Submitted:
19 December 2023
Posted:
20 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. High-density genome-wide markers
2.2. Phenotypic data
2.3. Data analysis
2.4. Marker trait association analysis
3. Results
3.1. Variation in biomass yield, plant height and feed quality traits of buffel grass accessions
3.2. Correlation of phenotypic and feed quality traits
3.3. Effect of genotype and seasonality on buffel grass forage performance
3.4. Quantitative genetic variation
3.5. Buffel grass accession clustering based on phenotypic and feed quality traits
3.6. Performance of genetic clusters identified using DArTSeq genome-wide markers
3.7. Genome-wide distribution and density of markers
3.8. Data filtering for association studies
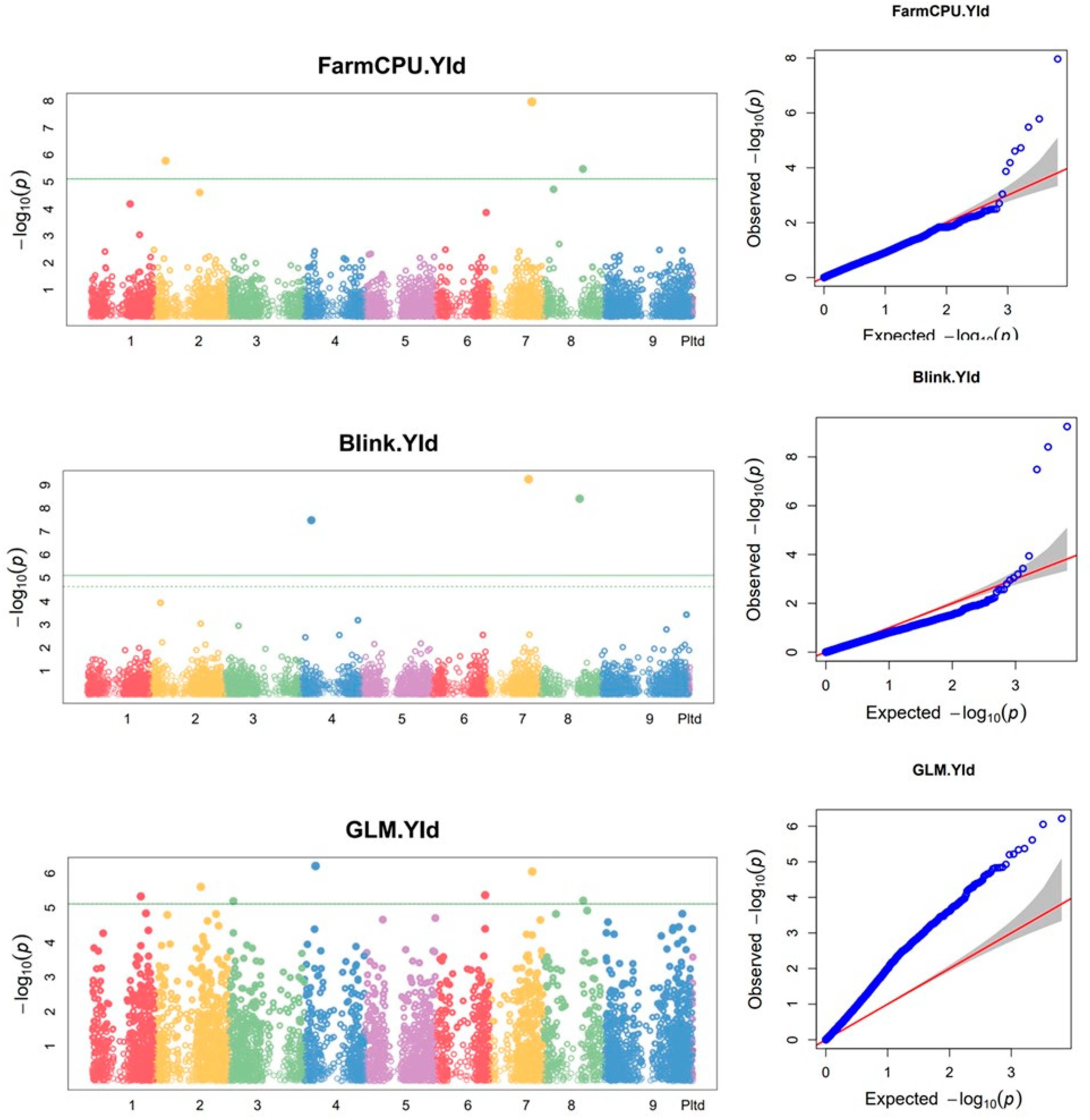
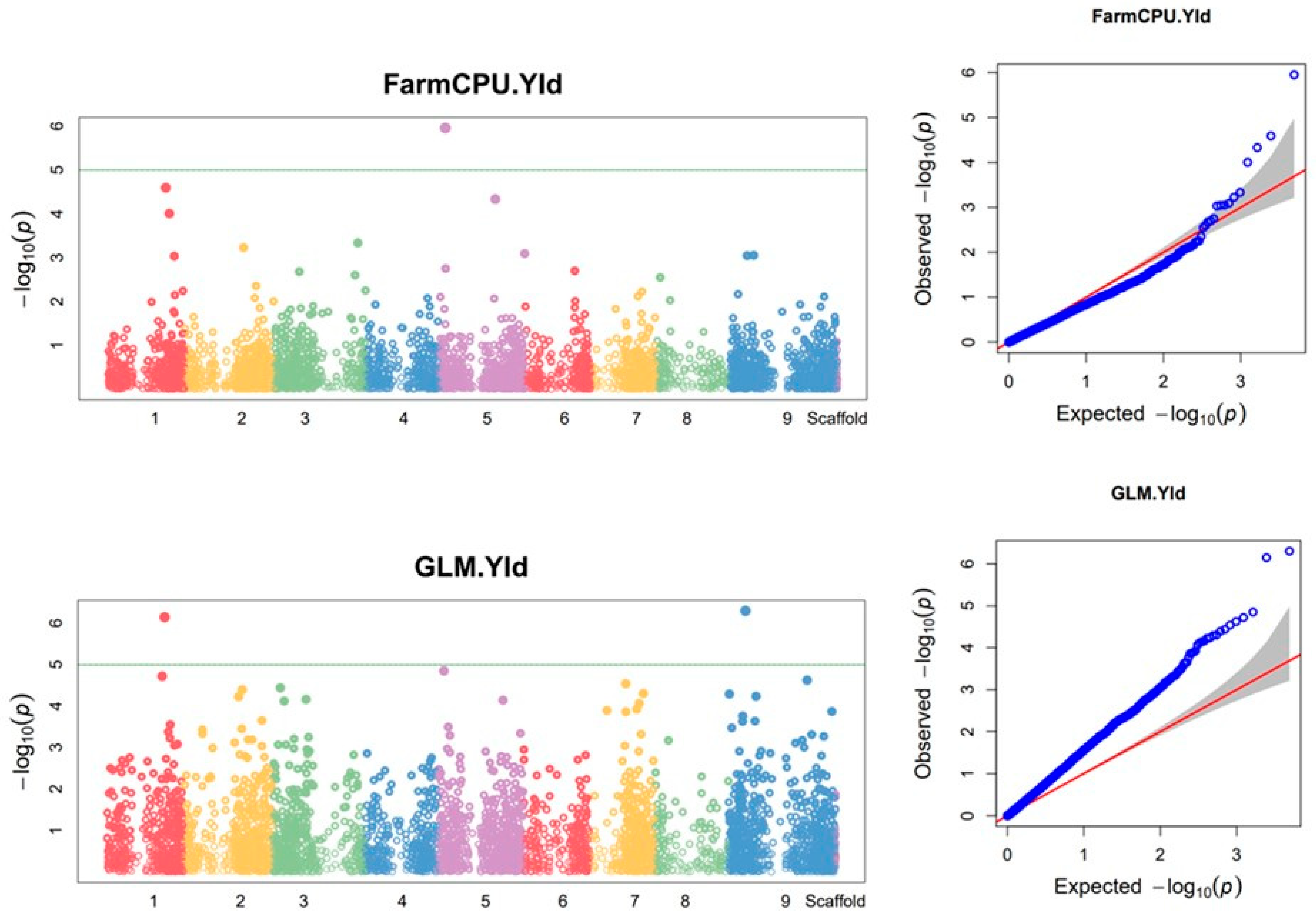
3.9. Markers associated with biomass yield and plant height
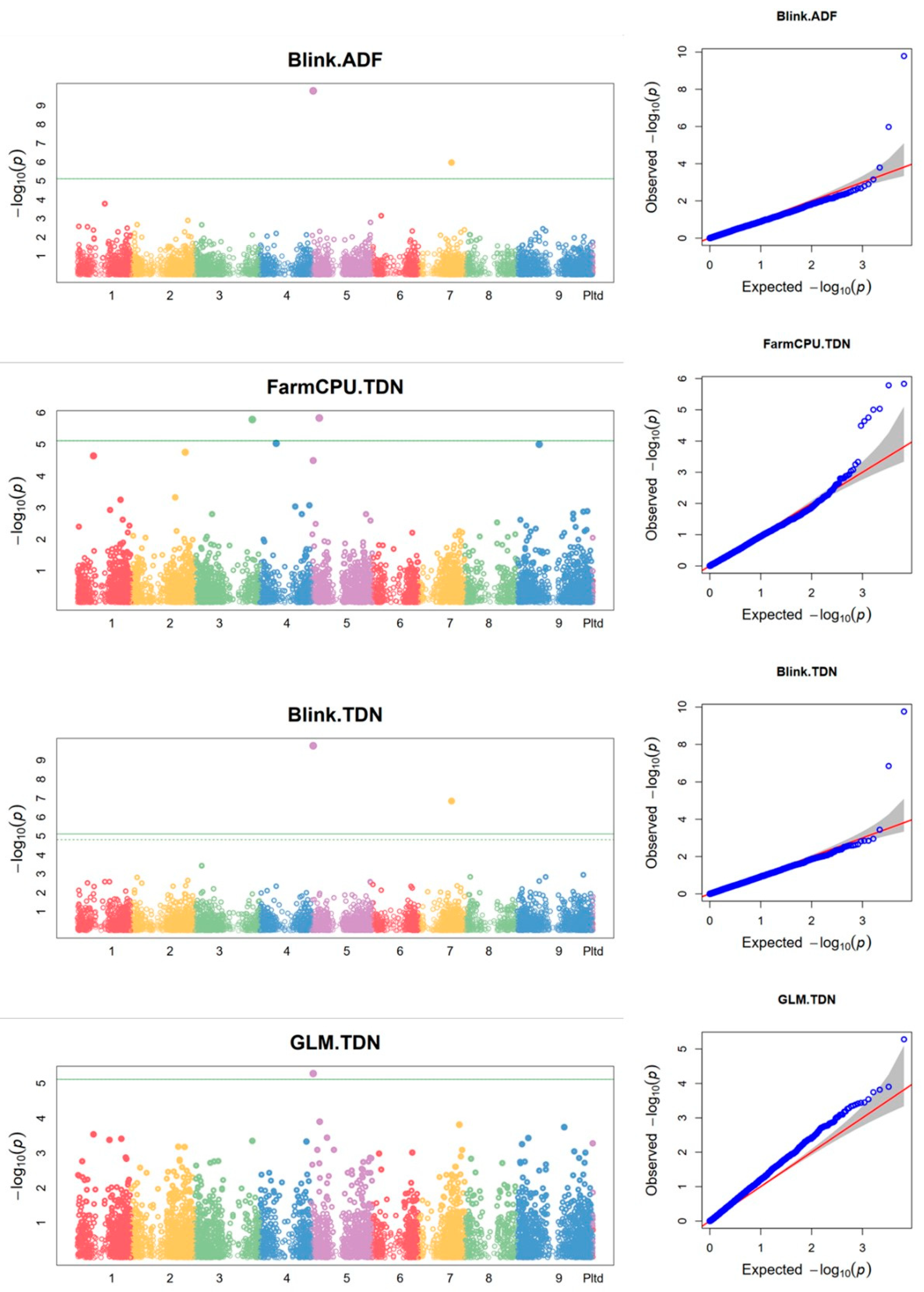
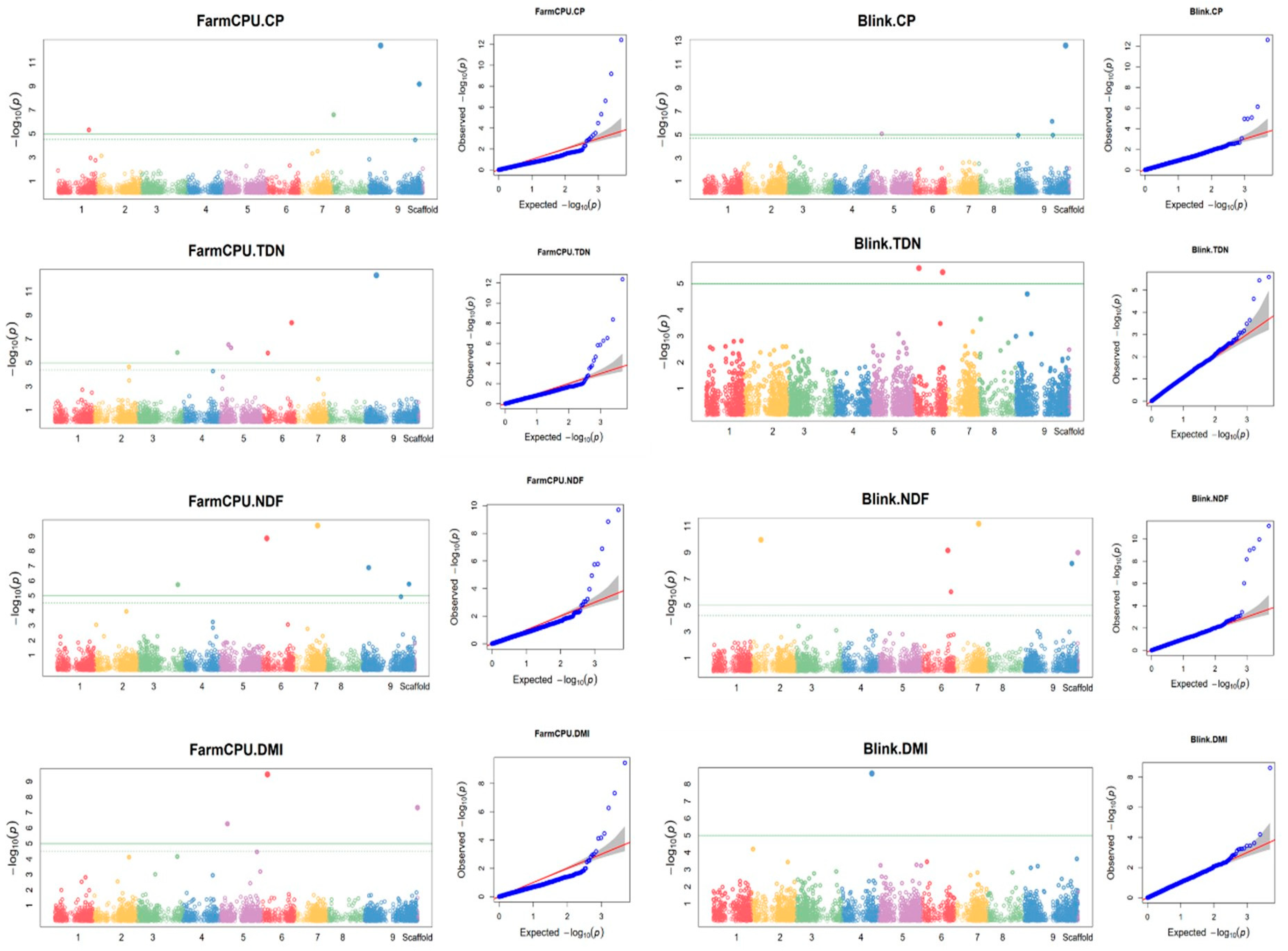
3.10. Markers associated with feed quality traits
4. Discussion
4.1. Markers associated with feed quality traits
4.2. Correlation of biomass yield, plant height and feed quality traits in buffel grass
4.3. Marker trait associations in buffel grass
4.4. Genome wide distribution and co-localization of the marker trait associations
4.5. Marker trait association in functional putative genomic regions
5. Conclusions and recommendations
- Developing a reference genome that can be used for marker mapping and genome wide association studies to identify major QTL for traits of interest with improved association accuracy.
- Buffel grass has different ploidy levels. Hence, determining the ploidy level, coupled with identification of sexually reproducing lines, will facilitate a breeding program for developing new improved varieties of this economically important forage species.
- Buffel grass is a drought tolerant grass species. Being an underutilized crop, little is known about the genetic basis of its drought tolerance trait. Hence, it is important to study the genetic and physiological basis of drought tolerance and other important traits to develop a climate resilient variety.
- The results of this study can also be used as a basis to develop a set of markers for future marker assisted selection and breeding.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herrero, M.; Grace, D.; Njuki, J.; Johnson, N.; Enahoro, D.; Silvestri, S.; Rufino, M.C. The roles of livestock in developing countries. Animal 2013, 7, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Moyo, S.; Swanepoel, F. Multifunctionality of livestock in developing communities. The role of livestock in developing communities: Enhancing multifunctionality 2010, 3, 69. [Google Scholar]
- Stroebel, A.; Swanepoel, F.; Pell, A. Sustainable smallholder livestock systems: A case study of Limpopo Province, South Africa. Livestock Science 2011, 139, 186–190. [Google Scholar] [CrossRef]
- Mekuriaw, Z.; Harris-Coble, L. Ethiopia’s livestock systems: Overview and areas of inquiry; Feed the Future Innovation Lab for Livestock Systems: Gainesville, FL, USA, 2021. [Google Scholar]
- Qaim, M. Role of new plant breeding technologies for food security and sustainable agricultural development. Applied Economic Perspectives and Policy 2020, 42, 129–150. [Google Scholar] [CrossRef]
- Nerkar, G.; Devarumath, S.; Purankar, M.; Kumar, A.; Valarmathi, R.; Devarumath, R.; Appunu, C. Advances in crop breeding through precision genome editing. Frontiers in Genetics 2022, 13, 880195. [Google Scholar] [CrossRef] [PubMed]
- Muktar, M.S.; Habte, E.; Teshome, A.; Assefa, Y.; Negawo, A.T.; Lee, K.W.; Zhang, J.Y.; Jones, C.S. Insights Into the Genetic Architecture of Complex Traits in Napier Grass (Cenchrus purpureus) and QTL Regions Governing Forage Biomass Yield, Water Use Efficiency and Feed Quality Traits. Frontiers in Plant Science 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Muktar, M.S.; Teshome, A.; Hanson, J.; Negawo, A.T.; Habte, E.; Entfellner, J.-B.D.; Lee, K.-W.; Jones, C.S. Genotyping by sequencing provides new insights into the diversity of Napier grass (Cenchrus purpureus) and reveals variation in genome-wide LD patterns between collections. Sci Rep-Uk 2019, 9, 6936. [Google Scholar] [CrossRef]
- Negawo, A.T.; Assefa, Y.; Hanson, J.; Abdena, A.; Muktar, M.S.; Habte, E.; Sartie, A.M.; Jones, C.S. Genotyping-By-Sequencing Reveals Population Structure and Genetic Diversity of a Buffelgrass (Cenchrus ciliaris L.) Collection. Diversity-Basel 2020, 12. [Google Scholar] [CrossRef]
- Negawo, A.T.; Muktar, M.S.; Assefa, Y.; Hanson, J.; Sartie, A.M.; Habte, E.; Jones, C.S. Genetic diversity and population structure of a Rhodes grass (Chloris gayana) collection. Genes 2021, 12, 1233. [Google Scholar] [CrossRef] [PubMed]
- Negawo, A.T.; Akinmade, H.O.; Muktar, M.S.; Habte, E.; Assefa, Y.; Muchugi, A.; Sartie, A.M.; Jones, C.S. Genetic Diversity, Population Structure and Subset Development in a Sesbania sesban Collection. Plants 2022, 12, 13. [Google Scholar] [CrossRef]
- Muktar, M.S.; Sartie, A.; Neawo, A.T.; Habte, E.; Jones, C.S. In Genetic diversity among and within accessions of a lablab (Lablab purpureus) collection maintained in the ILRI forage genebank, The XXIV International Grassland Congress / XI International Rangeland Congress, 2021; Kenya Agricultural and Livestock Research Organization.
- Higgins, J.; Tomaszewska, P.; Pellny, T.K.; Castiblanco, V.; Arango, J.; Tohme, J.; Schwarzacher, T.; Mitchell, R.A.; Heslop-Harrison, J.; De Vega, J.J. Diverged subpopulations in tropical Urochloa (Brachiaria) forage species indicate a role for facultative apomixis and varying ploidy in their population structure and evolution. Annals of Botany 2022, 130, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Sserumaga, J.P.; Kayondo, S.I.; Kigozi, A.; Kiggundu, M.; Namazzi, C.; Walusimbi, K.; Bugeza, J.; Molly, A.; Mugerwa, S. Genome-wide diversity and structure variation among lablab [Lablab purpureus (L.) Sweet] accessions and their implication in a Forage breeding program. Genetic Resources and Crop Evolution 2021, 68, 2997–3010. [Google Scholar] [CrossRef] [PubMed]
- Deo, T.G.; Ferreira, R.C.; Lara, L.A.; Moraes, A.C.; Alves-Pereira, A.; De Oliveira, F.A.; Garcia, A.A.; Santos, M.F.; Jank, L.; de Souza, A.P. High-resolution linkage map with allele dosage allows the identification of regions governing complex traits and apospory in guinea grass (Megathyrsus maximus). Frontiers in plant science 2020, 11, 15. [Google Scholar] [CrossRef]
- Carballo, J.; Santos, B.; Zappacosta, D.; Garbus, I.; Selva, J.P.; Gallo, C.A.; Díaz, A.; Albertini, E.; Caccamo, M.; Echenique, V. A high-quality genome of Eragrostis curvula grass provides insights into Poaceae evolution and supports new strategies to enhance forage quality. Sci Rep-Uk 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Njaci, I.; Waweru, B.; Kamal, N.; Muktar, M.S.; Fisher, D.; Gundlach, H.; Muli, C.; Muthui, L.; Maranga, M.; Kiambi, D.; Maass, B.L.; Emmrich, P.M.; Entfellner, J.-B.D.; Spannagl, M.; Chapman, M.A.; Shorinola, O.; Jones, C.S. Chromosome-scale assembly of the lablab genome - A model for inclusive orphan crop genomics. bioRxiv 2005. [Google Scholar] [CrossRef]
- Pessoa Filho, M.d.P.; Sousa Sobrinho, F.d.; Fragoso, R.d.R.; da Silva Junior, O.; Ferreira, M. In A draft genome assembly for the forage grass Urochloa ruziziensis based on single-molecule real-time sequencing, 2018; In: BRAZILIAN BIOTECHNOLOGY CONGRESS, 7.; BIOTECHNOLOGY IBERO-AMERICAN …. 2018. [Google Scholar]
- Worthington, M.; Perez, J.G.; Mussurova, S.; Silva-Cordoba, A.; Castiblanco, V.; Cardoso Arango, J.A.; Jones, C.; Fernandez-Fuentes, N.; Skot, L.; Dyer, S.; Tohme, J.; Di Palma, F.; Arango, J.; Armstead, I.; De Vega, J.J. A new genome allows the identification of genes associated with natural variation in aluminium tolerance in Brachiaria grasses. Journal of Experimental Botany 2020, 72, 302–319. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Wu, F.; Xu, P.; Sun, Z.Y.; Li, J.; Gao, L.J.; Lu, L.Y.; Chen, D.D.; Muktar, M.; Jones, C.; Yi, X.F.; Zhang, J.Y. The elephant grass (Cenchrus purpureus) genome provides insights into anthocyanidin accumulation and fast growth. Mol Ecol Resour 2021, 21, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xia, Z.; Li, C.; Wang, X.; Lu, X.; Zhang, W.; Ma, H.; Zhou, X.; Zhang, W.; Zhu, T. Chromosome-scale genome assembly provides insights into speciation of allotetraploid and massive biomass accumulation of elephant grass (Pennisetum purpureum Schum.). Mol Ecol Resour 2022, 22, 2363–2378. [Google Scholar] [CrossRef]
- Zheng, H.; Wang, B.; Hua, X.; Gao, R.; Wang, Y.; Zhang, Z.; Zhang, Y.; Mei, J.; Huang, Y.; Huang, Y. A near-complete genome assembly of the allotetrapolyploid Cenchrus fungigraminus (JUJUNCAO) provides insights into its evolution and C4 photosynthesis. Plant Communications 2023. [Google Scholar] [CrossRef] [PubMed]
- Marks, R.A.; Hotaling, S.; Frandsen, P.B.; VanBuren, R. Representation and participation across 20 years of plant genome sequencing. Nature plants 2021, 7, 1571–1578. [Google Scholar] [CrossRef] [PubMed]
- Marshall, V.M.; Lewis, M.M.; Ostendorf, B. Buffelgrass (Cenchrus ciliaris) as an invader and threat to biodiversity in arid environments: A review. Journal of Arid Environments 2012, 78, 1–12. [Google Scholar] [CrossRef]
- Cook, B.; Pengelly, B.; Schultze-Kraft, R.; Taylor, M.; Burkart, S.; Cardoso Arango, J.; González Guzmán, J.; Cox, K.; Jones, C.; Peters, M. Tropical Forages: An interactive selection tool. 2nd and Revised Edn. International Center for Tropical Agriculture (CIAT), Cali, Colombia and International Livestock Research Institute (ILRI), Nairobi, Kenya. www. tropicalforages. info 2020.
- Kharrat-Souissi, A.; Siljak-Yakovlev, S.; Brown, S.C.; Baumel, A.; Torre, F.; Chaieb, M. The polyploid nature of Cenchrus ciliaris L. (Poaceae) has been overlooked: new insights for the conservation and invasion biology of this species - a review. Rangeland J 2014, 36, 11–23. [Google Scholar] [CrossRef]
- Kharrat-Souissi, A.; Siljak-Yakovlev, S.; Brown, S.C.; Chaieb, M. Cytogeography of Cenchrus ciliaris (Poaceae) in Tunisia. Folia Geobot 2013, 48, 95–113. [Google Scholar] [CrossRef]
- Heuzé, V.; Tran, G.; Baumont, R.; Lebas, F. Buffel grass (Cenchrus ciliaris). Feedipedia, a programme by INRA, CIRAD, AFZ and FAO. 2016.
- Kisambo, B.K.; Wasonga, O.V.; Koech, O.K.; Karuku, G.N. Morphological and productivity responses of Buffel grass (Cenchrus ciliaris) and Guinea grass (Panicum maximum) ecotypes to simulated grazing in a semi-arid environment. Grassland Research 2022, 1, 290–300. [Google Scholar] [CrossRef]
- Alhammad, B.A.; Mohamed, A.; Raza, M.A.; Ngie, M.; Maitra, S.; Seleiman, M.F.; Wasonga, D.; Gitari, H.I. Optimizing productivity of Buffel and Sudan grasses using optimal nitrogen fertilizer application under arid conditions. Agronomy 2023, 13, 2146. [Google Scholar] [CrossRef]
- Sanderson, M.; Voigt, P.; Jones, R. Yield and quality of warm-season grasses in central Texas. Rangeland Ecology & Management/Journal of Range Management Archives 1999, 52, 145–150. [Google Scholar]
- Arshadullah, M.; Malik, M.A.; Rasheed, M.; Jilani, G.; Zahoor, F.; Kaleem, S. Seasonal and genotypic variations influence the biomass and nutritional ingredients of Cenchrus ciliaris grass forage. International Journal of Agriculture and Biology 2011, 13. [Google Scholar]
- Yigzaw, G.W. Effect of harvesting stage on yield and nutritive value of buffel grass (Cenchrus ciliaris linn) under irrigation at Gewane district, north eastern Ethiopia. J. Sci. Innov. Res 2019, 8, 7–12. [Google Scholar] [CrossRef]
- Simeão, R.M.; Resende, M.D.; Alves, R.S.; Pessoa-Filho, M.; Azevedo, A.L.S.; Jones, C.S.; Pereira, J.F.; Machado, J.C. Genomic selection in tropical forage grasses: Current status and future applications. Frontiers in Plant Science 2021, 12, 665195. [Google Scholar] [CrossRef] [PubMed]
- Jorge, M.A.B.; Van De Wouw, M.; Hanson, J.; Mohammed, J. Characterisation of a collection of buffel grass (Cenchrus ciliaris). Tropical Grasslands 2008, 42, 27–39. [Google Scholar]
- Sánchez Gutiérrez, R.A.; Morales Nieto, C.R.; Hanson, J.; Santellano Estrada, E.; Jurado Guerra, P.; Villanueva Avalos, J.F.; Melgoza Castillo, A. Forage characterization of ecotypes of buffel grass under temporary conditions in Debre Zeit, Ethiopia. Revista Mexicana De Ciencias Agrícolas 2017, 8, 14. [Google Scholar]
- Negawo, A.T.; Habte, E.; Muktar, M.S.; Sartie, A.; Jones, C.S. In Molecular characterization of apomixis in Cenchrus ciliaris and its application in genetic resources improvement, Conference on International Research on Food Security, 2021.
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C.; Aschenbrenner-Kilian, M.; Evers, M.; Peng, K.; Cayla, C.; Hok, P.; Uszynski, G. Diversity arrays technology: a generic genome profiling technology on open platforms. Methods in molecular biology 2012, 888, 67–89. [Google Scholar] [PubMed]
- Bennetzen, J.L.; Schmutz, J.; Wang, H.; Percifield, R.; Hawkins, J.; Pontaroli, A.C.; Estep, M.; Feng, L.; Vaughn, J.N.; Grimwood, J.; Jenkins, J.; Barry, K.; Lindquist, E.; Hellsten, U.; Deshpande, S.; Wang, X.W.; Wu, X.M.; Mitros, T.; Triplett, J.; Yang, X.H.; Ye, C.Y.; Mauro-Herrera, M.; Wang, L.; Li, P.H.; Sharma, M.; Sharma, R.; Ronald, P.C.; Panaud, O.; Kellogg, E.A.; Brutnell, T.P.; Doust, A.N.; Tuskan, G.A.; Rokhsar, D.; Devos, K.M. Reference genome sequence of the model plant Setaria. Nat Biotechnol 2012, 30, 555. [Google Scholar] [CrossRef] [PubMed]
- Habte, E.; Muktar, M.S.; Abdena, A.; Hanson, J.; Sartie, A.M.; Negawo, A.T.; Machado, J.C.; Ledo, F.J.d.S.; Jones, C.S. Forage performance and detection of marker trait associations with potential for Napier grass (Cenchrus purpureus) improvement. Agronomy 2020, 10, 542. [Google Scholar] [CrossRef]
- Endecott, R.L.; Mathis, C.P. Ration Balancing on the Ranch. New Mexico State University, Cooperative Extension Service. 2006.
- Juergen, G.; Uwe, L. nortest: Tests for Normality. R package version 1.0-4. 2015.
- Burton, G.W.; Devane, d.E. Estimating heritability in tall fescue (Festuca arundinacea) from replicated clonal material 1. Agronomy journal 1953, 45, 478–481. [Google Scholar] [CrossRef]
- Team, R.C. R: A language and environment for statistical computing; R Foundation for Statistical Computing: Vienna, Austria, URL https://www.R-project.org/. 2022.
- Liu, X.L.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z.W. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. Plos Genet 2016, 12. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, X.L.; Zhou, Y.; Summers, R.M.; Zhang, Z.W. BLINK: a package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2019, 8. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006, 38, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.B.; Zhang, Z.W. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom Proteom Bioinf 2021, 19, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Ouellette, L.A.; Reid, R.W.; Blanchard, S.G.; Brouwer, C.R. LinkageMapView—rendering high-resolution linkage and QTL maps. Bioinformatics 2018, 34, 306–307. [Google Scholar] [CrossRef] [PubMed]
- Hanselka, C.W. Forage quality of common buffelgrass as influenced by prescribed fire. Texas Journal of Agriculture and Natural Resources 1989, 3, 15–18. [Google Scholar]
- Sánchez-Gutiérrez, R.A.; Hanson, J.; Jones, C.; Jurado-Guerra, P.; Santellano-Estrada, E.; Melgoza-Castillo, A.; Morales-Nieto, C. Caracterización morfológica de genotipos de pasto Buffel con potencial para producción de forraje y semilla. Rev Fitotec Mex 2020, 43, 343–343. [Google Scholar] [CrossRef]
- Tyagi, V.C.; Singh, T.; Dikshit, N.; Singh, S.; Rana, M.; Kaldate, R.; Govindaswamy, P.; Halli, H.M.; Ghosh, A.; Singhal, R.K. Genetic and Genomic Resources of Range Grasses: Status and Future Prospects. Molecular Interventions for Developing Climate-Smart Crops: A Forage Perspective 2023, 3–34. [Google Scholar]
- Gurikar, C.; Gowda, N.N.; Hanumantharaju, K.; Netravati, B. Role of Bacillus species in soil fertility with reference to rhizosphere engineering. In Rhizosphere Engineering; Elsevier, 2022; pp. 65–76. [Google Scholar]
- Shine, M.; Yang, J.W.; El-Habbak, M.; Nagyabhyru, P.; Fu, D.Q.; Navarre, D.; Ghabrial, S.; Kachroo, P.; Kachroo, A. Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytologist 2016, 212, 627–636. [Google Scholar] [CrossRef]
- Cass, C.L.; Peraldi, A.; Dowd, P.F.; Mottiar, Y.; Santoro, N.; Karlen, S.D.; Bukhman, Y.V.; Foster, C.E.; Thrower, N.; Bruno, L.C. Effects of PHENYLALANINE AMMONIA LYASE (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. Journal of Experimental Botany 2015, 66, 4317–4335. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Song, B.; Dai, Y.; Zhang, S.; Huang, X. Genome-wide identification and functional analysis of U-box E3 ubiquitin ligases gene family related to drought stress response in Chinese white pear (Pyrus bretschneideri). BMC Plant Biology 2021, 21, 235. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-Y.; Lu, Z.-W.; Sun, Y.; Fang, Z.-W.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z.; Xu, Z.-S.; Min, D.-H. The ankyrin-repeat gene GmANK114 confers drought and salt tolerance in Arabidopsis and soybean. Frontiers in Plant Science 2020, 11, 584167. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Ortiz, C.; Peña-Garcia, Y.; Natarajan, P.; Bhandari, M.; Abburi, V.; Dutta, S.K.; Yadav, L.; Stommel, J.; Nimmakayala, P.; Reddy, U.K. The ankyrin repeat gene family in Capsicum spp: Genome-wide survey, characterization and gene expression profile. Sci Rep-Uk 2020, 10, 4044. [Google Scholar] [CrossRef] [PubMed]
- Kolodziej, M.C.; Singla, J.; Sánchez-Martín, J.; Zbinden, H.; Šimková, H.; Karafiátová, M.; Doležel, J.; Gronnier, J.; Poretti, M.; Glauser, G. A membrane-bound ankyrin repeat protein confers race-specific leaf rust disease resistance in wheat. Nature communications 2021, 12, 956. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhao, J.; Chen, D.; Wang, Y. E3 ubiquitin ligases: styles, structures and functions. Molecular Biomedicine 2021, 2, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Al-Saharin, R.; Hellmann, H.; Mooney, S. Plant E3 Ligases and Their Role in Abiotic Stress Response. Cells-Basel 2022, 11. [Google Scholar] [CrossRef]
- van Amerongen, H.; van Bolhuis, B.M.; Betts, S.; Mei, R.; van Grondelle, R.; Yocum, C.F.; Dekker, J.P. Spectroscopic characterization of CP26, a chlorophyll ab binding protein of the higher plant Photosystem II complex. Biochimica et Biophysica Acta (BBA)-Bioenergetics 1994, 1188, 227–234. [Google Scholar] [CrossRef]
- Huang, Q.; Li, L.; Zheng, M.; Chen, F.; Long, H.; Deng, G.; Pan, Z.; Liang, J.; Li, Q.; Yu, M. The tryptophan decarboxylase 1 gene from Aegilops variabilis No. 1 regulate the resistance against cereal cyst nematode by altering the downstream secondary metabolite contents rather than auxin synthesis. Frontiers in Plant Science 2018, 9, 1297. [Google Scholar] [CrossRef] [PubMed]
- Facchini, P.J.; Huber-Allanach, K.L.; Tari, L.W. Plant aromatic L-amino acid decarboxylases: evolution, biochemistry, regulation, and metabolic engineering applications. Phytochemistry 2000, 54, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; An, Y.; Xu, P.; Xiao, J. Functioning of PPR proteins in organelle RNA metabolism and chloroplast biogenesis. Frontiers in plant science 2021, 12, 627501. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; He, Z.; Pandey, G.K.; Tsuchiya, T.; Luan, S. Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. Journal of Biological Chemistry 2002, 277, 5360–5368. [Google Scholar] [CrossRef]
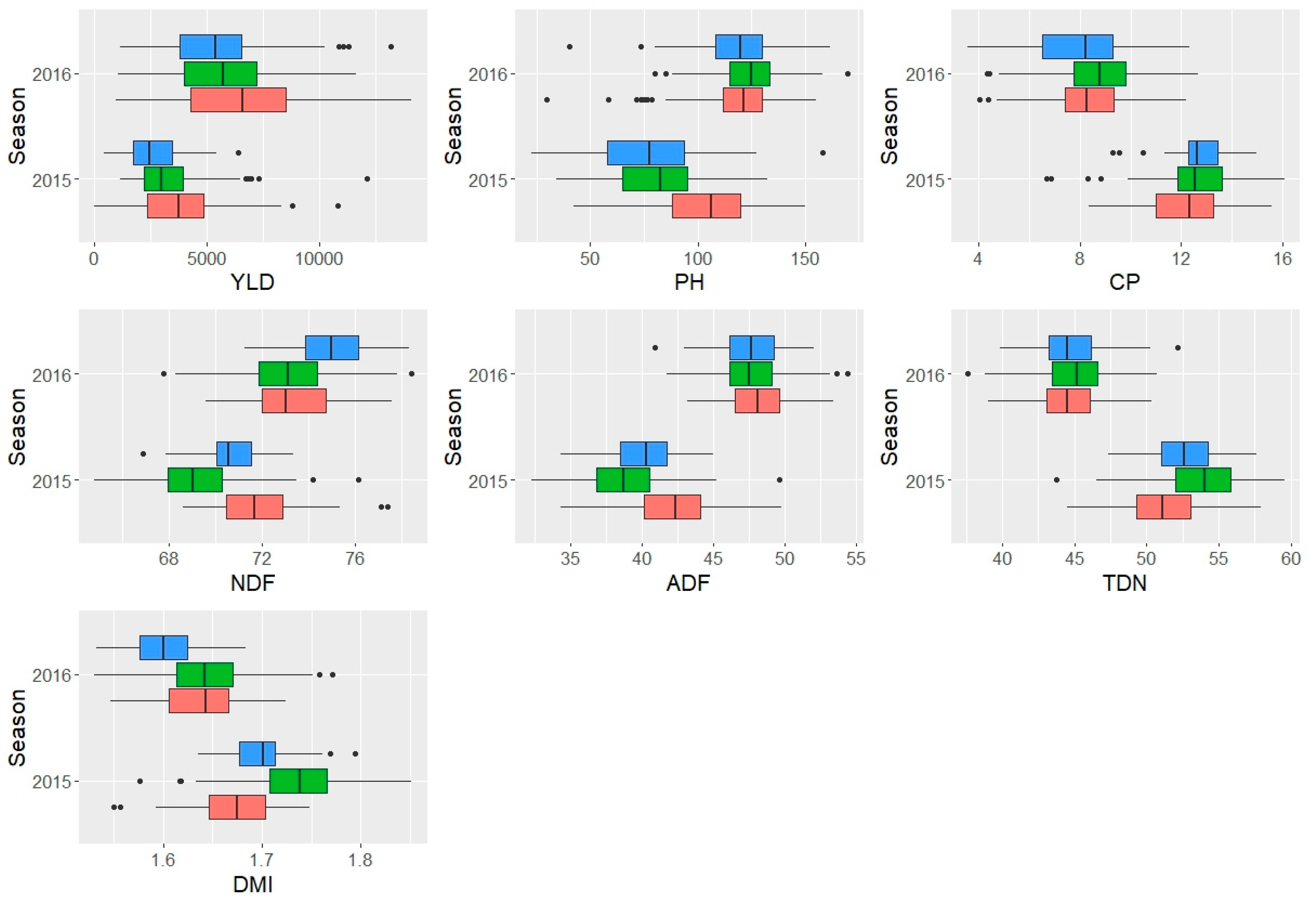
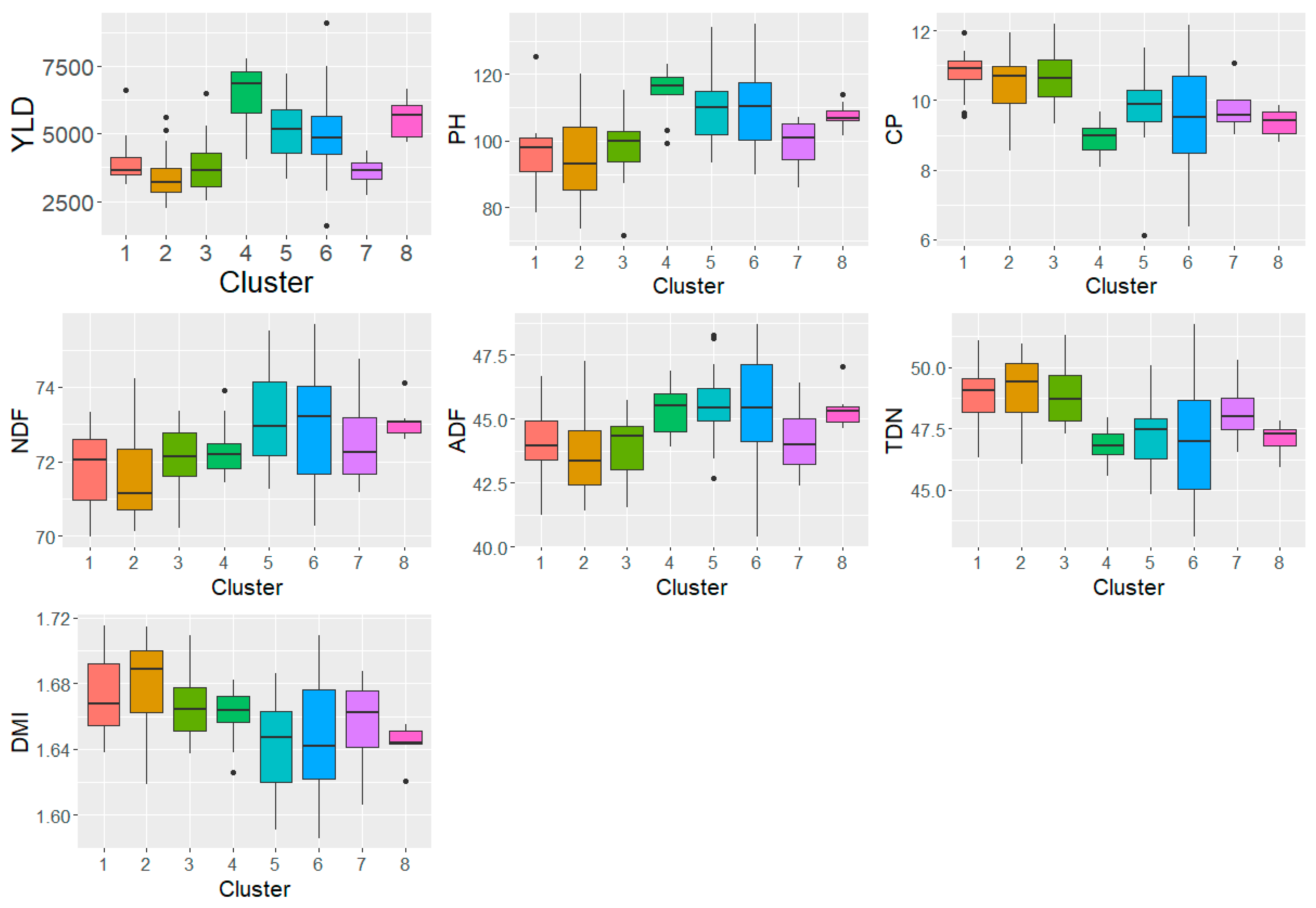
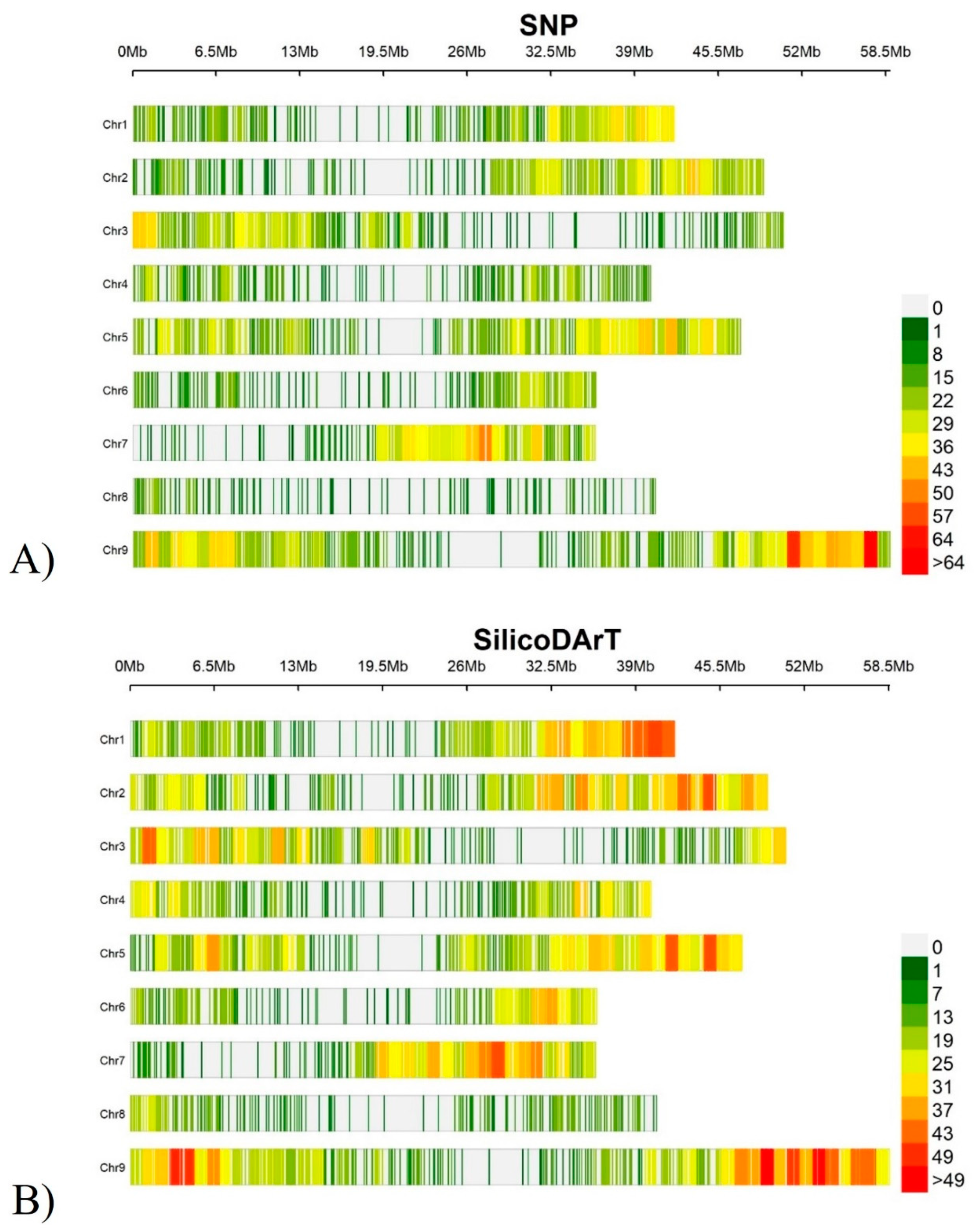

| Traits /Sources of variation | YLD | PH | NDF | ADF | CP | TND | DMI |
| Genotype | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 |
| Replication | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 |
| Season | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 |
| Genotype: Season | NS | NS | NS | 0.001 | NS | NS | NS |
| CV% | 34.9 | 17.9 | 2.3 | 4.7 | 13.4 | 4.1 | 2.3 |
| R-square % | 73 | 73 | 77 | 88 | 85 | 89 | 78 |
| Traits/statistics | Minimum | Maximum | Mean | PCV | GCV | H2 (%) |
| YLD (Kg/ha) | 1609.65 | 9097.54 | 4562.22 | 28.1 | 28.1 | 99.9 |
| PH (cm) | 71.50 | 135.22 | 103.31 | 13.9 | 9.9 | 70.9 |
| CP (%) | 6.11 | 12.21 | 10.02 | 32.8 | 8.9 | 27.0 |
| NDF (%) | 69.98 | 75.68 | 72.37 | 11.8 | 1.2 | 10.2 |
| ADF (%) | 40.40 | 48.69 | 44.62 | 15.2 | 2.8 | 18.5 |
| TND (%) | 43.10 | 51.73 | 48.03 | 14.7 | 2.9 | 19.7 |
| DMI (%) | 1.59 | 1.72 | 1.66 | 77.6 | 1.2 | 1.6 |
| No. | Model | Marker ID | Marker sequence | RefSeq sequence | Chr | pos | P.value | maf | R2.without.SNP | R2.with.SNP | R2* | FDR_Adjusted_P-values | Effect |
| 1 | GLM | 30838261 | TGCAGGTTTGAGGCTTGTCAGTGTGCTCGTCCCCTTGTGCCGACCTTTCCCAGGCGTCCCTGTCCGAGA | NC_028450.1 | 1 | 32744739 | 4.60E-06 | 0.0789 | 0.296 | 0.445 | 0.149 | 0.0059 | 2009.3639 |
| 2 | FarmCPU | 30921428 | TGCAGCAAATACTTACCAGAGCACAGGTTGCCAGAAAATATTGTTGCAACAACAAGTGCTGCTGATGCT | NC_028451.1 | 2 | 8049803 | 1.65E-06 | 0.0965 | NA | NA | NA | 0.0054 | -969.2590 |
| 3 | GLM | 30912865 | TGCAGAGAGTTGCAAAACGTATCGAAACAAATGTTGGAGACTTGCCGTGGGGTGAGGTGAAGACGGACT | NC_028451.1 | 2 | 30749442 | 2.45E-06 | 0.0965 | 0.296 | 0.455 | 0.158 | 0.0053 | 1608.4054 |
| 4 | GLM | 30829864 | TGCAGGCCGATCACGCTGTACGCCATGTGACCCAGCCGCGACGCCACCTGCACCGCGAACCGCAAAATG | NC_028452.1 | 3 | 3213526 | 6.32E-06 | 0.1184 | 0.296 | 0.441 | 0.144 | 0.0059 | -1985.8887 |
| 5 | GLM | 30944290 | TGCAGCTGCTCCACTGTTTTCGCACTGCTGAACTGTTCTTCTCTAACTGAAGAATATTTGTGGGCAACC | NC_028453.1 | 4 | 7437264 | 6.10E-07 | 0.0746 | 0.296 | 0.476 | 0.180 | 0.0029 | 1779.3729 |
| 6 | Blink | 30944290 | TGCAGCTGCTCCACTGTTTTCGCACTGCTGAACTGTTCTTCTCTAACTGAAGAATATTTGTGGGCAACC | NC_028453.1 | 4 | 7437264 | 3.27E-08 | 0.0746 | NA | NA | NA | 0.0001 | NA |
| 7 | GLM | 30846885 | TGCAGAGAGAGGGAGAGAGAGGCTATCCTACTATGCAACGGTCAAAAGGCTTCAAAGGAGGAGAAATCA | NC_028455.1 | 6 | 33041360 | 4.25E-06 | 0.1053 | 0.296 | 0.447 | 0.150 | 0.0059 | -1877.6778 |
| 8 | GLM | 30838332 | TGCAGTCCTAAACACCAGCACAGCACTCTCCTCTCCTTCCATCCCTAACATACATCATCAGCGATACAG | NC_028456.1 | 7 | 28411310 | 8.87E-07 | 0.0789 | 0.296 | 0.470 | 0.174 | 0.0029 | 1764.3647 |
| 9 | FarmCPU | 30838332 | TGCAGTCCTAAACACCAGCACAGCACTCTCCTCTCCTTCCATCCCTAACATACATCATCAGCGATACAG | NC_028456.1 | 7 | 28411310 | 1.08E-08 | 0.0789 | NA | NA | NA | 0.0001 | 1528.8742 |
| 10 | Blink | 30838332 | TGCAGTCCTAAACACCAGCACAGCACTCTCCTCTCCTTCCATCCCTAACATACATCATCAGCGATACAG | NC_028456.1 | 7 | 28411310 | 5.72E-10 | 0.0789 | NA | NA | NA | 0.0000 | NA |
| 11 | GLM | 30846154 | TGCAGTCTCCCAATCTCCCGTGGGAGCTCTGTGATTTGATCGCAGTCCTTGAGATCCAGATACCTAAGC | NC_028457.1 | 8 | 26442566 | 6.10E-06 | 0.0877 | 0.296 | 0.441 | 0.145 | 0.0059 | -1463.6930 |
| 12 | FarmCPU | 30846154 | TGCAGTCTCCCAATCTCCCGTGGGAGCTCTGTGATTTGATCGCAGTCCTTGAGATCCAGATACCTAAGC | NC_028457.1 | 8 | 26442566 | 3.30E-06 | 0.0877 | NA | NA | NA | 0.0072 | -853.8910 |
| 13 | Blink | 30846154 | TGCAGTCTCCCAATCTCCCGTGGGAGCTCTGTGATTTGATCGCAGTCCTTGAGATCCAGATACCTAAGC | NC_028457.1 | 8 | 26442566 | 3.91E-09 | 0.0877 | NA | NA | NA | 0.0000 | NA |
| No. | Model | Marker ID | Marker sequence | Alleles | RefSeq sequence | Chr | pos | P.value | maf | R2without.SNP | R2.with.SNP | R2* | FDR_Adjusted_P-values | Effect |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GLM | 30964292-59-G/A | TGCAGCTCAGAGCAGTACGACGCCATGGCGATCTCGGCGCCCTTGAACCCGTAGTCCAGGCTCGGGTTG | G/A | NC_028450.1 | 1 | 31786540 | 7.11E-07 | 0.179 | 0.437 | 0.571 | 0.314 | 0.0017 | -1149.0389 |
| 2 | FarmCPU | 30935961-51-C/T | TGCAGATCTACTAAAATCTAGCCGCGCCAGCAGCGACGCGAACCGCTAAATCCACCCAAACCTAGCACC | C/T | NC_028454.1 | 5 | 3450894 | 1.12E-06 | 0.058 | NA | NA | NA | 0.0055 | 992.5899 |
| 3 | GLM | 30882610-38-G/A | TGCAGCGTGCGGCAGCAGACCAGATCCGTCGGGTTGAAGTTCACCG | G/A | NC_028458.1 | 9 | 9428069 | 4.99E-07 | 0.079 | 0.437 | 0.575 | 0.138 | 0.0017 | 1629.3846 |
| No. | Trait | Model | Marker ID | Marker sequence | RefSeq sequence | Chr | pos | P.value | maf | R2.without.SNP | R2.with.SNP | R2* | FDR_Adjusted_P-values | Effect |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TDN | FarmCPU | 30930072 | TGCAGCTGGCGTCGGCGACGGCGTGCGTCGCGCTGTCGGCGGCGCGGCTCGCCCG | NC_028452.1 | 3 | 44894692 | 1.64E-06 | 0.32 | NA | NA | NA | 0.0054 | -0.8184 |
| 2 | ADF | Blink | 30879386 | TGCAGTAGTGGCGGTGGACTACGACGCCTCCCCCTGCGAGCACATCATATCCCAGACGCCTGCTCGACG | NC_028454.1 | 5 | 1406759 | 1.64E-10 | 0.272 | NA | NA | NA | 1.07E-06 | NA |
| TDN | GLM | 30879386 | TGCAGTAGTGGCGGTGGACTACGACGCCTCCCCCTGCGAGCACATCATATCCCAGACGCCTGCTCGACG | NC_028454.1 | 5 | 1406759 | 5.22E-06 | 0.272 | 0.202 | 0.368 | 0.167 | 0.0341 | 1.4635 | |
| TDN | Blink | 30879386 | TGCAGTAGTGGCGGTGGACTACGACGCCTCCCCCTGCGAGCACATCATATCCCAGACGCCTGCTCGACG | NC_028454.1 | 5 | 1406759 | 1.74E-10 | 0.272 | NA | NA | NA | 0.0000 | NA | |
| ADF | GLM | 30879386 | TGCAGTAGTGGCGGTGGACTACGACGCCTCCCCCTGCGAGCACATCATATCCCAGACGCCTGCTCGACG | NC_028454.1 | 5 | 1406759 | 3.65E-06 | 0.272 | 0.169 | 0.349 | 0.180 | 0.0238 | -1.5094 | |
| 3 | TDN | FarmCPU | 30841580 | TGCAGAACGTTCAGACTTCAAACCACATGCTGCCGTGCGCATCAGCACATGTGCTTGACTTGTGACCTG | NC_028454.1 | 5 | 6158000 | 1.47E-06 | 0.145 | NA | NA | NA | 0.0054 | -1.1296 |
| 4 | ADF | Blink | 30930612 | TGCAGCTCCCGCCGTGGCAGCACTCCAGCGCGTCCCAGCCG | NC_028456.1 | 7 | 25606103 | 1.06E-06 | 0.18 | NA | NA | NA | 0.0034 | NA |
| TDN | Blink | 30930612 | TGCAGCTCCCGCCGTGGCAGCACTCCAGCGCGTCCCAGCCG | NC_028456.1 | 7 | 25606103 | 1.40E-07 | 0.18 | NA | NA | NA | 0.0005 | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





