Submitted:
15 December 2023
Posted:
19 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Field Experiments
2.2. Phenotypic Investigation for Eight Lodging Resistance Traits
2.2.1. Stem Antithrust
2.2.2. Plant Height
2.2.3. Stem Diameter and Internode Length
2.2.4. SSR Marker Genotyping
2.2.5. Heritability
2.2.6. Genetic Phylogenetic and Population Structure Analysis
2.2.7. Linkage Disequilibrium Analysis
2.2.8. Association Mapping
3. Results
3.1. Phenotypic Evaluations
3.2. Genetic Diversity
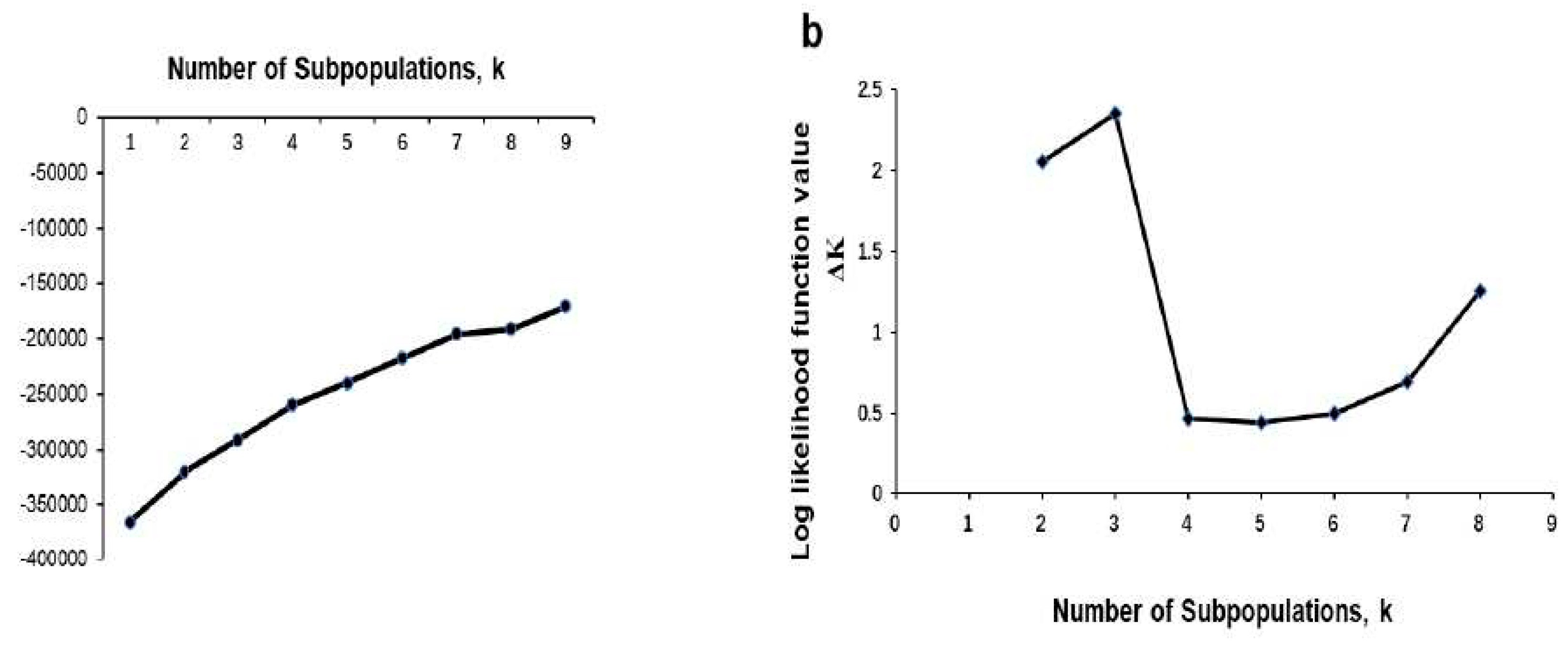
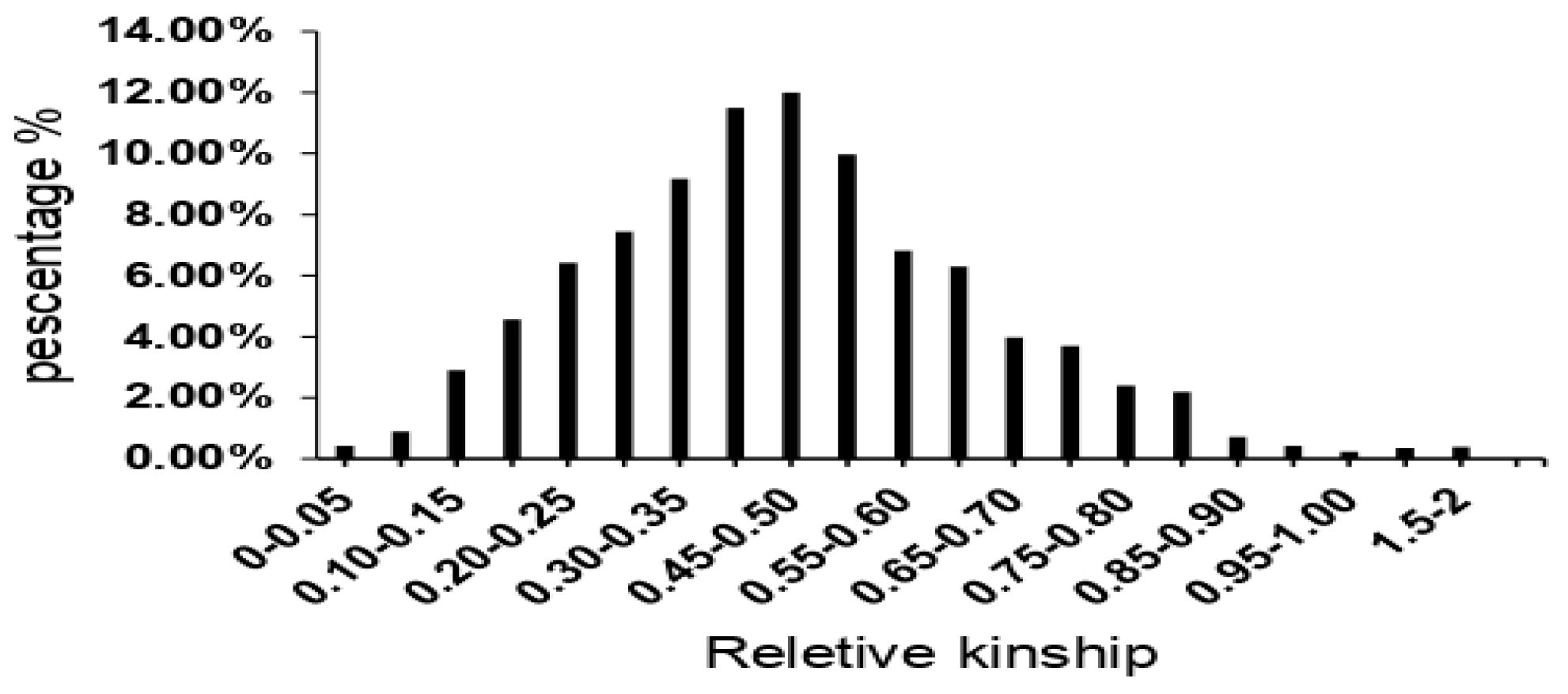
3.3. Population Structure and Genetic Relatedness
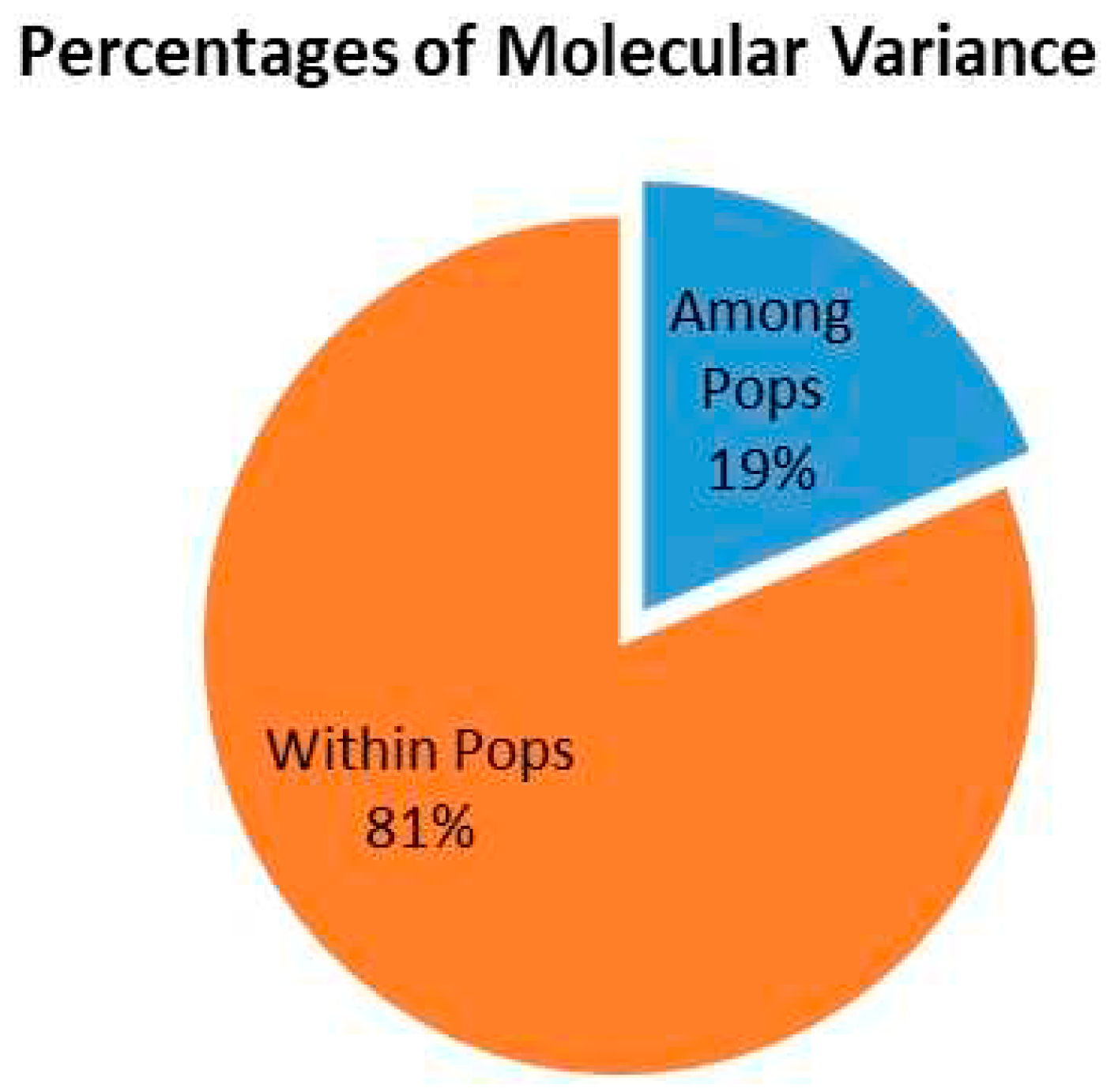
3.4. Genetic Differentiation among Subpopulation
3.5. Linkage Disequilibrium
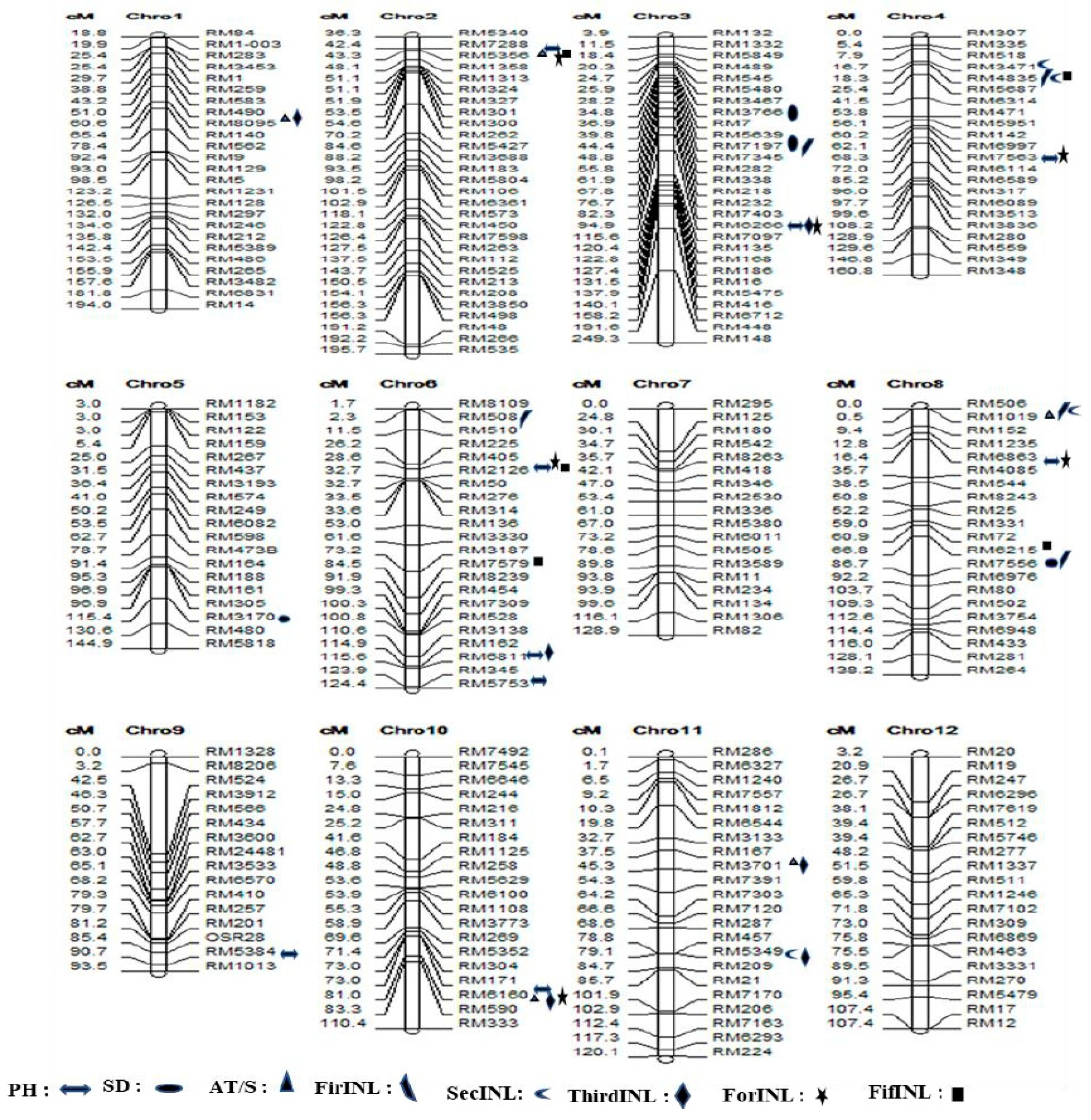
3.6. Discovery of Marker-Trait Associations and Favorable Alleles for the Eight Traits in a Natural Population
3.7. SSR Association Loci and Favorable Alleles for Various Plant Traits
3.7.1. Plant Height in the Natural Population
3.7.2. Stem Diameter in the Natural Population
3.7.3. Stem Antithrust in the Natural Population
3.7.4. First Internode Length Trait (FirINL) in the Natural Population
3.7.5. Second Internode Length Trait (SecINL) in the Natural Population
3.7.6. Third Internode Length Trait (ThirINL) in the Natural Population
3.7.7. Fourth Internode Length (ForINL) in the Natural Population
3.7.8. Fifth Internode Length Trait (FifINL) in the Natural Population
3.8. New Qtls Detected for the 8 Traits
3.9. Parental Combinations Predicted for Lodging-Resistant Improvement
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
References
- Peña, D.; Martín, C.; Fernández-Rodríguez, D.; Terrón-Sánchez, J.; Vicente, L.A.; Albarrán, Á.; Rato-Nunes, J.M.; López-Piñeiro, A. Medium-Term Effects of Sprinkler Irrigation Combined with a Single Compost Application on Water and Rice Productivity and Food Safety. Plants 2023, 12, 456. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, T.; Sasaki, H.; Ishimaru, K. Factors Responsible for Decreasing Sturdiness of the Lower Part in Lodging of Rice (Oryza sativa L.). Plant Production Science 2005, 8, 166–172. [Google Scholar] [CrossRef]
- Shah, L.; Yahya, M.; Shah, S.M.A.; Nadeem, M.; Ali, A.; Ali, A.; Wang, J.; Riaz, M.W.; Rehman, S.; Wu, W.; et al. Improving Lodging Resistance: Using Wheat and Rice as Classical Examples. Int J Mol Sci 2019, 20, 4211. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Chen, T.; Zhao, C.; Zhou, M. Lodging Prevention in Cereals: Morphological, Biochemical, Anatomical Traits and Their Molecular Mechanisms, Management and Breeding Strategies. Field Crops Res 2022, 289, 108733. [Google Scholar] [CrossRef]
- Gardiner, B.; Berry, P.; Moulia, B. Review: Wind Impacts on Plant Growth, Mechanics and Damage. Plant Sci 2016, 245, 94–118. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Laza, Ma.R.C.; Mendez, K.V.; Bhosale, S.; Dingkuhn, M. The Blaster: A Methodology to Induce Rice Lodging at Plot Scale to Study Lodging Resistance. Field Crops Res 2020, 245, 107663. [Google Scholar] [CrossRef]
- Zsögön, A.; Peres, L.E.P.; Xiao, Y.; Yan, J.; Fernie, A.R. Enhancing Crop Diversity for Food Security in the Face of Climate Uncertainty. Plant J 2022, 109, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; He, L.; Li, Y.; Wang, Y.; Ashraf, U.; Hamoud, Y.A.; Hu, X.; Wu, T.; Tang, X.; Pan, S. Deep Fertilization Combined with Straw Incorporation Improved Rice Lodging Resistance and Soil Properties of Paddy Fields. Eur J Agron 2023, 142, 126659. [Google Scholar] [CrossRef]
- Banan, D. Phenotypic and Genetic Variation in the Architectural Responses of a C4 Grass to Drought Stress. Ph.D. Dissertation, University of Illinois at Urbana-Champaign: Urbana, Illinois, 2019. [Google Scholar]
- Sowadan, O.; Li, D.; Zhang, Y.; Zhu, S.; Hu, X.; Bhanbhro, L.B.; Edzesi, W.M.; Dang, X.; Hong, D. Mining of Favorable Alleles for Lodging Resistance Traits in Rice (Oryza sativa) through Association Mapping. Planta 2018, 248, 155–169. [Google Scholar] [CrossRef]
- Cai, W.; Hong, J.; Liu, Z.; Wang, W.; Zhang, J.; An, G.; Liang, W.; Persson, S.; Zhang, D. A Receptor-like Kinase Controls the Amplitude of Secondary Cell Wall Synthesis in Rice. Curr Biol 2023, 33, 498–506. [Google Scholar] [CrossRef]
- Huang, N.; Courtois, B.; Khush, G.S.; Lin, H.; Wang, G.; Wu, P.; Zheng, K. Association of Quantitative Trait Loci for Plant Height with Major Dwarfing Genes in Rice. Heredity 1996, 77, 130–137. [Google Scholar] [CrossRef]
- Cao, G.; Zhu, J.; He, C.; Gao, Y.; Yan, J.; Wu, P. Impact of Epistasis and QTL×environment Interaction on the Developmental Behavior of Plant Height in Rice (Oryza sativa L.). Theor Appl Genet 2001, 103, 153–160. [Google Scholar] [CrossRef]
- Hittalmani, S.; Shashidhar, H.E.; Bagali, P.G.; Huang, N.; Sidhu, J.S.; Singh, V.P.; Khush, G.S. Molecular Mapping of Quantitative Trait Loci for Plant Growth, Yield and Yield Related Traits across Three Diverse Locations in a Doubled Haploid Rice Population. Euphytica 2002, 125, 207–214. [Google Scholar] [CrossRef]
- Han, Y.; Teng, W.; Sun, D.; Du, Y.; Qiu, L.; Xu, X.; Li, W. Impact of Epistasis and QTL×environment Interaction on the Accumulation of Seed Mass of Soybean (Glycine max L. Merr.). Genet Res 2008, 90, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.M.; Wu, Y.J.; Fu, X.D.; Qian, Q. Characterizations and Molecular Mapping of a Novel Dominant Semi-dwarf Gene Sdd(t) in Rice ( Oryza sativa ). Plant Breeding 2008, 127, 125–130. [Google Scholar] [CrossRef]
- Miura, K.; Wu, J.; Sunohara, H.; Wu, X.; Matsumoto, T.; Matsuoka, M.; Ashikari, M.; Kitano, H. High-Resolution Mapping Revealed a 1.3-Mbp Genomic Inversion in Ssi1, a Dominant Semidwarf Gene in Rice (Oryza sativa). Plant Breed 2009, 128, 63–69. [Google Scholar] [CrossRef]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of Polyploid Wheat Genomic Diversity Using a High-Density 90 000 Single Nucleotide Polymorphism Array. Plant Biotechnol J 2014, 12, 787–796. [Google Scholar] [CrossRef]
- Wang, J.; Gang, S.; Yang, L.; Zheng, H.; Sun, J.; Liu, H.; Zhao, H.; Zou, D. Markers Associated with Culm Length and Elongated Internode Length in Japonica Rice. Crop Sci 2017, 57, 2329–2344. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Yu, Y.; Ji, H.; Lee, G.-S.; Hyung, N.-I.; Lee, K.; Kim, T.-H. Fine-Mapping of a Major Quantitative Trait Locus q2ID1 for Rice Stem Diameter. Plant Breed Biotech 2021, 9, 298–309. [Google Scholar] [CrossRef]
- Kashiwagi, T.; Ishimaru, K. Identification and Functional Analysis of a Locus for Improvement of Lodging Resistance in Rice. Plant Physiol 2004, 134, 676–683. [Google Scholar] [CrossRef]
- Desai, H.; Hamid, R.; Ghorbanzadeh, Z.; Bhut, N.; Padhiyar, S.M.; Kheni, J.; Tomar, R.S. Genic Microsatellite Marker Characterization and Development in Little Millet (Panicum Sumatrense) Using Transcriptome Sequencing. Sci Rep 2021, 11, 20620. [Google Scholar] [CrossRef] [PubMed]
- Temnykh, S.; DeClerck, G.; Lukashova, A.; Lipovich, L.; Cartinhour, S.; McCouch, S. Computational and Experimental Analysis of Microsatellites in Rice (Oryza sativa L.): Frequency, Length Variation, Transposon Associations, and Genetic Marker Potential. Genome Res 2001, 11, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- McCouch, S.R.; Teytelman, L.; Xu, Y.; Lobos, K.B.; Clare, K.; Walton, M.; Fu, B.; Maghirang, R.; Li, Z.; Xing, Y.; et al. Development and Mapping of 2240 New SSR Markers for Rice (Oryza sativa L.). DNA Res 2002, 9, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic Microsatellite Markers in Plants: Features and Applications. Trends Biotechnol 2005, 23, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Su, X.; Ma, H.; Du, K.; Yang, M.; Chen, B.; Fu, S.; Fu, T.; Xiang, C.; Zhao, Q.; et al. Development of Genic SSR Marker Resources from RNA-Seq Data in Camellia Japonica and Their Application in the Genus Camellia. Sci Rep 2021, 11, 9919. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.G.; Thompson, W.F. Rapid Isolation of High Molecular Weight Plant DNA. Nucleic Acids Res 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Wang, Y.; Ni, H.; Li, H.; Chen, J.; Zhang, D.; Fu, L. Plasmonic Microneedle Arrays for Rapid Extraction, SERS Detection, and Inactivation of Bacteria. Chem Eng J 2022, 442, 136140. [Google Scholar] [CrossRef]
- Dries, R.; Zhu, Q.; Dong, R.; Eng, C.-H.L.; Li, H.; Liu, K.; Fu, Y.; Zhao, T.; Sarkar, A.; Bao, F.; et al. Giotto: A Toolbox for Integrative Analysis and Visualization of Spatial Expression Data. Genome Biol 2021, 22, 78. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol Biol Evol 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Chiquito-Almanza, E.; Caballero-Pérez, J.; Acosta-Gallegos, J.A.; Montero-Tavera, V.; Mariscal-Amaro, L.A.; Anaya-López, J.L. Diversity and Distribution of Viruses Infecting Wild and Domesticated Phaseolus Spp. in the Mesoamerican Center of Domestication. Viruses 2021, 13, 1153. [Google Scholar] [CrossRef]
- Zheng, Y.; Han, X.; Zhao, Y.; Zhu, L.; Huang, Y.; Jia, X.; Zhang, Z.; Chen, J.; Guo, J. Association Mapping for General Combining Ability with Yield, Plant Height and Ear Height Using F1 Population in Maize. PLoS ONE 2021, 16, e0258327. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Muse, S.V. PowerMarker: An Integrated Analysis Environment for Genetic Marker Analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of Population Structure Using Multilocus Genotype Data: Dominant Markers and Null Alleles. Mol Ecol Notes 2007, 7, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Tajima, F.; Tateno, Y. Accuracy of Estimated Phylogenetic Trees from Molecular Data. J Mol Evol 1983, 19, 153–170. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Wang, J.; Zhao, H.; Liu, H.; Sun, J.; Guo, L.; Zou, D. Genetic Structure, Linkage Disequilibrium and Association Mapping of Salt Tolerance in Japonica Rice Germplasm at the Seedling Stage. Mol Breeding 2015, 35, 152. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (Version 3.0): An Integrated Software Package for Population Genetics Data Analysis. Evol Bioinform Online 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A Unified Mixed-Model Method for Association Mapping That Accounts for Multiple Levels of Relatedness. Nat Genet 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for Association Mapping of Complex Traits in Diverse Samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Salas Fernandez, M.G.; Bao, Y.; Tang, L.; Schnable, P.S. A High-Throughput, Field-Based Phenotyping Technology for Tall Biomass Crops. Plant Physiol. 2017, 174, 2008–2022. [Google Scholar] [CrossRef] [PubMed]
- Breseghello, F.; Sorrells, M.E. Association Mapping of Kernel Size and Milling Quality in Wheat (Triticum Aestivum L.) Cultivars. Genetics 2006, 172, 1165–1177. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Song, J.; Wang, Y.; Huang, X.; Zhang, F.; Wang, W.; Xu, J.; Zhang, Y.; Yu, H.; Pang, Y.; et al. Rapid Prediction of Head Rice Yield and Grain Shape for Genome-Wide Association Study in Indica Rice. J Cereal Sci 2020, 96, 103091. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software Structure: A Simulation Study. Mol Ecol 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Guo, J.; Jing, S.; Zhu, L.; He, G. Fine Mapping of the Rice Brown Planthopper Resistance Gene BPH7 and Characterization of Its Resistance in the 93-11 Background. Euphytica 2014, 198, 369–379. [Google Scholar] [CrossRef]
- Dang, X.; Fang, B.; Chen, X.; Li, D.; Sowadan, O.; Dong, Z.; Liu, E.; She, D.; Wu, G.; Liang, Y.; et al. Favorable Marker Alleles for Panicle Exsertion Length in Rice (Oryza sativa L.) Mined by Association Mapping and the RSTEP-LRT Method. Front Plant Sci 2017, 8, 2112. [Google Scholar] [CrossRef] [PubMed]
- Multani, D.S.; Jiao, S.; Jung, M.T.; Simcox, K.D. Stalk Strength Improvement in Crop Plants: A Progress Report. Annu Plant Rev Online 2021, 4, 357–396. [Google Scholar] [CrossRef]
- Kashiwagi, T. Novel QTL for Lodging Resistance, PRL4, Improves Physical Properties with High Non-Structural Carbohydrate Accumulation of Basal Culms in Rice (Oryza sativa L.). Euphytica 2022, 218, 83. [Google Scholar] [CrossRef]
- Wang, J.; Yang, W.; Zhang, S.; Hu, H.; Yuan, Y.; Dong, J.; Chen, L.; Ma, Y.; Yang, T.; Zhou, L.; et al. A Pangenome Analysis Pipeline Provides Insights into Functional Gene Identification in Rice. Genome Biol 2023, 24, 19. [Google Scholar] [CrossRef]
- Gutiérrez, A.G.; Carabalí, S.J.; Giraldo, O.X.; Martínez, C.P.; Correa, F.; Prado, G.; Tohme, J.; Lorieux, M. Identification of a Rice Stripe Necrosis Virus Resistance Locus and Yield Component QTLs Using Oryza sativa × O. glaberrima Introgression Lines. BMC Plant Biol 2010, 10, 6. [Google Scholar] [CrossRef]
- Li, J.; Xiao, J.; Grandillo, S.; Jiang, L.; Wan, Y.; Deng, Q.; Yuan, L.; McCouch, S.R. QTL Detection for Rice Grain Quality Traits Using an Interspecific Backcross Population Derived from Cultivated Asian (O. sativa L.) and African (O. Glaberrima S.) Rice. Genome 2004, 47, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, K.; Ono, K.; Kashiwagi, T. Identification of a New Gene Controlling Plant Height in Rice Using the Candidate-Gene Strategy. Planta 2004, 218, 388–395. [Google Scholar] [CrossRef]
- Kashiwagi, T.; Togawa, E.; Hirotsu, N.; Ishimaru, K. Improvement of Lodging Resistance with QTLs for Stem Diameter in Rice (Oryza sativa L.). Theor Appl Genet 2008, 117, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Madoka, Y.; Kashiwagi, T.; Hirotsu, N.; Ishimaru, K. Indian Rice “Kasalath” Contains Genes That Improve Traits of Japanese Premium Rice “Koshihikari. ” Theor Appl Genet 2008, 116, 603–612. [Google Scholar] [CrossRef]
- Yadav, S.; Singh, U.M.; Naik, S.M.; Venkateshwarlu, C.; Ramayya, P.J.; Raman, K.A.; Sandhu, N.; Kumar, A. Molecular Mapping of QTLs Associated with Lodging Resistance in Dry Direct-Seeded Rice (Oryza sativa L.). Front Plant Sci 2017, 8, 01431. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Jeong, I.-S.; Ji, H.; Lee, G.-S.; Yoon, U.-H.; Kim, T.-H. Development of New CAPS Markers and Their Application in QTL Analysis of Stem Diameter in Rice. Korean J Breed Sci 2014, 46, 116–128. [Google Scholar] [CrossRef]
- Luo, A.; Qian, Q.; Yin, H.; Liu, X.; Yin, C.; Lan, Y.; Tang, J.; Tang, Z.; Cao, S.; Wang, X.; et al. EUI1, Encoding a Putative Cytochrome P450 Monooxygenase, Regulates Internode Elongation by Modulating Gibberellin Responses in Rice. Plant Cell Physiol 2006, 47, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.H.; Zhong, D.B.; Xu, J.L.; Yu, S.B.; Li, Z.K. Differential Expression of Lodging Resistance Related QTLs in Rice (Oryza sativa L.). Plant Sci 2008, 175, 898–905. [Google Scholar] [CrossRef]
- Zha, R.; Yang, Z.; Zhao, F.; Sang, X.; Ling, Y.; Xie, R.; He, G. Prediction of F1 yield using genetic effects of molecular marker in indica rice (Oryza sativa L.). J Plant Genet Resour 2010, 11, 72–77. [Google Scholar] [CrossRef]
- Jiang, C.-J.; Shimono, M.; Maeda, S.; Inoue, H.; Mori, M.; Hasegawa, M.; Sugano, S.; Takatsuji, H. Suppression of the Rice Fatty-Acid Desaturase Gene OsSSI2 Enhances Resistance to Blast and Leaf Blight Diseases in Rice. Mol Plant Microbe Interact 2009, 22, 820–829. [Google Scholar] [CrossRef]
- Yamamoto, T.; Taguchi-Shiobara, F.; Ukai, Y.; Sasaki, T.; Yano, M. Mapping Quantitative Trait Loci for Days-to-Heading, and Culm, Panicle and Internode Lengths in a BC1F3 Population Using an Elite Rice Variety, Koshihikari, as the Recurrent Parent. Breed Sci 2001, 51, 63–71. [Google Scholar] [CrossRef]
- Nagai, K.; Kuroha, T.; Ayano, M.; Kurokawa, Y.; Angeles-Shim, R.B.; Shim, J.-H.; Yasui, H.; Yoshimura, A.; Ashikari, M. Two Novel QTLs Regulate Internode Elongation in Deepwater Rice during the Early Vegetative Stage. Breed Sci 2012, 62, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Zhu, X.; Wang, Q.; Zhang, J.; Chen, H.; Dong, G.; Zhu, L.; Zheng, H.; Xie, Q.; Nian, J.; et al. Rice TUTOU1 Encodes a Suppressor of cAMP Receptor-like Protein That Is Important for Actin Organization and Panicle Development. Plant Physiol. 2015, 169, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Nagai, K.; Kondo, Y.; Kitaoka, T.; Noda, T.; Kuroha, T.; Angeles-Shim, R.B.; Yasui, H.; Yoshimura, A.; Ashikari, M. QTL Analysis of Internode Elongation in Response to Gibberellin in Deepwater Rice. AoB Plants 2014, 6, plu028. [Google Scholar] [CrossRef] [PubMed]
- Qiao, B.-J.; Wang, Y.-Y.; Zhu, X.-B.; Hong, D.-L. QTL analysis of the uppermost internode length in rice under different growing environments. Yi Chuan 2007, 29, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Nagai, K.; Mori, H.; Kitano, H.; Matsuoka, M.; Ashikari, M. Mapping of Three QTLs That Regulate Internode Elongation in Deepwater Rice. Breed Sci 2008, 58, 39–46. [Google Scholar] [CrossRef]
- Park, J.-R.; Jang, Y.-H.; Kim, E.-G.; Hur, S.-S.; Kim, K.-M. Quantitative Trait Loci Mapping Identified Candidate Genes Involved in Plant Height Regulation in Rice. Int J Mol Sci 2023, 24, 16895. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Xue, W.-Y.; Luo, L.-J.; Xing, Y.-Z. QTL Analysis for Flag Leaf Characteristics and Their Relationships with Yield and Yield Traits in Rice. Acta Genetica Sinica 2006, 33, 824–832. [Google Scholar] [CrossRef]
- Fu, F.-F.; Xue, H.-W. Coexpression Analysis Identifies Rice Starch Regulator1, a Rice AP2/EREBP Family Transcription Factor, as a Novel Rice Starch Biosynthesis Regulator. Plant Physiol 2010, 154, 927–938. [Google Scholar] [CrossRef]
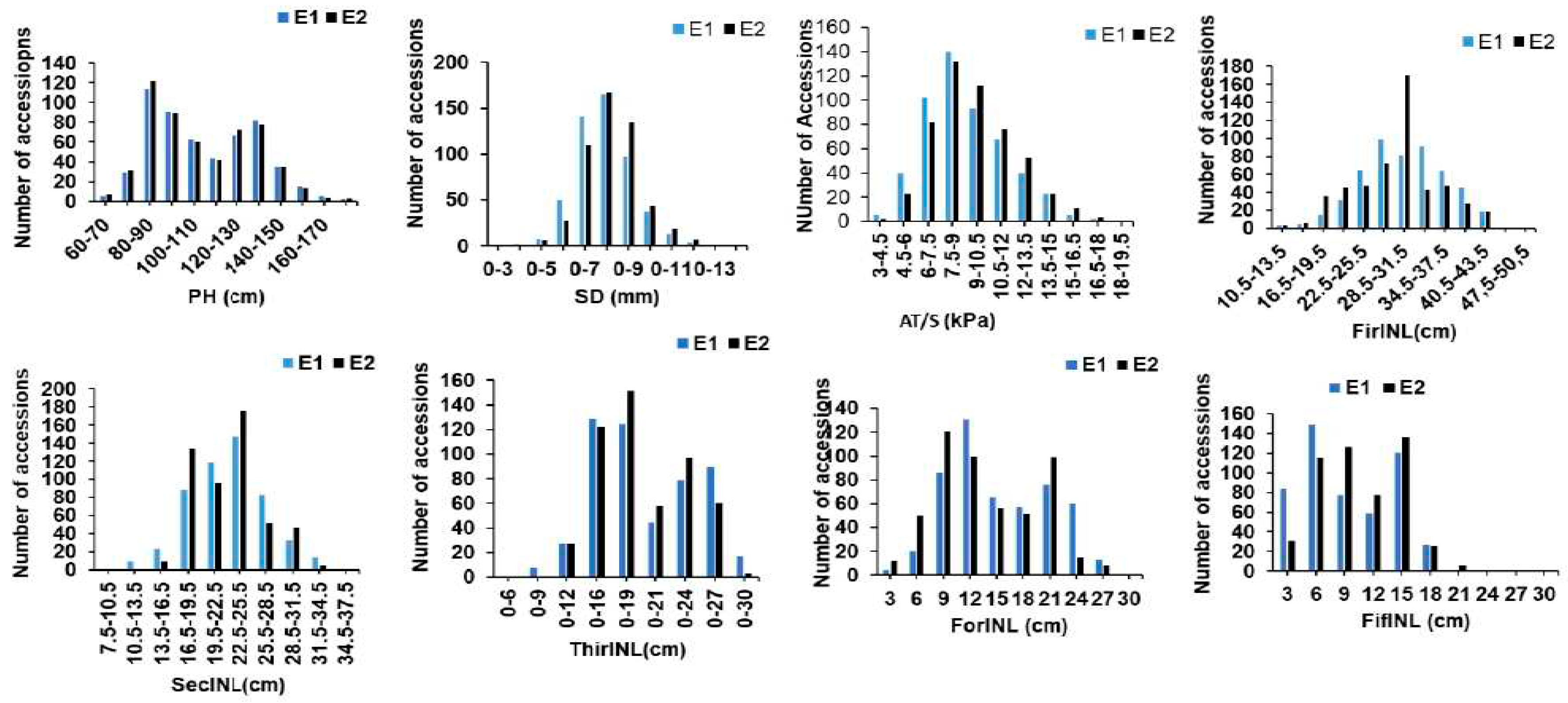
| Traits | Year | Mean ± SD | Min | Max | Skewness | Kurtosis | CV(%) | h2(%) |
| PH(cm) | 2021 | 111.23±22.93 | 62.22 | 175.5 | 0.22 | -0.89 | 20.62 | 99.15 |
| 2022 | 112.20±23.13 | 62.35 | 175 | 0.25 | -0.89 | 21.79 | 99.09 | |
| SD(cm) | 2021 | 7.4 1±1.28 | 3.69 | 13.1 | 0.44 | 0.84 | 17.29 | 80.04 |
| 2022 | 7.40 ±1.29 | 3.69 | 12.9 | 0.41 | 0.82 | 15.38 | 80.31 | |
| AT/S(Kpa) | 2021 | 9.06 ±2.47 | 3.42 | 17.79 | 0.57 | 0.04 | 28.34 | 80.06 |
| 2022 | 9.05 ±2.48 | 3.42 | 17.79 | 0.56 | 0.03 | 27.44 | 80.16 | |
| FirINL(cm) | 2021 | 34.86±7.93 | 12.43 | 95.67 | 1.59 | 8.87 | 22.75 | 65.2 |
| 2022 | 34.85±7.93 | 12.4 | 95.66 | 1.59 | 8.87 | 20.95 | 64.25 | |
| SedINL(cm) | 2021 | 23.46±5.24 | 10.18 | 40.38 | 0.47 | 0.06 | 21.35 | 92.25 |
| 2022 | 23.58±5.22 | 10.32 | 38.06 | 0.43 | -0.4 | 22.16 | 97.34 | |
| ThirINL(cm) | 2021 | 19.31±5.45 | 6.82 | 32.93 | 0.29 | -0.87 | 28.35 | 94.15 |
| 2022 | 19±5.48 | 6.5 | 32.61 | 0.27 | -0.87 | 28.85 | 94.17 | |
| ForINL(cm) | 2021 | 14.1±6.05 | 1.96 | 31.44 | 0.42 | -0.75 | 42.91 | 93.77 |
| 2022 | 14.49±6.05 | 2.43 | 31.73 | 0.41 | -0.77 | 41.73 | 92.83 | |
| FifINL(cm) | 2021 | 8.42±5.42 | 0.94 | 28.88 | 0.59 | -0.66 | 45.35 | 92.57 |
| 2022 | 9.13±5.44 | 0.71 | 29.58 | 0.57 | -0.67 | 46.63 | 92.66 |
| Source | df | SS | MS | Est. Var. | PMV% | P-Value |
|---|---|---|---|---|---|---|
| Among Pops | 2 | 13681.113 | 6840.557 | 19.919 | 19% | P<0.01 |
| Within Pops | 466 | 87608.867 | 85.140 | 85.140 | 81% | P<0.01 |
| Total | 468 | 101289.981 | 105.058 | 100% |
| Subpopulation | Pop1 | Pop2 | Pop3 |
|---|---|---|---|
| Pop1 | 0.52 | 0.69 | |
| Pop2 | 0.56 | 0.58 | |
| Pop3 | 0.48 | 0.44 |
| Cluster | No. of LDa | Ratiob | Frequency of D′c value (P < 0.05) | Means of D′ | ||||
| locus pairs | (%) | 0-0.2 | 0.2-0.4 | 0.4-0.6 | 0.6-0.8 | 0.8-1.0 | ||
| POP1 | 1240 | 2.7 | 160 | 250 | 271 | 370 | 302 | 0.64 |
| POP2 | 725 | 4.7 | 96 | 266 | 265 | 145 | 193 | 0.61 |
| POP3 | 1437 | 2.4 | 49 | 227 | 361 | 335 | 190 | 0.53 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





