18 | 11.84 | 2.75 | 0.58 | 1.01 | 6.21 | 6.44 | G05.16 | 3.42 | 5.92 | 0.00 | 0.00 | 1.71 | 4.19 |
| G02.19 | 5.04 | 0.08 | 0.00 | 0.00 | 2.52 | 2.76 | G05.17 | 11.56 | 5.38 | 0.00 | 0.00 | 5.78 | 7.19 |
| G02.20 | 17.68 | 12.84 | 1.72 | 2.99 | 9.70 | 12.08 | G05.18 | 6.10 | 4.08 | 0.00 | 0.00 | 3.05 | 4.22 |
| G02.21 | 3.42 | 2.97 | 0.00 | 0.00 | 1.71 | 2.65 | G05.19 | 12.65 | 5.23 | 4.97 | 2.72 | 8.81 | 5.62 |
| G03.01 | 5.00 | 4.33 | 3.02 | 2.20 | 4.01 | 3.26 | G05.20 | 17.50 | 9.01 | 2.21 | 1.92 | 9.85 | 10.21 |
| G03.02 | 10.94 | 8.37 | 0.63 | 1.09 | 5.79 | 7.77 | G05.21 | 10.92 | 1.37 | 2.35 | 2.67 | 6.64 | 5.07 |
| G03.03 | 11.82 | 10.24 | 0.00 | 0.00 | 5.91 | 9.15 | G06.01 | 9.34 | 10.38 | 1.82 | 3.15 | 5.58 | 8.00 |
| G03.04 | 1.67 | 2.89 | 1.17 | 2.03 | 1.42 | 2.25 | G06.02 | 6.84 | 5.93 | 0.00 | 0.00 | 3.42 | 5.30 |
| G03.05 | 19.57 | 10.48 | 6.28 | 8.07 | 12.93 | 11.09 | G06.03 | 9.43 | 5.65 | 1.73 | 1.73 | 5.58 | 5.63 |
| G03.06 | 19.62 | 13.23 | 0.00 | 0.00 | 9.81 | 13.62 | G06.04 | 20.21 | 13.80 | 0.00 | 0.00 | 10.11 | 14.10 |
| G03.07 | 31.18 | 16.18 | 2.98 | 5.16 | 17.08 | 18.81 | G06.05 | 7.54 | 6.59 | 0.00 | 0.00 | 3.77 | 5.87 |
| G03.08 | 24.32 | 7.44 | 0.00 | 0.00 | 12.16 | 14.13 | G06.06 | 17.50 | 5.00 | 1.13 | 0.98 | 9.32 | 9.53 |
| G03.09 | 14.92 | 8.73 | 0.00 | 0.00 | 7.46 | 9.86 | G06.07 | 9.38 | 6.49 | 0.57 | 0.99 | 4.98 | 6.36 |
| G03.10 | 4.23 | 3.90 | 0.57 | 0.99 | 2.40 | 3.24 | G06.08 | 10.00 | 2.50 | 1.15 | 1.00 | 5.58 | 5.14 |
| G03.11 | 13.33 | 8.04 | 0.00 | 0.00 | 6.67 | 8.90 | G06.09 | 9.19 | 7.09 | 0.00 | 0.00 | 4.59 | 6.74 |
| G03.12 | 5.92 | 5.31 | 1.74 | 1.76 | 3.83 | 4.21 | G06.10 | 14.98 | 2.36 | 1.80 | 1.85 | 8.39 | 7.47 |
| G03.13 | 15.61 | 14.03 | 4.55 | 7.88 | 10.08 | 11.84 | G06.11 | 14.17 | 11.55 | 0.00 | 0.00 | 7.08 | 10.66 |
| G03.14 | 15.46 | 4.85 | 0.00 | 0.00 | 7.73 | 9.00 | G06.12 | 25.15 | 7.28 | 0.60 | 1.03 | 12.87 | 14.23 |
| G03.15 | 15.00 | 5.00 | 5.32 | 6.03 | 10.16 | 7.26 | G06.13 | 14.49 | 7.79 | 0.00 | 0.00 | 7.25 | 9.34 |
| G03.16 | 5.88 | 1.41 | 0.00 | 0.00 | 2.94 | 3.34 | G06.14 | 11.84 | 8.17 | 0.00 | 0.00 | 5.92 | 8.29 |
| G03.17 | 13.55 | 1.30 | 6.16 | 9.14 | 9.86 | 7.11 | G06.15 | 10.92 | 6.28 | 0.00 | 0.00 | 5.46 | 7.18 |
| G03.18 | 5.04 | 2.50 | 0.00 | 0.00 | 2.52 | 3.18 | G06.16 | 5.77 | 1.34 | 0.00 | 0.00 | 2.89 | 3.27 |
| G03.19 | 15.28 | 9.29 | 3.64 | 3.57 | 9.46 | 8.96 | G06.17 | 5.83 | 2.89 | 0.00 | 0.00 | 2.92 | 3.68 |
| Location | 11.39 | 8.33 | 1.20 | 2.79 | |||||||||
| LSD0.05 – Genotype: 6.06; Location: 0.78; Genotype x Location: 8.58 | |||||||||||||
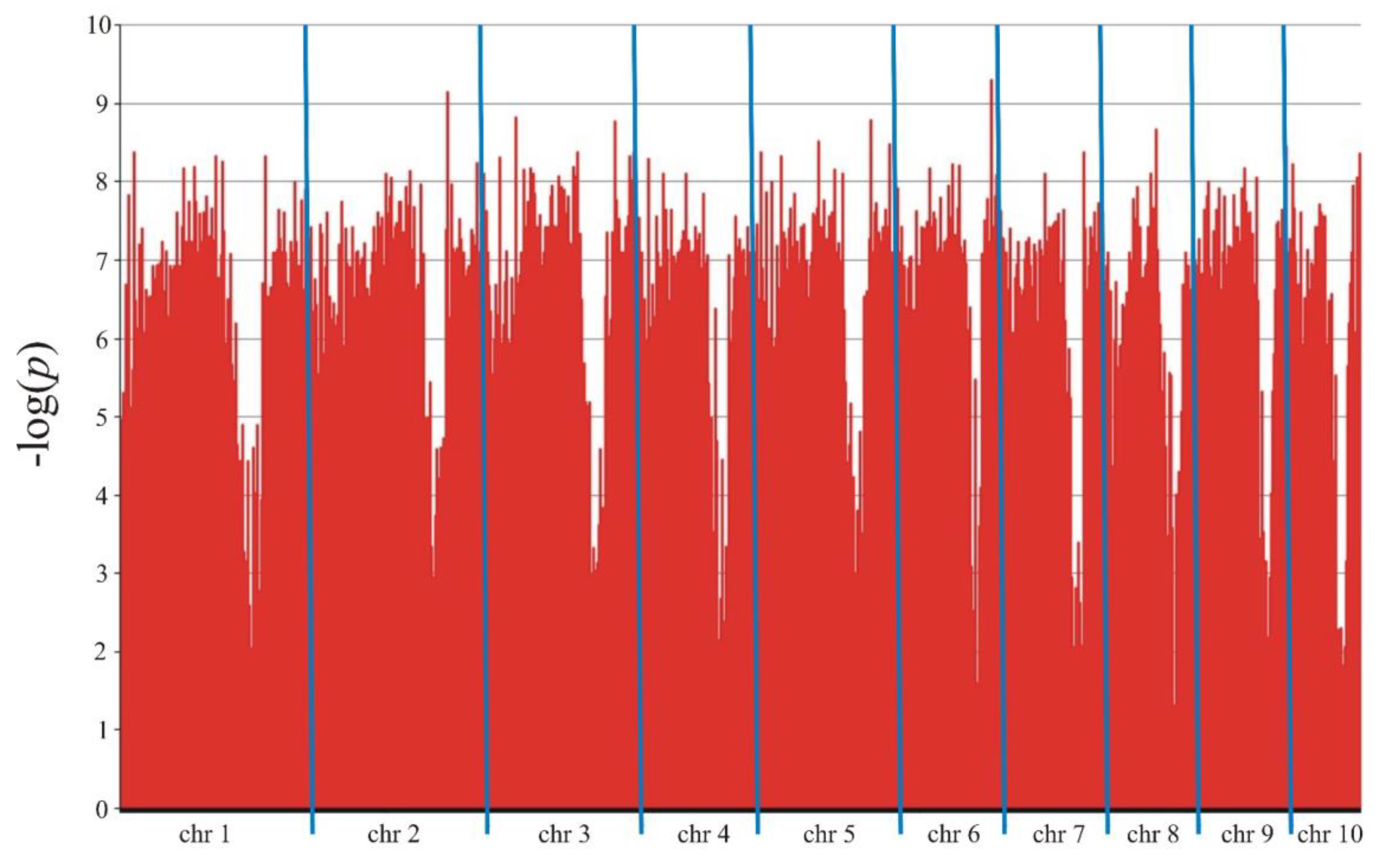
| Type of markers | All | SilicoDArT | SNP | |
|---|---|---|---|---|
| The number of markers | 11282 | 8405 | 2877 | |
| Negative | Numbers | 5800 | 4336 | 1464 |
| Effects | -17.06 – -2.14 | -13.88 – -2.14 | -17.06 – -2.14 | |
| Percentage variance accounted for | 2.4 – 31.4 | 2.4 – 31.4 | 2.4 – 30.5 | |
| LOD | 1.30 – 12.70 | 1.30 – 16.70 | 1.30 – 11.10 | |
| Positive | Numbers | 5482 | 4069 | 1413 |
| Effects | 2.14 – 8.55 | 2.14 – 8.55 | 2.17 – 7.37 | |
| Percentage variance accounted for | 2.4 – 34.4 | 2.4 – 33.7 | 2.4 – 34.4 | |
| LOD | 1.30 – 14.40 | 1.30 – 14.40 | 1.30 – 13.53 | |
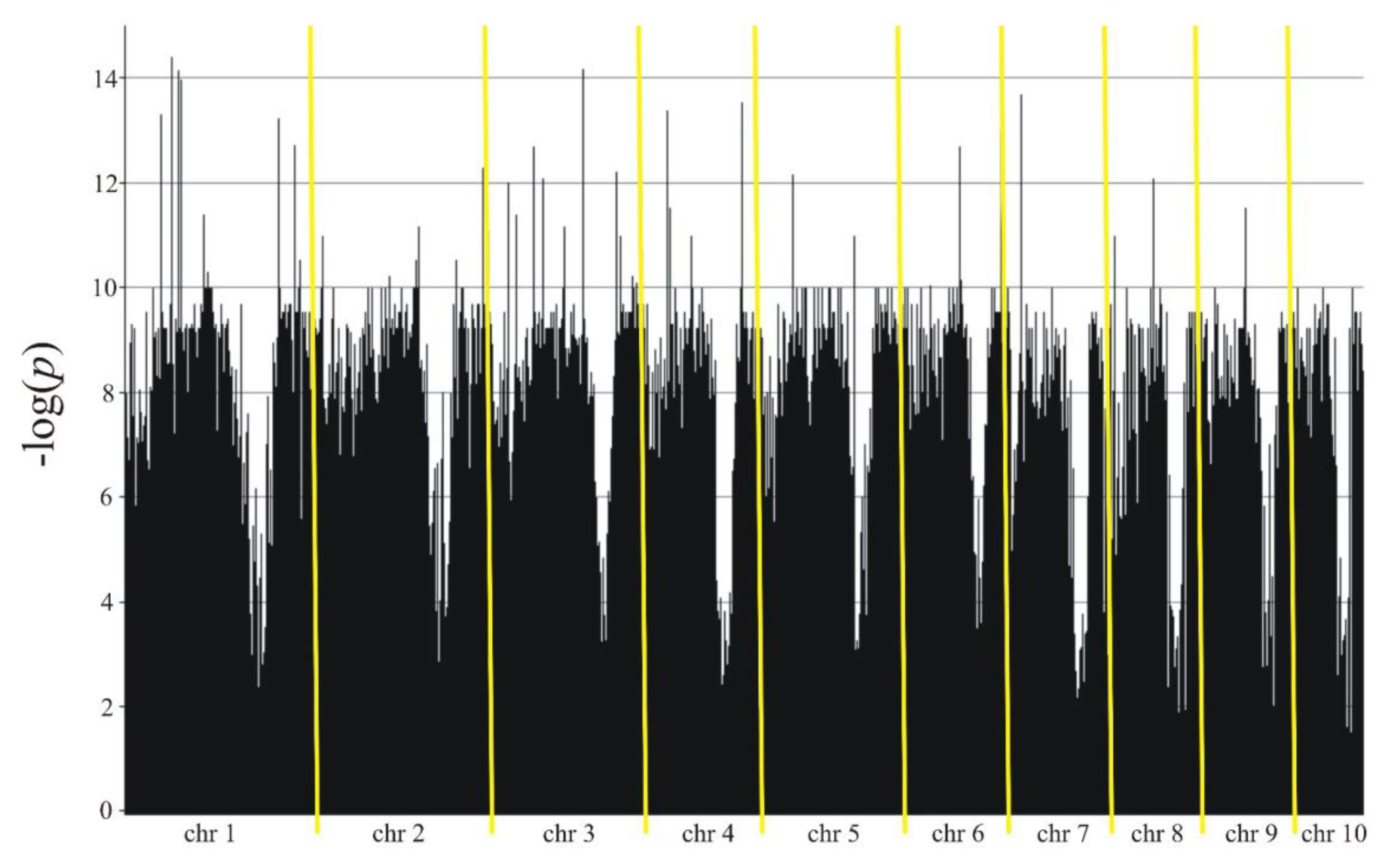
| Type of markers | All | SilicoDArT | SNP | |
|---|---|---|---|---|
| The number of markers | 9439 | 7148 | 2291 | |
| Negative | Numbers | 4523 | 3437 | 1086 |
| Effects | -6.49 – -0.78 | -6.79 – -0.777 | -4.38 – -0.784 | |
| Percentage variance accounted for | 2.4 – 27.0 | 2.4 – 25.7 | 2.4 – 27.0 | |
| LOD | 1.30 – 9.30 | 1.30 – 8.82 | 1.30 – 9.30 | |
| Positive | Numbers | 4916 | 3711 | 1205 |
| Effects | 0.779 – 2.214 | 0.779 – 2.214 | 0.788 – 2.173 | |
| Percentage variance accounted for | 2.4 – 25.3 | 2.4 – 25.3 | 2.4 – 23.9 | |
| LOD | 1.30 – 8.68 | 1.30 – 8.68 | 1.31 – 8.17 | |
| Chromosome | Marker type | CloneID | Kobierzyce | Smolice | ||||
|---|---|---|---|---|---|---|---|---|
| Estimate | Percen1 | LOD | Estimate | Percen | LOD | |||
| 1 | SilicoDArT | 29628241 | 5.902 | 23.4 | 8.02 | 2.167 | 24.3 | 8.33 |
| 1 | SilicoDArT | 82348823 | 6.031 | 24.4 | 8.34 | 2.167 | 24.2 | 8.27 |
| 1 | SilicoDArT | 24026805 | 6.219 | 26.0 | 8.92 | 2.156 | 24.0 | 8.20 |
| 1 | SNP | 4772298|F|0-68:A>G-68:A>G | -6.032 | 24.5 | 8.40 | -2.127 | 23.4 | 8.00 |
| 1 | SilicoDArT | 2488934 | -5.894 | 22.1 | 7.54 | -2.166 | 22.9 | 7.84 |
| 1 | SNP | 2439850|F|0-12:A>C-12:A>C | -6.061 | 24.9 | 8.52 | -2.092 | 22.7 | 7.76 |
| 1 | SilicoDArT | 4774875 | 5.823 | 22.9 | 7.83 | 2.092 | 22.7 | 7.76 |
| 1 | SilicoDArT | 9626410 | 6.623 | 29.9 | 10.30 | 2.073 | 22.3 | 7.62 |
| 1 | SNP | 2529315|F|0-56:T>G-56:T>G | -6.163 | 25.8 | 8.82 | -2.073 | 22.3 | 7.62 |
| 1 | SilicoDArT | 7054095 | -6.361 | 27.5 | 9.52 | -2.073 | 22.3 | 7.61 |
| 1 | SilicoDArT | 7051768 | 6.122 | 25.4 | 8.70 | 2.071 | 22.2 | 7.60 |
| 2 | SNP | 4772836|F|0-17:C>A-17:C>A | -6.722 | 30.5 | 11.10 | -2.148 | 23.7 | 8.11 |
| 2 | SNP | 4582743|F|0-42:G>A-42:G>A | -6.073 | 24.6 | 8.42 | -2.135 | 23.3 | 7.97 |
| 2 | SilicoDArT | 9694283 | 5.932 | 23.8 | 8.12 | 2.100 | 22.8 | 7.81 |
| 2 | SilicoDArT | 82349036 | 6.234 | 26.4 | 9.05 | 2.092 | 22.7 | 7.76 |
| 2 | SilicoDArT | 2395963 | 5.823 | 22.9 | 7.83 | 2.092 | 22.7 | 7.76 |
| 2 | SilicoDArT | 2382023 | 5.823 | 22.9 | 7.83 | 2.092 | 22.7 | 7.76 |
| 2 | SilicoDArT | 9703016 | 5.823 | 22.9 | 7.83 | 2.092 | 22.7 | 7.76 |
| 2 | SilicoDArT | 25942787 | 5.764 | 22.3 | 7.63 | 2.088 | 22.5 | 7.69 |
| 2 | SilicoDArT | 9633940 | 6.000 | 24.1 | 8.24 | 2.093 | 22.5 | 7.68 |
| 2 | SilicoDArT | 9718212 | 5.965 | 24.1 | 8.24 | 2.073 | 22.3 | 7.61 |
| 2 | SilicoDArT | 4778784 | -6.347 | 27.4 | 9.40 | -2.072 | 22.3 | 7.61 |
| 2 | SilicoDArT | 29619311 | 5.742 | 22.2 | 7.59 | 2.066 | 22.1 | 7.54 |
| 3 | SNP | 4772456|F|0-52:T>G-52:T>G | -5.802 | 22.5 | 7.68 | -2.227 | 25.6 | 8.77 |
| 3 | SNP | 4585365|F|0-34:T>C-34:T>C | -5.979 | 23.8 | 8.15 | -2.186 | 24.5 | 8.39 |
| 3 | SilicoDArT | 9682713 | -5.718 | 22.0 | 7.52 | -2.138 | 23.7 | 8.11 |
| 3 | SilicoDArT | 9717799 | 5.981 | 24.0 | 8.22 | 2.140 | 23.6 | 8.08 |
| 3 | SilicoDArT | 77157803 | 5.877 | 23.2 | 7.92 | 2.140 | 23.6 | 8.08 |
| 3 | SNP | 2433795|F|0-30:G>C-30:G>C | 5.996 | 24.2 | 8.29 | 2.129 | 23.4 | 8.02 |
| 3 | SilicoDArT | 77158337 | 5.768 | 22.5 | 7.68 | 2.111 | 23.1 | 7.91 |
| 3 | SilicoDArT | 29621917 | -5.998 | 24.4 | 8.36 | -2.101 | 23.0 | 7.85 |
| 3 | SilicoDArT | 5583810 | 6.071 | 24.9 | 8.54 | 2.100 | 22.8 | 7.81 |
| 3 | SNP | 4774080|F|0-65:C>T-65:C>T | -5.868 | 23.3 | 7.95 | -2.092 | 22.7 | 7.76 |
| 3 | SNP | 4774088|F|0-45:G>A-45:G>A | -5.868 | 23.3 | 7.95 | -2.092 | 22.7 | 7.76 |
| 3 | SNP | 7048352|F|0-24:A>G-24:A>G | -5.737 | 22.2 | 7.59 | -2.092 | 22.7 | 7.76 |
| 3 | SNP | 5586725|F|0-40:C>A-40:C>A | -5.761 | 22.2 | 7.60 | -2.099 | 22.7 | 7.75 |
| 3 | SilicoDArT | 82349016 | 6.017 | 24.5 | 8.38 | 2.074 | 22.3 | 7.60 |
| 3 | SilicoDArT | 4768318 | -5.845 | 23.1 | 7.90 | -2.067 | 22.2 | 7.58 |
| 3 | SilicoDArT | 34685358 | 6.096 | 25.2 | 8.64 | 2.066 | 22.2 | 7.57 |
| 3 | SNP | 2403483|F|0-62:C>A-62:C>A | -5.881 | 23.4 | 8.00 | -2.066 | 22.2 | 7.57 |
| 3 | SNP | 4771426|F|0-54:A>G-54:A>G | 6.129 | 25.3 | 8.66 | 2.072 | 22.1 | 7.55 |
| 3 | SNP | 4592970|F|0-41:T>C-41:T>C | -5.924 | 23.6 | 8.08 | -2.067 | 22.1 | 7.54 |
| 3 | SNP | 4586493|F|0-10:T>C-10:T>C | -5.734 | 22.1 | 7.56 | -2.063 | 22.0 | 7.52 |
| 4 | SilicoDArT | 70092308 | 5.823 | 22.9 | 7.83 | 2.092 | 22.7 | 7.76 |
| 4 | SilicoDArT | 9680684 | 5.823 | 22.9 | 7.83 | 2.092 | 22.7 | 7.76 |
| 4 | SilicoDArT | 25001071 | 5.854 | 23.1 | 7.90 | 2.078 | 22.4 | 7.64 |
| 4 | SilicoDArT | 25942566 | 5.747 | 22.2 | 7.60 | 2.078 | 22.4 | 7.64 |
| 4 | SilicoDArT | 25004669 | -6.261 | 26.3 | 9.05 | -2.078 | 22.1 | 7.56 |
| 5 | SNP | 4583014|F|0-63:A>G-63:A>G | -5.998 | 24.0 | 8.20 | -2.235 | 25.6 | 8.80 |
| 5 | SNP | 2536415|F|0-25:C>T-25:C>T | -5.926 | 23.6 | 8.09 | -2.187 | 24.8 | 8.48 |
| 5 | SilicoDArT | 4578971 | -5.881 | 23.2 | 7.93 | -2.177 | 24.5 | 8.38 |
| 5 | SilicoDArT | 2401113 | -6.205 | 26.0 | 8.92 | -2.168 | 24.3 | 8.33 |
| 5 | SilicoDArT | 29628894 | 6.294 | 26.8 | 9.22 | 2.148 | 23.9 | 8.17 |
| 5 | SilicoDArT | 2432042 | -5.722 | 22.0 | 7.51 | -2.128 | 23.4 | 8.01 |
| 5 | SilicoDArT | 2499631 | -6.401 | 27.7 | 9.52 | -2.108 | 23.0 | 7.85 |
| 5 | SilicoDArT | 9690308 | 6.326 | 27.2 | 9.40 | 2.092 | 22.7 | 7.76 |
| 5 | SilicoDArT | 9681187 | 6.145 | 25.6 | 8.77 | 2.092 | 22.7 | 7.76 |
| 5 | SilicoDArT | 2539842 | 6.016 | 24.5 | 8.38 | 2.081 | 22.4 | 7.66 |
| 5 | SilicoDArT | 4776114 | 6.062 | 24.8 | 8.51 | 2.081 | 22.4 | 7.66 |
| 5 | SilicoDArT | 4779143 | 6.465 | 28.5 | 9.70 | 2.066 | 22.2 | 7.57 |
| 5 | SilicoDArT | 2532640 | 6.355 | 27.5 | 9.52 | 2.066 | 22.2 | 7.57 |
| 6 | SNP | 4771464|F|0-65:C>T-65:C>T | -6.251 | 25.7 | 8.82 | -2.309 | 27.0 | 9.30 |
| 6 | SilicoDArT | 5587687 | 5.989 | 24.1 | 8.24 | 2.158 | 24.0 | 8.22 |
| 6 | SilicoDArT | 4770044 | -6.073 | 24.6 | 8.42 | -2.160 | 23.9 | 8.17 |
| 6 | SilicoDArT | 5588627 | 5.855 | 23.1 | 7.90 | 2.119 | 23.3 | 7.96 |
| 6 | SNP | 4770865|F|0-46:C>T-46:C>T | -6.028 | 24.6 | 8.41 | -2.101 | 22.9 | 7.81 |
| 6 | SilicoDArT | 2530960 | 5.794 | 22.6 | 7.73 | 2.098 | 22.8 | 7.79 |
| 6 | SilicoDArT | 9698143 | 6.459 | 28.2 | 9.70 | 2.084 | 22.4 | 7.67 |
| 6 | SNP | 4764810|F|0-20:T>C-20:T>C | -6.424 | 27.2 | 9.40 | -2.107 | 22.3 | 7.63 |
| 6 | SilicoDArT | 4775726 | 6.185 | 26.0 | 8.92 | 2.073 | 22.3 | 7.63 |
| 6 | SilicoDArT | 24029725 | 5.850 | 23.1 | 7.90 | 2.073 | 22.3 | 7.61 |
| 6 | SNP | 25947704|F|0-38:C>T-38:C>T | -6.129 | 25.4 | 8.72 | -2.061 | 22.0 | 7.51 |
| 7 | SNP | 67225764|F|0-56:C>G-56:C>G | -6.228 | 26.3 | 9.05 | -2.073 | 22.3 | 7.61 |
| 7 | SNP | 2523212|F|0-36:C>T-36:C>T | -5.778 | 22.5 | 7.70 | -2.072 | 22.3 | 7.61 |
| 8 | SilicoDArT | 9694332 | 5.899 | 23.5 | 8.03 | 2.081 | 22.4 | 7.66 |
| 8 | SilicoDArT | 9698173 | 5.991 | 24.3 | 8.33 | 2.066 | 22.2 | 7.57 |
| 8 | SilicoDArT | 4766257 | 5.708 | 22.0 | 7.52 | 2.060 | 22.0 | 7.52 |
| 9 | SNP | 2475427|F|0-37:A>G-37:A>G | -6.020 | 24.2 | 8.27 | -2.195 | 24.7 | 8.46 |
| 9 | SilicoDArT | 77157588 | 5.888 | 22.8 | 7.81 | 2.156 | 23.6 | 8.06 |
| 9 | SilicoDArT | 4584767 | -5.958 | 23.6 | 8.09 | -2.140 | 23.4 | 8.01 |
| 9 | SilicoDArT | 7049648 | -6.051 | 24.7 | 8.46 | -2.083 | 22.4 | 7.66 |
| 9 | SilicoDArT | 2384252 | 5.712 | 22.0 | 7.52 | 2.072 | 22.3 | 7.61 |
| 9 | SilicoDArT | 25947915 | 5.811 | 22.5 | 7.71 | 2.082 | 22.2 | 7.60 |
| 9 | SilicoDArT | 73750965 | 5.912 | 23.5 | 8.05 | 2.065 | 22.0 | 7.52 |
| 10 | SNP | 9667431|F|0-38:G>C-38:G>C | 6.403 | 27.8 | 9.52 | 2.120 | 23.3 | 7.96 |
| 10 | SilicoDArT | 2575390 | 5.817 | 22.9 | 7.82 | 2.085 | 22.6 | 7.72 |
| 10 | SilicoDArT | 9669953 | -5.857 | 22.8 | 7.80 | -2.095 | 22.4 | 7.66 |
| 10 | SilicoDArT | 5584917 | 6.512 | 28.9 | 10.00 | 2.073 | 22.3 | 7.62 |
| 10 | SilicoDArT | 2408858 | -5.816 | 22.6 | 7.72 | -2.084 | 22.3 | 7.61 |
| 10 | SilicoDArT | 7054499 | 6.179 | 25.9 | 8.89 | 2.066 | 22.2 | 7.57 |
| 10 | SilicoDArT | 25002574 | 6.455 | 28.2 | 9.70 | 2.070 | 22.1 | 7.56 |
| Marker | Marker Type | Chromosome | Marker Location | Candidate Genes |
|---|---|---|---|---|
| 4772836 | SNP | Chr 2 | 1002990 | serine/threonine-protein kinase bsk3 |
| 9626410 | SilicoDArT | Chr 1 | 147389719 | 45560 bp at 5' side: uncharacterized protein loc100276421 isoform x1 74882 bp at 3' side: ubiquitin carboxyl-terminal hydrolase 15 isoform x1 |
| 5584917 | SilicoDArT | Chr 10 | 137279593 | uncharacterized protein loc103641914 isoform x1 uncharacterized protein loc103641914 isoform x2 |
| 157434 | SilicoDArT | Chr 2 | 210848643 | 14188 bp at 5' side: formin-like protein 11 23892 bp at 3' side: uncharacterized protein loc100273879 |
| 9698143 | SilicoDArT | Chr 6 | 168628814 | 440 bp at 5' side: uncharacterized protein loc100273604 86823 bp at 3' side: uncharacterized protein loc100382826 precursor |
| 2499631 | SilicoDArT | Chr 5 | 223354035 | 33662 bp at 5' side: putative protein of unknown function (duf640) domain fami 10443 bp at 3' side: nac domain-containing protein 92 |
| 21699135 | SilicoDArT | Chr 5 | 148291902 | 3967 bp at 5' side: uncharacterized protein loc103627005 11771 bp at 3' side: uncharacterized protein loc100279673 precursor |
| 7054095 | SilicoDArT | Chr 1 | 192290825 | 1016 bp at 5' side: abc transporter c family member 10 24169 bp at 3' side: probable lipoxygenase 8, chloroplastic |
| 4779143 | SilicoDArT | Chr 5 | 32830342 | 20659 bp at 5' side: putative disease resistance protein rga4 517 bp at 3' side: uncharacterized protein loc101027116 precursor |
| 4765764 | SNP | Chr 5 | 213770951 | histidine kinase 1 |
| Marker | Primer sequence | Melting temperature (°C) | Product size (bp) | |
|---|---|---|---|---|
| Forward | Reverse | |||
| 4772836 | GGTGGTTTTACCCCCTGCAG | TATTTGCAGGCCCTTGACCT | 59 | 281 |
| 9626410 | AGCAATTTCTCCAGAGTCTGATG | CATGCATTTTTCTGCATTGGGC | 61 | 50 |
| 5584917 | TATTGAAGAGAGATATGATAATCGCTGCAG | GTTCAAATAACTCGCAAAAGACTCG | 58 | 78 |
| 157434 | CGGACCGTATTACCCGGTTA | AATTTCCGCGGTACCGAGGC | 55 | 270 |
| 9698143 | ACCGTGGCTAATCCGGTTAT | GGCATTCCGGGTAATCCGTT | 59 | 350 |
| 2499631 | CGGTTCCAATTGGGATTACC | CCTGGACCGGCTTTACAATC | 61 | 145 |
| 21699135 | CCGATACTGCATGCTCTGCG | CCTCTGTTTGGCGTAGGTGA | 59 | 64 |
| 7054095 | GTCGACGACGAACCCTGCAG | CCAATATCCGGCGGACAGAC | 61 | 55 |
| 4779143 | AATACCCTGGGTCCGGTAA | TTACCGGGTCCAACCTGGC | 58 | 180 |
| 4765764 | TTTTTTCCTTCTTGCTGCAG | CCTCGTTCTGTGAACTGGAA | 61 | 263 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





