Submitted:
13 December 2023
Posted:
14 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Transcription factors regulating ripening
3. Epigenetic modifications as regulators of ripening
4. Hormonal control of ripening
5. Abiotic ripening factors
6. System of regulation of tomato fruit ripening process
7. Future Prospects and Challenges
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, R.; Tavano, E.C. da R.; Lammers, M.; Martinelli, A.P.; Angenent, G.C.; de Maagd, R.A. Re-Evaluation of Transcription Factor Function in Tomato Fruit Development and Ripening with CRISPR/Cas9-Mutagenesis. Sci. Rep. 2019, 9. [CrossRef]
- Zhao, X.; Yuan, X.; Chen, S.; Meng, L.; Fu, D. Role of the Tomato TAGL1 Gene in Regulating Fruit Metabolites Elucidated Using RNA Sequence and Metabolomics Analyses. PLoS ONE 2018, 13, e0199083. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yuan, X.; Chen, S.; Fu, D.-Q.; Jiang, C.-Z. Metabolomic and Transcriptomic Analyses Reveal That a MADS-Box Transcription Factor TDR4 Regulates Tomato Fruit Quality. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Nishizawa-Yokoi, A.; Endo, M.; Mikami, M.; Shima, Y.; Nakamura, N.; Kotake-Nara, E.; Kawasaki, S.; Toki, S. Re-Evaluation of the Rin Mutation and the Role of RIN in the Induction of Tomato Ripening. Nature Plants 2017, 3, 866–874. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, H.; Ju, Z.; Cao, D.; Zhu, H.; Fu, D.; Grierson, D.; Qin, G.; Luo, Y.; Zhu, B. The RIN-MC Fusion of MADS-Box Transcription Factors Has Transcriptional Activity and Modulates Expression of Many Ripening Genes. Plant Physiol. 2017, 176, 891–909. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhu, B.; Pirrello, J.; Xu, C.; Zhang, B.; Bouzayen, M.; Chen, K.; Grierson, D. Roles of RIN and Ethylene in Tomato Fruit Ripening and Ripening-associated Traits. New Phytol. 2019, 226, 460–475. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, R.K.; Tucker, M.L.; Mattoo, A.K. Ethylene and RIPENING INHIBITOR Modulate Expression of SlHSP17.7A, B Class I Small Heat Shock Protein Genes During Tomato Fruit Ripening. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Sekiyama, Y.; Nakayama, H.; Nishizawa-Yokoi, A.; Endo, M.; Shima, Y.; Nakamura, N.; Kotake-Nara, E.; Kawasaki, S.; Hirose, S.; et al. Allelic Mutations in the Ripening-Inhibitor Locus Generate Extensive Variation in Tomato Ripening. Plant Physiol. 2020, 183, 80–95. [Google Scholar] [CrossRef]
- Yu, T.; Tzeng, D.T.W.; Li, R.; Chen, J.; Zhong, S.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. Genome-Wide Identification of Long Non-Coding RNA Targets of the Tomato MADS Box Transcription Factor RIN and Function Analysis. Annals of Botany 2018, 123, 469–482. [Google Scholar] [CrossRef]
- Kang, J.; Gong, J.; Zhang, L.; Gao, Z.; Xie, Q.; Hu, Z.; Chen, G. A Novel E6-like Gene, E6-2, Affects Fruit Ripening in Tomato. Plant Science 2021, 313, 111066. [Google Scholar] [CrossRef]
- Osakabe, K.; Wada, N.; Miyaji, T.; Murakami, E.; Marui, K.; Ueta, R.; Hashimoto, R.; Abe-Hara, C.; Kong, B.; Yano, K.; et al. Genome Editing in Plants Using CRISPR Type I-D Nuclease. Commun. Biol. 2020, 3. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Wu, S.; Li, Y.; Yang, X.; Liu, P.; Xu, Y.; Lang, Z. Expanding the Scope of CRISPR/Cas9-mediated Genome Editing in Plants Using an xCas9 and Cas9-NG Hybrid. JIPB 2020, 62, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Wu, S.; Xie, H.; Wu, Q.; Liu, P.; Xu, Y.; Lang, Z. Efficient A·T to G·C Base Conversions in Dicots Using Adenine Base Editors Expressed under the Tomato EF1α Promoter. Plant Biotechnol. J. 2022, 21, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wei, W.; Zhao, X.; Tan, X.; Fan, Z.; Zhang, Y.; Jing, Y.; Meng, L.; Zhu, B.; Zhu, H.; et al. A NAC Transcription Factor, NOR-Like1, Is a New Positive Regulator of Tomato Fruit Ripening. Hortic. Res. 2018, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Chen, H.; Nonaka, S.; Sato-Izawa, K.; Kusano, M.; Ezura, H. Ethylene Biosynthesis Controlled by NON-RIPENING: A Regulatory Conflict between Wounding and Ripening. Plant Physiol. Biochem. 2018, 132, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wei, W.; Fan, Z.; Zhao, X.; Zhang, Y.; Jing, Y.; Zhu, B.; Zhu, H.; Shan, W.; Chen, J.; et al. Re-Evaluation of the nor Mutation and the Role of the NAC-NOR Transcription Factor in Tomato Fruit Ripening. J. Exp. Bot. 2020, 71, 3560–3574. [Google Scholar] [CrossRef]
- Yu, Q.; Wang, B.; Li, N.; Tang, Y.; Yang, S.; Yang, T.; Xu, J.; Guo, C.; Yan, P.; Wang, Q.; et al. CRISPR/Cas9-Induced Targeted Mutagenesis and Gene Replacement to Generate Long-Shelf Life Tomato Lines. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Wei, C.; Duan, W.; Gao, Y.; Kuang, J.; Liu, M.; Chen, K.; Klee, H.; Zhang, B. Transcriptional and Epigenetic Analysis Reveals That NAC Transcription Factors Regulate Fruit Flavor Ester Biosynthesis. Plant J. 2021, 106, 785–800. [Google Scholar] [CrossRef]
- Jian, W.; Zheng, Y.; Yu, T.; Cao, H.; Chen, Y.; Cui, Q.; Xu, C.; Li, Z. SlNAC6, A NAC Transcription Factor, Is Involved in Drought Stress Response and Reproductive Process in Tomato. J. Plant Physiol. 2021, 264, 153483. [Google Scholar] [CrossRef]
- Gong, J.; Zeng, Y.; Meng, Q.; Guan, Y.; Li, C.; Yang, H.; Zhang, Y.; Ampomah-Dwamena, C.; Liu, P.; Chen, C.; et al. Red Light-Induced Kumquat Fruit Coloration Is Attributable to Increased Carotenoid Metabolism Regulated by FcrNAC22. J. Exp. Bot. 2021, 72, 6274–6290. [Google Scholar] [CrossRef]
- Gao, Y.; Fan, Z.; Zhang, Q.; Li, H.; Liu, G.; Jing, Y.; Zhang, Y.; Zhu, B.; Zhu, H.; Chen, J.; et al. A Tomato NAC Transcription Factor, SlNAM1, Positively Regulates Ethylene Biosynthesis and the Onset of Tomato Fruit Ripening. Plant J. 2021, 108, 1317–1331. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Vishwakarma, A.; Kenea, H.D.; Galsurker, O.; Cohen, H.; Aharoni, A.; Arazi, T. CRISPR/Cas9 Mutants of Tomato MICRORNA164 Genes Uncover Their Functional Specialization in Development. Plant Physiol. 2021, 187, 1636–1652. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Zhu, X.; Qi, B.; Gao, Z.; Tian, P.; Li, Z.; Lin, Z.; Zhang, Y.; Huang, T. SlMIR164A Regulates Fruit Ripening and Quality by Controlling SlNAM2 and SlNAM3 in Tomato. Plant Biotechnol. J. 2022, 20, 1456–1469. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Tang, M.; Huang, Z.; Song, J.; Xu, J.; Ahammed, G.J.; Yu, J.; Zhou, Y. The miR164a-NAM3 Module Confers Cold Tolerance by Inducing Ethylene Production in Tomato. Plant J. 2022, 111, 440–456. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Kawasaki, S.; Abdellatif, I.M.Y.; Nishida, K.; Kondo, A.; Ariizumi, T.; Ezura, H.; Miura, K. Efficient Base Editing in Tomato Using a Highly Expressed Transient System. Plant Cell Rep. 2021, 40, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Y.; Tureckova, V.; Xue, G.-P.; Fernie, A.R.; Mueller-Roeber, B.; Balazadeh, S. The NAC Transcription Factor SlNAP2 Regulates Leaf Senescence and Fruit Yield in Tomato. Plant Physiol. 2018, 177, 1286–1302. [Google Scholar] [CrossRef]
- Forlani, S.; Cozzi, C.; Rosa, S.; Tadini, L.; Masiero, S.; Mizzotti, C. HEBE, a Novel Positive Regulator of Senescence in Solanum lycopersicum. Sci Rep 2020, 10. [Google Scholar] [CrossRef]
- Gao, Y.; Zhu, N.; Zhu, X.; Wu, M.; Jiang, C.-Z.; Grierson, D.; Luo, Y.; Shen, W.; Zhong, S.; Fu, D.-Q.; et al. Diversity and Redundancy of the Ripening Regulatory Networks Revealed by the fruitENCODE and the New CRISPR/Cas9 CNR and NOR Mutants. Hortic. Res. 2019, 6. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Zeng, J.; Li, Z.; Song, Y.; Yan, H.; He, J.; Jiang, Y.; Duan, X. Redox Regulation of the NOR Transcription Factor Is Involved in the Regulation of Fruit Ripening in Tomato. Plant Physiol. 2020, 183, 671–685. [Google Scholar] [CrossRef]
- Lai, T.; Wang, X.; Ye, B.; Jin, M.; Chen, W.; Wang, Y.; Zhou, Y.; Blanks, A.M.; Gu, M.; Zhang, P.; et al. Molecular and Functional Characterization of the SBP-Box Transcription Factor SPL-CNR in Tomato Fruit Ripening and Cell Death. J. Exp. Bot. 2020, 71, 2995–3011. [Google Scholar] [CrossRef]
- Yin, W.; Hu, Z.; Cui, B.; Guo, X.; Hu, J.; Zhu, Z.; Chen, G. Suppression of the MADS-Box Gene SlMBP8 Accelerates Fruit Ripening of Tomato (Solanum lycopersicum). Plant Physiol. Biochem. 2017, 118, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Yu, X.; Chen, G.; Tang, B.; Wang, Y.; Liao, C.; Zhang, Y.; Hu, Z. Suppression of SlMBP15 Inhibits Plant Vegetative Growth and Delays Fruit Ripening in Tomato. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, X.; Li, C.; Li, H.; Zhang, J.; Ye, Z. Silencing GRAS2 Reduces Fruit Weight in Tomato. JIPB 2018, 60, 498–513. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shi, Y.; Su, D.; Lu, W.; Li, Z. SlGRAS4 Accelerates Fruit Ripening by Regulating Ethylene Biosynthesis Genes and SlMADS1 in Tomato. Hortic. Res. 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Peng, S.; Xian, Z.; Lin, D.; Hu, G.; Yang, L.; Ren, M.; Li, Z. Overexpression of a Tomato miR171 Target Gene SlGRAS24 Impacts Multiple Agronomical Traits via Regulating Gibberellin and Auxin Homeostasis. Plant Biotechnol. J. 2016, 15, 472–488. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Li, Y.; Fan, S.; Wen, T.; Wang, M.; Zhang, L.; Zhao, L. The Transcription Factor WRKY32 Affects Tomato Fruit Colour by Regulating YELLOW FRUITED-TOMATO 1, a Core Component of Ethylene Signal Transduction. J. Exp. Bot. 2021, 72, 4269–4282. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, X.; Wang, L.; Tian, Y.; Jia, N.; Chen, S.; Shi, N.; Huang, X.; Zhou, C.; Yu, Y.; et al. Regulation of Ethylene-Responsive SlWRKYs Involved in Color Change during Tomato Fruit Ripening. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Lang, Z.; Wang, Y.; Tang, K.; Tang, D.; Datsenka, T.; Cheng, J.; Zhang, Y.; Handa, A.K.; Zhu, J.-K. Critical Roles of DNA Demethylation in the Activation of Ripening-Induced Genes and Inhibition of Ripening-Repressed Genes in Tomato Fruit. Proc. Natl. Acad. Sci. USA 2017, 114. [Google Scholar] [CrossRef]
- Li, Z.; Pi, Y.; Fan, J.; Yang, X.; Zhai, C.; Chen, H.; Wang, F.; Ding, J.; Gu, T.; Li, Y.; et al. High Mobility Group A3 Enhances Transcription of the DNA Demethylase Gene SlDML2 to Promote Tomato Fruit Ripening. Plant Physiol. 2022, 189, 315–328. [Google Scholar] [CrossRef]
- Hollwey, E.; Out, S.; Watson, M.R.; Heidmann, I.; Meyer, P. TET3-Mediated Demethylation in Tomato Activates Expression of a CETS Gene That Stimulates Vegetative Growth. Plant Direct 2017, 1. [Google Scholar] [CrossRef]
- Yang, Y.; Tang, K.; Datsenka, T.U.; Liu, W.; Lv, S.; Lang, Z.; Wang, X.; Gao, J.; Wang, W.; Nie, W.; et al. Critical Function of DNA Methyltransferase 1 in Tomato Development and Regulation of the DNA Methylome and Transcriptome. JIPB 2019, 61, 1224–1242. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Chen, W.; Kong, J.; Zhang, X.; Shi, N.; Zhong, S.; Ma, P.; Gallusci, P.; Jackson, S.; Liu, Y.; et al. METHYLTRANSFERASE1 and Ripening Modulate Vivipary during Tomato Fruit Development. Plant Physiol. 2020, 183, 1883–1897. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.X.; Wang, J.Y.; Zhu, H.H.; Han, G.H.; Huang, R.N.; Huang, L.; Hong, Y.G.; Zheng, S.J.; Yang, J.L.; Chen, W.W. Potential Role of Domains Rearranged Methyltransferase7 in Starch and Chlorophyll Metabolism to Regulate Leaf Senescence in Tomato. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Jia, H.; Lu, S.; Zhang, Z.; Su, Z.; Sadeghnezhad, E.; Li, T.; Xiao, X.; Wang, M.; Pervaiz, T.; et al. DNA and Histone Methylation Regulates Different Types of Fruit Ripening by Transcriptome and Proteome Analyses. J. Agric. Food Chem. 2022, 70, 3541–3556. [Google Scholar] [CrossRef] [PubMed]
- Corem, S.; Doron-Faigenboim, A.; Jouffroy, O.; Maumus, F.; Arazi, T.; Bouché, N. Redistribution of CHH Methylation and Small Interfering RNAs across the Genome of Tomato Ddm1 Mutants. Plant Cell 2018, 30, 1628–1644. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhao, J.; Chen, Z.; Qiao, J.; Zhang, Y.; Shen, H.; Hu, Z. CRISPR/Cas9-Targeted Mutagenesis ofSlCMT4Causes Changes in Plant Architecture and Reproductive Organs in Tomato. Hortic. Res. 2022, 9. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Huang, B.; Wang, K.; Frasse, P.; Maza, E.; Djari, A.; Benhamed, M.; Gallusci, P.; Li, Z.; Zouine, M.; et al. Histone Posttranslational Modifications Rather than DNA Methylation Underlie Gene Reprogramming in Pollination-dependent and Pollination-independent Fruit Set in Tomato. New Phytol. 2020, 229, 902–919. [Google Scholar] [CrossRef] [PubMed]
- Bvindi, C.; Tang, L.; Lee, S.; Patrick, R.M.; Yee, Z.R.; Mengiste, T.; Li, Y. Histone Methyltransferases SDG33 and SDG34 Regulate Organ-Specific Nitrogen Responses in Tomato. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, G.; Liu, X.; Ding, X.; Zhang, D.; Wang, X.; Zhou, Y.; Yan, H.; Li, T.; Wu, K.; et al. Histone Demethylase SlJMJ6 Promotes Fruit Ripening by Removing H3K27 Methylation of Ripening-related Genes in Tomato. New Phytol. 2020, 227, 1138–1156. [Google Scholar] [CrossRef]
- Ding, X.; Zhang, D.; Gu, D.; Li, Z.; Liang, H.; Zhu, H.; Jiang, Y.; Duan, X. The Histone H3K27 Demethylase SlJMJ4 Promotes Dark- and ABA-Induced Leaf Senescence in Tomato. Hortic. Res. 2022, 9. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, X.; Yang, Y.; Zhang, H.; Zhu, W.; Nie, W.-F. The Histone Variant Sl_H2A.Z Regulates Carotenoid Biosynthesis and Gene Expression during Tomato Fruit Ripening. Hortic. Res. 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.-E.; Hu, Z.; Li, F.; Zhang, L.; Yu, X.; Tang, B.; Chen, G. Silencing of Histone Deacetylase SlHDT3 Delays Fruit Ripening and Suppresses Carotenoid Accumulation in Tomato. Plant Sci. 2017, 265, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.-E.; Hu, Z.; Zhu, M.; Li, F.; Zhu, Z.; Lu, Y.; Chen, G. The Tomato Histone Deacetylase SlHDA1 Contributes to the Repression of Fruit Ripening and Carotenoid Accumulation. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.-E. Histone Deacetylase Gene SlHDT1 Regulates Tomato Fruit Ripening by Affecting Carotenoid Accumulation and Ethylene Biosynthesis. Plant Sci. 2022, 318, 111235. [Google Scholar] [CrossRef]
- Hawar, A.; Xiong, S.; Yang, Z.; Sun, B. Histone Acetyltransferase SlGCN5 Regulates Shoot Meristem and Flower Development in Solanum lycopersicum. Front. Plant Sci. 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Bollier, N.; Sicard, A.; Leblond, J.; Latrasse, D.; Gonzalez, N.; Gévaudant, F.; Benhamed, M.; Raynaud, C.; Lenhard, M.; Chevalier, C.; et al. At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a Conserved Missing Link in the Regulation of Floral Meristem Termination in Arabidopsis and Tomato. Plant Cell 2018, 30, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Ai, G.; Huang, R.; Zhang, D.; Li, M.; Li, G.; Li, W.; Ahiakpa, J.K.; Wang, Y.; Hong, Z.; Zhang, J. SlGH3.15, a Member of the GH3 Gene Family, Regulates Lateral Root Development and Gravitropism Response by Modulating Auxin Homeostasis in Tomato. Plant Sci. 2023, 330, 111638. [Google Scholar] [CrossRef]
- Sun, M.; Li, H.; Li, Y.; Xiang, H.; Liu, Y.; He, Y.; Qi, M.; Li, T. Tomato YABBY2b Controls Plant Height through Regulating Indole-3-Acetic Acid-Amido Synthetase (GH3.8) Expression. Plant Scie. 2020, 297, 110530. [Google Scholar] [CrossRef]
- Chen, X.; Liao, D.; Yang, X.; Ji, M.; Wang, S.; Gu, M.; Chen, A.; Xu, G. Three Cis-Regulatory Motifs, AuxRE, MYCRS1 and MYCRS2, Are Required for Modulating the Auxin- and Mycorrhiza-Responsive Expression of a Tomato GH3 Gene. Plant Cell Physiol. 2017, 58, 770–778. [Google Scholar] [CrossRef]
- Chen, X.; Chen, J.; Liao, D.; Ye, H.; Li, C.; Luo, Z.; Yan, A.; Zhao, Q.; Xie, K.; Li, Y.; et al. Auxin-mediated Regulation of Arbuscular Mycorrhizal Symbiosis: A Role of SlGH3.4 in Tomato. Plant Cell Environ. 2021, 45, 955–968. [Google Scholar] [CrossRef]
- Sravankumar, T.; Akash; Naik, N.; Kumar, R. A Ripening-Induced SlGH3-2 Gene Regulates Fruit Ripening via Adjusting Auxin-Ethylene Levels in Tomato (Solanum lycopersicum L.). Plant Mol. Biol. 2018, 98, 455–469. [CrossRef] [PubMed]
- Shi, Z.; Jiang, Y.; Han, X.; Liu, X.; Cao, R.; Qi, M.; Xu, T.; Li, T. SlPIN1 Regulates Auxin Efflux to Affect Flower Abscission Process. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Chen, G.; Yu, X.; Zhu, Z.; Zhang, L.; Zhou, S.; Hu, Z. The Tomato MADS-Box Gene SlMBP9 Negatively Regulates Lateral Root Formation and Apical Dominance by Reducing Auxin Biosynthesis and Transport. Plant Cell Rep. 2019, 38, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, Y.; Feng, Q.; Qin, L.; Pan, C.; Lamin-Samu, A.T.; Lu, G. Tomato AUXIN RESPONSE FACTOR 5 Regulates Fruit Set and Development via the Mediation of Auxin and Gibberellin Signaling. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Mei, L.; Wu, M.; Wei, W.; Shan, W.; Gong, Z.; Zhang, Q.; Yang, F.; Yan, F.; Zhang, Q.; et al. SlARF10, an Auxin Response Factor, Is Involved in Chlorophyll and Sugar Accumulation during Tomato Fruit Development. J. Exp. Bot. 2018. [Google Scholar] [CrossRef]
- Israeli, A.; Capua, Y.; Shwartz, I.; Tal, L.; Meir, Z.; Levy, M.; Bar, M.; Efroni, I.; Ori, N. Multiple Auxin-Response Regulators Enable Stability and Variability in Leaf Development. Curr. Biol. 2019, 29, 1746–1759.e5. [Google Scholar] [CrossRef] [PubMed]
- Abe-Hara, C.; Yamada, K.; Wada, N.; Ueta, R.; Hashimoto, R.; Osakabe, K.; Osakabe, Y. Effects of the Sliaa9 Mutation on Shoot Elongation Growth of Tomato Cultivars. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Wu, L.; Tian, Z.; Zhang, J. Functional Dissection of Auxin Response Factors in Regulating Tomato Leaf Shape Development. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Damodharan, S.; Corem, S.; Gupta, S.K.; Arazi, T. Tuning of SlARF10A Dosage by sly-miR160a Is Critical for Auxin-mediated Compound Leaf and Flower Development. Plant J. 2018, 96, 855–868. [Google Scholar] [CrossRef]
- Tomlinson, L.; Yang, Y.; Emenecker, R.; Smoker, M.; Taylor, J.; Perkins, S.; Smith, J.; MacLean, D.; Olszewski, N.E.; Jones, J.D.G. Using CRISPR/Cas9 Genome Editing in Tomato to Create a Gibberellin-responsive Dominant Dwarf DELLA Allele. Plant Biotechnol. J. 2018, 17, 132–140. [Google Scholar] [CrossRef]
- Shinozaki, Y.; Ezura, K.; Hu, J.; Okabe, Y.; Bénard, C.; Prodhomme, D.; Gibon, Y.; Sun, T.; Ezura, H.; Ariizumi, T. Identification and Functional Study of a Mild Allele of SlDELLA Gene Conferring the Potential for Improved Yield in Tomato. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Yin, S.; Tu, Y.; Mei, H.; Wang, Y.; Yang, Y. SlCAND1, Encoding Cullin-Associated Nedd8-Dissociated Protein 1, Regulates Plant Height, Flowering Time, Seed Germination, and Root Architecture in Tomato. Plant Mol. Biol. 2020, 102, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Illouz-Eliaz, N.; Ramon, U.; Shohat, H.; Blum, S.; Livne, S.; Mendelson, D.; Weiss, D. Multiple Gibberellin Receptors Contribute to Phenotypic Stability under Changing Environments. Plant Cell 2019, 31, 1506–1519. [Google Scholar] [CrossRef] [PubMed]
- Nir, I.; Shohat, H.; Panizel, I.; Olszewski, N.; Aharoni, A.; Weiss, D. The Tomato DELLA Protein PROCERA Acts in Guard Cells to Promote Stomatal Closure. Plant Cell 2017, 29, 3186–3197. [Google Scholar] [CrossRef] [PubMed]
- Shohat, H.; Illouz-Eliaz, N.; Kanno, Y.; Seo, M.; Weiss, D. The Tomato DELLA Protein PROCERA Promotes Abscisic Acid Responses in Guard Cells by Upregulating an Abscisic Acid Transporter. Plant Physiol. 2020, 184, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.F.F.; Silva, E.M.; Correa, J.P.O.; Vicente, M.H.; Jiang, N.; Notini, M.M.; Junior, A.C.; De Jesus, F.A.; Castilho, P.; Carrera, E.; et al. Tomato Floral Induction and Flower Development Are Orchestrated by the Interplay between Gibberellin and Two Unrelated microRNA-controlled Modules. New Phytol. 2018, 221, 1328–1344. [Google Scholar] [CrossRef]
- Naeem, M.; Waseem, M.; Zhu, Z.; Zhang, L. Downregulation of SlGRAS15 Manipulates Plant Architecture in Tomato (Solanum lycopersicum). Dev. Genes. Evol. 2019, 230, 1–12. [Google Scholar] [CrossRef]
- Zhu, Z.; Liang, H.; Chen, G.; Li, F.; Wang, Y.; Liao, C.; Hu, Z. The bHLH Transcription Factor SlPRE2 Regulates Tomato Fruit Development and Modulates Plant Response to Gibberellin. Plant Cell Rep. 2019, 38, 1053–1064. [Google Scholar] [CrossRef]
- Zhu, Z.; Chen, G.; Guo, X.; Yin, W.; Yu, X.; Hu, J.; Hu, Z. Overexpression of SlPRE2, an Atypical bHLH Transcription Factor, Affects Plant Morphology and Fruit Pigment Accumulation in Tomato. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Li, J.; Gong, J.; Zhang, L.; Shen, H.; Chen, G.; Xie, Q.; Hu, Z. Overexpression of SlPRE5, an Atypical bHLH Transcription Factor, Affects Plant Morphology and Chlorophyll Accumulation in Tomato. Journal of Plant Physiol. 2022, 273, 153698. [Google Scholar] [CrossRef]
- Glanz-Idan, N.; Lach, M.; Tarkowski, P.; Vrobel, O.; Wolf, S. Delayed Leaf Senescence by Upregulation of Cytokinin Biosynthesis Specifically in Tomato Roots. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef]
- Gan, L.; Song, M.; Wang, X.; Yang, N.; Li, H.; Liu, X.; Li, Y. Cytokinins Are Involved in Regulation of Tomato Pericarp Thickness and Fruit Size. Hortic. Res. 2022, 9. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Du, P.; Ding, N.; Zhang, Q.; Xing, S.; Wei, L.; Zhao, Y.; Mao, W.; Li, J.; Li, B.; et al. Two FERONIA-Like Receptor Kinases Regulate Apple Fruit Ripening by Modulating Ethylene Production. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Yanping, Z.; Yuqing, H.; Chen, W.; Qian, M.; Songtao, J.; Xudong, Z.; Ting, Z.; Kekun, Z.; Haifeng, J.; Tariq, P.; et al. Characterization and Identification of PpEIN3 during the Modulation of Fruit Ripening Process by Ectopic Expressions in Tomato. Plant Genome 2019, 12. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, H.; Liang, F.; Cong, L.; Song, L.; Li, X.; Zhai, R.; Yang, C.; Wang, Z.; Ma, F.; et al. PbEIL1 Acts Upstream ofPbCysp1to Regulate Ovule Senescence in Seedless Pear. Hortic. Res. 2021, 8. [Google Scholar] [CrossRef]
- Althiab-Almasaud, R.; Chen, Y.; Maza, E.; Djari, A.; Frasse, P.; Mollet, J.; Mazars, C.; Jamet, E.; Chervin, C. Ethylene Signaling Modulates Tomato Pollen Tube Growth through Modifications of Cell Wall Remodeling and Calcium Gradient. Plant J. 2021, 107, 893–908. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, G.; Rodriguez, C.; Liu, M.; Binder, B.M.; Chervin, C. Roles of SlETR7, a Newly Discovered Ethylene Receptor, in Tomato Plant and Fruit Development. Hortic. Res. 2020, 7. [Google Scholar] [CrossRef]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted Base Editing in Rice and Tomato Using a CRISPR-Cas9 Cytidine Deaminase Fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef]
- Kashojiya, S.; Lu, Y.; Takayama, M.; Komatsu, H.; Minh, L.H.T.; Nishida, K.; Shirasawa, K.; Miura, K.; Nonaka, S.; Masuda, J.; et al. Modification of Tomato Breeding Traits and Plant Hormone Signaling by Target-AID, the Genome-Editing System Inducing Efficient Nucleotide Substitution. Hortic. Res. 2022, 9. [Google Scholar] [CrossRef]
- Guo, H.; Mao, M.; Deng, Y.; Sun, L.; Chen, R.; Cao, P.; Lai, J.; Zhang, Y.; Wang, C.; Li, C.; et al. Multi-Omics Analysis Reveals That SlERF.D6 Synergistically Regulates SGAs and Fruit Development. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef]
- Gambhir, P.; Singh, V.; Parida, A.; Raghuvanshi, U.; Kumar, R.; Sharma, A.K. Ethylene Response Factor ERF.D7 Activates Auxin Response Factor 2 Paralogs to Regulate Tomato Fruit Ripening. Plant Physiol. 2022, 190, 2775–2796. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gao, M.; Zhao, Y.; Chen, Y.; Wu, L.; Yin, H.; Yang, J.; Xiong, S.; Wang, S.; Wang, J.; et al. LcERF19, an AP2/ERF Transcription Factor from Litsea Cubeba, Positively Regulates Geranial and Neral Biosynthesis. Hortic. Res. 2022, 9. [Google Scholar] [CrossRef]
- Li, G.; Wang, J.; Zhang, C.; Ai, G.; Zhang, D.; Wei, J.; Cai, L.; Li, C.; Zhu, W.; Larkin, R.M.; et al. L2, a Chloroplast Metalloproteinase, Regulates Fruit Ripening by Participating in Ethylene Autocatalysis under the Control of Ethylene Response Factors. J. Exp. Bot. 2021, 72, 7035–7048. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Feng, P.; Tang, B.; Hu, Z.; Xie, Q.; Zhou, S.; Chen, G. The AP2/ERF Transcription Factor SlERF.F5 Functions in Leaf Senescence in Tomato. Plant Cell Rep. 2022, 41, 1181–1195. [Google Scholar] [CrossRef]
- Hu, C.; Wu, S.; Li, J.; Dong, H.; Zhu, C.; Sun, T.; Hu, Z.; Foyer, C.H.; Yu, J. Herbivore-induced Ca2+ Signals Trigger a Jasmonate Burst by Activating ERF16-mediated Expression in Tomato. New Phytol. 2022, 236, 1796–1808. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Hu, K.; Wei, S.; Yao, G.; Zhang, H. ETHYLENE RESPONSE FACTORS 4.1/4.2 with an EAR Motif Repress Anthocyanin Biosynthesis in Red-Skinned Pears. Plant Physiol. 2023, 192, 1892–1912. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Cheng, C. Pathogen-induced ERF68 Regulates Hypersensitive Cell Death in Tomato. Molecular Plant Pathology 2016, 18, 1062–1074. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yuan, L.; Guo, W.; Wu, W. Transcription Factor TERF1 Promotes Seed Germination under Osmotic Conditions by Activating Gibberellin Acid Signaling. Plant Sci. 2022, 322, 111350. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, H.; Tang, B.; Li, F.; Xie, Q.; Chen, G.; Hu, Z. The AP2/ERF Transcription Factor SlERF.J2 Functions in Hypocotyl Elongation and Plant Height in Tomato. Plant Cell Rep. 2022. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, X.; Tang, B.; Xie, Q.; Chen, G.; Chen, X.; Hu, Z. SlERF.J2 Reduces Chlorophyll Accumulation and Inhibits Chloroplast Biogenesis and Development in Tomato Leaves. Plant Sci. 2023, 328, 111578. [Google Scholar] [CrossRef]
- Gupta, A.; Upadhyay, R.K.; Prabhakar, R.; Tiwari, N.; Garg, R.; Sane, V.A.; Sane, A.P. SlDREB3, a Negative Regulator of ABA Responses, Controls Seed Germination, Fruit Size and the Onset of Ripening in Tomato. Plant Sci. 2022, 319, 111249. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Wang, Y.; Wang, M.; Wang, F.; Chi, C.; Zhou, Y.; Zhou, J.; Shi, K.; Xia, X.; Foyer, C.H.; et al. Crosstalk between Brassinosteroid and Redox Signaling Contributes to the Activation of CBF Expression during Cold Responses in Tomato. Antioxidants 2021, 10, 509. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, J.-J.; Wang, K.-X.; Xia, X.-J.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q.; Zhou, J. BZR1 Mediates Brassinosteroid-Induced Autophagy and Nitrogen Starvation in Tomato. Plant Physiol. 2018, 179, 671–685. [Google Scholar] [CrossRef] [PubMed]
- Sang, K.; Li, J.; Qian, X.; Yu, J.; Zhou, Y.; Xia, X. The APETALA2a/DWARF/BRASSINAZOLE-RESISTANT 1 Module Contributes to Carotenoid Synthesis in Tomato Fruits. Plant J. 2022, 112, 1238–1251. [Google Scholar] [CrossRef]
- Hu, S.; Liu, L.; Li, S.; Shao, Z.; Meng, F.; Liu, H.; Duan, W.; Liang, D.; Zhu, C.; Xu, T.; et al. Regulation of Fruit Ripening by the Brassinosteroid Biosynthetic Gene SlCYP90B3 via an Ethylene-Dependent Pathway in Tomato. Hortic. Res. 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Huang, S.; Wang, S.; Cheng, D.; Liu, J.; Lv, S.; Li, Q.; Wang, X. Enhancing Brassinosteroid Signaling via Overexpression of Tomato (Solanum lycopersicum) SlBRI1 Improves Major Agronomic Traits. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, J.; Zhao, T.; Du, C.; Nie, S.; Zhang, Y.; Lv, S.; Huang, S.; Wang, X. Modification of Threonine-1050 of SlBRI1 Regulates BR Signalling and Increases Fruit Yield of Tomato. BMC Plant Biol. 2019, 19. [Google Scholar] [CrossRef]
- Wang, D.; Yang, Z.; Wu, M.; Wang, W.; Wang, Y.; Nie, S. Enhanced Brassinosteroid Signaling via the Overexpression of SlBRI1 Positively Regulates the Chilling Stress Tolerance of Tomato. Plant Sci. 2022, 320, 111281. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Luo, B.; Hu, M.; Fu, S.; Liu, J.; Jiang, M.; Zhao, Y.; Huang, S.; Wang, S.; Wang, X. Brassinosteroid Signaling Downstream Suppressor BIN2 Interacts with SLFRIGIDA-LIKE to Induce Early Flowering in Tomato. IJMS 2022, 23, 11264. [Google Scholar] [CrossRef]
- Lee, J.; Han, S.; Lee, H.-Y.; Jeong, B.; Heo, T.-Y.; Hyun, T.K.; Kim, K.; Je, B.I.; Lee, H.; Shim, D.; et al. Brassinosteroids Facilitate Xylem Differentiation and Wood Formation in Tomato. Planta 2019, 249, 1391–1403. [Google Scholar] [CrossRef]
- Mori, K.; Lemaire-Chamley, M.; Jorly, J.; Carrari, F.; Conte, M.; Asamizu, E.; Mizoguchi, T.; Ezura, H.; Rothan, C. The Conserved Brassinosteroid-Related Transcription Factor BIM1a Negatively Regulates Fruit Growth in Tomato. J. Exp. Bot. 2020, 72, 1181–1197. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Ai, G.; Xie, Q.; Wang, W.; Song, J.; Wang, J.; Tao, J.; Zhang, X.; Hong, Z.; Lu, Y.; et al. Regulation of Tomato Fruit Elongation by Transcription Factor BZR1.7 through Promotion of SUN Gene Expression. Hortic. Res. 2022, 9. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Dong, H.; Yin, Y.; Song, X.; Gu, X.; Sang, K.; Zhou, J.; Shi, K.; Zhou, Y.; Foyer, C.H.; et al. Brassinosteroid Signaling Integrates Multiple Pathways to Release Apical Dominance in Tomato. Proc. Natl. Acad. Sci. U.S.A. 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, L.; Liang, D.; Zhang, M.; Jia, C.; Qi, M.; Liu, Y.; Shao, Z.; Meng, F.; Hu, S.; et al. SlBES1 Promotes Tomato Fruit Softening through Transcriptional Inhibition of PMEU1. iScience 2021, 24, 102926. [Google Scholar] [CrossRef] [PubMed]
- Sierra-Orozco, E.; Shekasteband, R.; Illa-Berenguer, E.; Snouffer, A.; van der Knaap, E.; Lee, T.G.; Hutton, S.F. Identification and Characterization of GLOBE, a Major Gene Controlling Fruit Shape and Impacting Fruit Size and Marketability in Tomato. Hortic Res 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Sun, Q.; Li, J.; Ahammed, G.J.; Yu, J.; Fang, H.; Xia, X. Glutaredoxin S25 and Its Interacting TGACG Motif-Binding Factor TGA2 Mediate Brassinosteroid-Induced Chlorothalonil Metabolism in Tomato Plants. Environ. Pollut. 2019, 255, 113256. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Zhang, Q.; Zhou, Y.; Ahammed, G.J.; Zhou, Y.; Yu, J.; Fang, H.; Xia, X. Glutaredoxin GRXS16 Mediates Brassinosteroid-Induced Apoplastic H2O2 Production to Promote Pesticide Metabolism in Tomato. Environ. Pollut. 2018, 240, 227–234. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Liu, Y.; Sang, K.; Wang, T.; Yu, J.; Zhou, Y.; Xia, X. Brassinosteroid Signaling Positively Regulates Abscisic Acid Biosynthesis in Response to Chilling Stress in Tomato. JIPB 2023, 65, 10–24. [Google Scholar] [CrossRef]
- Liang, B.; Sun, Y.; Wang, J.; Zheng, Y.; Zhang, W.; Xu, Y.; Li, Q.; Leng, P. Tomato Protein Phosphatase 2C Influences the Onset of Fruit Ripening and Fruit Glossiness. J. Exp. Bot. 2020, 72, 2403–2418. [Google Scholar] [CrossRef]
- Wang, J.; Xu, Y.; Zhang, W.; Zheng, Y.; Yuan, B.; Li, Q.; Leng, P. Tomato SlPP2C5 Is Involved in the Regulation of Fruit Development and Ripening. Plant Cell Physiol. 2021, 62, 1760–1769. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Q.; Jiang, L.; Kai, W.; Liang, B.; Wang, J.; Du, Y.; Zhai, X.; Wang, J.; Zhang, Y.; et al. Suppressing Type 2C Protein Phosphatases Alters Fruit Ripening and the Stress Response in Tomato. Plant Cell Physiol. 2017, 59, 142–154. [Google Scholar] [CrossRef]
- Sun, Y.; Ji, K.; Liang, B.; Du, Y.; Jiang, L.; Wang, J.; Kai, W.; Zhang, Y.; Zhai, X.; Chen, P.; et al. Suppressing ABA Uridine Diphosphate Glucosyltransferase (SlUGT75C1) Alters Fruit Ripening and the Stress Response in Tomato. Plant J. 2017, 91, 574–589. [Google Scholar] [CrossRef] [PubMed]
- Kai, W.; Wang, J.; Liang, B.; Fu, Y.; Zheng, Y.; Zhang, W.; Li, Q.; Leng, P. PYL9 Is Involved in the Regulation of ABA Signaling during Tomato Fruit Ripening. J. Exp. Bot. 2019, 70, 6305–6319. [Google Scholar] [CrossRef]
- Lei, L.; Zhang, J.; Pu, D.; Liu, B.; Meng, X.; Shang, Q.; Duan, Y.; Zhang, F.; Zhang, M.; Dong, C. ABA-responsive AREB1/ABI3-1/ABI5 Cascade Regulates IAA Oxidase Gene SlDAO2 to Inhibit Hypocotyl Elongation in Tomato. Plant Cell Environ. 2022, 46, 498–517. [Google Scholar] [CrossRef]
- Song, J.; Shang, L.; Wang, X.; Xing, Y.; Xu, W.; Zhang, Y.; Wang, T.; Li, H.; Zhang, J.; Ye, Z. MAPK11 Regulates Seed Germination and ABA Signaling in Tomato by Phosphorylating SnRKs. J. Exp. Bot. 2020, 72, 1677–1690. [Google Scholar] [CrossRef]
- Wang, W.; Wang, X.; Wang, Y.; Zhou, G.; Wang, C.; Hussain, S.; Adnan; Lin, R.; Wang, T.; Wang, S. SlEAD1, an EAR Motif-Containing ABA down-Regulated Novel Transcription Repressor Regulates ABA Response in Tomato. GM Crops Food 2020, 11, 275–289. [CrossRef]
- Hu, K.-D.; Zhang, X.-Y.; Yao, G.-F.; Rong, Y.-L.; Ding, C.; Tang, J.; Yang, F.; Huang, Z.-Q.; Xu, Z.-M.; Chen, X.-Y.; et al. A Nuclear-Localized Cysteine Desulfhydrase Plays a Role in Fruit Ripening in Tomato. Hortic. Res. 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-Q.; Hu, K.-D.; Yao, G.-F.; Wang, S.-Y.; Peng, X.-J.; Zhang, H. A D-Cysteine Desulfhydrase, SlDCD2, Participates in Tomato Fruit Ripening by Modulating ROS Homoeostasis and Ethylene Biosynthesis. Hortic. Res. 2023, 10. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yao, G.; Li, L.; Li, T.; Zhao, Y.; Hu, K.; Zhang, C.; Zhang, H. E3 Ligase BRG3 Persulfidation Delays Tomato Ripening by Reducing Ubiquitination of the Repressor WRKY71. Plant Physiol. 2023, 192, 616–632. [Google Scholar] [CrossRef]
- Hu, K.; Peng, X.; Yao, G.; Zhou, Z.; Yang, F.; Li, W.; Zhao, Y.; Li, Y.; Han, Z.; Chen, X.; et al. Roles of a Cysteine Desulfhydrase LCD1 in Regulating Leaf Senescence in Tomato. IJMS 2021, 22, 13078. [Google Scholar] [CrossRef]
- Ernesto Bianchetti, R.; Silvestre Lira, B.; Santos Monteiro, S.; Demarco, D.; Purgatto, E.; Rothan, C.; Rossi, M.; Freschi, L. Fruit-Localized Phytochromes Regulate Plastid Biogenesis, Starch Synthesis, and Carotenoid Metabolism in Tomato. J. Exp. Bot. 2018, 69, 3573–3586. [Google Scholar] [CrossRef]
- Naeem, M.; Muqarab, R.; Waseem, M. The Solanum Melongena COP1 Delays Fruit Ripening and Influences Ethylene Signaling in Tomato. Journal of Plant Physiol. 2019, 240, 152997. [Google Scholar] [CrossRef] [PubMed]
- Hunziker, J.; Nishida, K.; Kondo, A.; Kishimoto, S.; Ariizumi, T.; Ezura, H. Multiple Gene Substitution by Target-AID Base-Editing Technology in Tomato. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Gramegna, G.; Rosado, D.; Sánchez Carranza, A.P.; Cruz, A.B.; Simon-Moya, M.; Llorente, B.; Rodríguez-Concepcíon, M.; Freschi, L.; Rossi, M. PHYTOCHROME-INTERACTING FACTOR 3 Mediates Light-dependent Induction of Tocopherol Biosynthesis during Tomato Fruit Ripening. Plant Cell Environ. 2018, 42, 1328–1339. [Google Scholar] [CrossRef] [PubMed]
- Rosado, D.; Trench, B.; Bianchetti, R.; Zuccarelli, R.; Rodrigues Alves, F.R.; Purgatto, E.; Segal Floh, E.I.; Silveira Nogueira, F.T.; Freschi, L.; Rossi, M. Downregulation of PHYTOCHROME-INTERACTING FACTOR 4 Influences Plant Development and Fruit Production. Plant Physiol. 2019, 181, 1360–1370. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Liu, Y.; Ali, M.; Ye, L.; Pan, C.; Li, M.; Zhao, X.; Yu, F.; Zhao, X.; Lu, G. Phytochrome Interacting Factor 3 Regulates Pollen Mitotic Division through Auxin Signalling and Sugar Metabolism Pathways in Tomato. New Phytol. 2021, 234, 560–577. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Meng, L.; Gao, Y.; Grierson, D.; Fu, D. Manipulation of Light Signal Transduction Factors as a Means of Modifying Steroidal Glycoalkaloids Accumulation in Tomato Leaves. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, X.; Zhang, Y.; Yan, J.; Ahammed, G.J.; Bu, X.; Sun, X.; Liu, Y.; Xu, T.; Qi, H.; et al. SlFHY3 and SlHY5 Act Compliantly to Enhance Cold Tolerance through the Integration of Myo-inositol and Light Signaling in Tomato. New Phytol. 2022, 233, 2127–2143. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhang, C.; Dong, H.; Liu, X.; Guo, H.; Tong, B.; Fang, F.; Zhao, Y.; Yu, Y.; Liu, Y.; et al. Activation and Negative Feedback Regulation ofSlHY5Transcription by the SlBBX20/21–SlHY5 Transcription Factor Module in UV-B Signaling. Plant Cell 2022, 34, 2038–2055. [Google Scholar] [CrossRef]
- Qiu, Z.; Wang, H.; Li, D.; Yu, B.; Hui, Q.; Yan, S.; Huang, Z.; Cui, X.; Cao, B. Identification of Candidate HY5-Dependent and -Independent Regulators of Anthocyanin Biosynthesis in Tomato. Plant Cell Physiol. 2018, 60, 643–656. [Google Scholar] [CrossRef]
- Balderrama, D.; Barnwell, S.; Carlson, K.D.; Salido, E.; Guevara, R.; Nguyen, C.; Madlung, A. Phytochrome F Mediates Red Light Responsiveness Additively with Phytochromes B1 and B2 in Tomato. Plant Physiol. 2023, 191, 2353–2366. [Google Scholar] [CrossRef]
- Liu, C.; Chi, C.; Jin, L.; Zhu, J.; Yu, J.; Zhou, Y. The bZip Transcription Factor HY5 Mediates CRY1a-induced Anthocyanin Biosynthesis in Tomato. Plant Cell Environ. 2018, 41, 1762–1775. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Hu, C.; Liu, C.; Wang, J.; Zhou, Y.; Yu, J. ELONGATED HYPOCOTYL 5 Mediates Blue Light-Induced Starch Degradation in Tomato. J. Exp. Bot. 2020, 72, 2627–2641. [Google Scholar] [CrossRef]
- Zhi, J.; Liu, X.; Li, D.; Huang, Y.; Yan, S.; Cao, B.; Qiu, Z. CRISPR/Cas9-Mediated SlAN2 Mutants Reveal Various Regulatory Models of Anthocyanin Biosynthesis in Tomato Plant. Plant Cell Rep. 2020, 39, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Jian, W.; Cao, H.; Yuan, S.; Liu, Y.; Lu, J.; Lu, W.; Li, N.; Wang, J.; Zou, J.; Tang, N.; et al. SlMYB75, an MYB-Type Transcription Factor, Promotes Anthocyanin Accumulation and Enhances Volatile Aroma Production in Tomato Fruits. Hortic. Res. 2019, 6. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.; Bang, W.Y.; Jeong, J.C.; Park, S.-C.; Lee, J.M.; Choi, S.; Lee, B.; Lee, Y.K.; Kim, K.; Park, S.J. The Comparisons of Expression Pattern Reveal Molecular Regulation of Fruit Metabolites in S. nigrum and S. lycopersicum. Sci. Rep. 2022, 12. [Google Scholar] [CrossRef]
- Cerqueira, J.V.A.; Zhu, F.; Mendes, K.; Nunes-Nesi, A.; Martins, S.C.V.; Benedito, V.; Fernie, A.R.; Zsogon, A. Promoter Replacement of ANT1 Induces Anthocyanin Accumulation and Triggers the Shade Avoidance Response through Developmental, Physiological and Metabolic Reprogramming in Tomato. Hortic. Res. 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Wang, H.; Sun, C.; Li, Q.; Jiang, H.; Du, M.; Li, C.-B.; Li, C. Efficient Generation of Pink-Fruited Tomatoes Using CRISPR/Cas9 System. J. Genet. Genomics 2018, 45, 51–54. [Google Scholar] [CrossRef]
- Sun, C.; Deng, L.; Du, M.; Zhao, J.; Chen, Q.; Huang, T.; Jiang, H.; Li, C.-B.; Li, C. A Transcriptional Network Promotes Anthocyanin Biosynthesis in Tomato Flesh. Mol. Plant 2020, 13, 42–58. [Google Scholar] [CrossRef]
- Yan, S.; Chen, N.; Huang, Z.; Li, D.; Zhi, J.; Yu, B.; Liu, X.; Cao, B.; Qiu, Z. Anthocyanin Fruit Encodes an R2R3-MYB Transcription Factor, SlAN2-like, Activating the Transcription of SlMYBATV to Fine-tune Anthocyanin Content in Tomato Fruit. New Phytol. 2019, 225, 2048–2063. [Google Scholar] [CrossRef]
- Colanero, S.; Perata, P.; Gonzali, S. The Atroviolacea Gene Encodes an R3-MYB Protein Repressing Anthocyanin Synthesis in Tomato Plants. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, Z.; Xu, Z.; Wang, L.; Chen, C.; Ren, Z. Overexpression of SlMYB75 Enhances Resistance to Botrytis Cinerea and Prolongs Fruit Storage Life in Tomato. Plant Cell Rep. 2020, 40, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Danilo, B.; Perrot, L.; Botton, E.; Nogué, F.; Mazier, M. The DFR Locus: A Smart Landing Pad for Targeted Transgene Insertion in Tomato. PLoS ONE 2018, 13, e0208395. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Xiong, C.; Lin, A.; Zhang, C.; Sun, W.; Zhang, J.; Yang, C.; Lu, Y.; Li, H.; Ye, Z.; et al. SlBBX20 Interacts with the COP9 Signalosome Subunit SlCSN5-2 to Regulate Anthocyanin Biosynthesis by Activating SlDFR Expression in Tomato. Hortic. Res. 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Quinet, M.; Angosto, T.; Yuste-Lisbona, F.J.; Blanchard-Gros, R.; Bigot, S.; Martinez, J.-P.; Lutts, S. Tomato Fruit Development and Metabolism. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Li, S.; Chen, K.; Grierson, D. Molecular and Hormonal Mechanisms Regulating Fleshy Fruit Ripening. Cells 2021, 10, 1136. [Google Scholar] [CrossRef] [PubMed]
- Fenn, M.A.; Giovannoni, J.J. Phytohormones in Fruit Development and Maturation. Plant J. 2021, 105, 446–458. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Andersson, M.; Granell, A.; Cardi, T.; Hofvander, P.; Nicolia, A. Establishment of a DNA-Free Genome Editing and Protoplast Regeneration Method in Cultivated Tomato (Solanum lycopersicum). Plant Cell Rep. 2022, 41, 1843–1852. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, X.; Li, Q.; Peng, W.; Zhang, Z.; Chu, P.; Guo, S.; Fan, Y.; Lyu, S. AtGCS Promoter-Driven Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Highly Efficiently Generates Homozygous/Biallelic Mutations in the Transformed Roots by Agrobacterium rhizogenes–Mediated Transformation. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef]
- Zhang, N.; Roberts, H.M.; Van Eck, J.; Martin, G.B. Generation and Molecular Characterization of CRISPR/Cas9-Induced Mutations in 63 Immunity-Associated Genes in Tomato Reveals Specificity and a Range of Gene Modifications. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Vu, T.V.; Doan, D.T.H.; Tran, M.T.; Sung, Y.W.; Song, Y.J.; Kim, J.-Y. Improvement of the LbCas12a-crRNA System for Efficient Gene Targeting in Tomato. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Kirke, J.; Kaplan, N.; Velez, S.; Jin, X.-L.; Vichyavichien, P.; Zhang, X.-H. Tissue-Preferential Activity and Induction of the Pepper Capsaicin Synthase PUN1 Promoter by Wounding, Heat and Metabolic Pathway Precursor in Tobacco and Tomato Plants. Mol. Biotechnol. 2018, 60, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.V.; Sivankalyani, V.; Kim, E.; Doan, D.T.H.; Tran, M.T.; Kim, J.; Sung, Y.W.; Park, M.; Kang, Y.J.; Kim, J. Highly Efficient Homology-directed Repair Using CRISPR/Cpf1-geminiviral Replicon in Tomato. Plant Biotechnol. J. 2020, 18, 2133–2143. [Google Scholar] [CrossRef] [PubMed]
- de Maagd, R.A.; Loonen, A.; Chouaref, J.; Pele, A.; Meijer-Dekens, F.; Fransz, P.; Bai, Y. CRISPR/Cas Inactivation of RECQ4 Increases Homeologous Crossovers in an Interspecific Tomato Hybrid. Plant Biotechnol. J. 2019, 18, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Wada, N.; Osakabe, K.; Osakabe, Y. Type I-D CRISPR System-Mediated Genome Editing in Plants. Methods Mol. Biol. 2023, 21–38. [Google Scholar] [CrossRef]
- Veillet, F.; Perrot, L.; Guyon-Debast, A.; Kermarrec, M.-P.; Chauvin, L.; Chauvin, J.-E.; Gallois, J.-L.; Mazier, M.; Nogue, F. Expanding the CRISPR Toolbox in P. Patens Using SpCas9-NG Variant and Application for Gene and Base Editing in Solanaceae Crops. IJMS 2020, 21, 1024. [Google Scholar] [CrossRef] [PubMed]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted Base Editing in Rice and Tomato Using a CRISPR-Cas9 Cytidine Deaminase Fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; He, Y.; Sretenovic, S.; Liu, S.; Cheng, Y.; Han, Y.; Liu, G.; Bao, Y.; Fang, Q.; Zheng, X.; et al. CRISPR-BETS: A Base-editing Design Tool for Generating Stop Codons. Plant Biotechnol. J. 2021, 20, 499–510. [Google Scholar] [CrossRef]
- Matsumoto, A.; Schlüter, T.; Melkonian, K.; Takeda, A.; Nakagami, H.; Mine, A. A Versatile Tn7 Transposon-Based Bioluminescence Tagging Tool for Quantitative and Spatial Detection of Bacteria in Plants. Plant Commun. 2022, 3, 100227. [Google Scholar] [CrossRef]
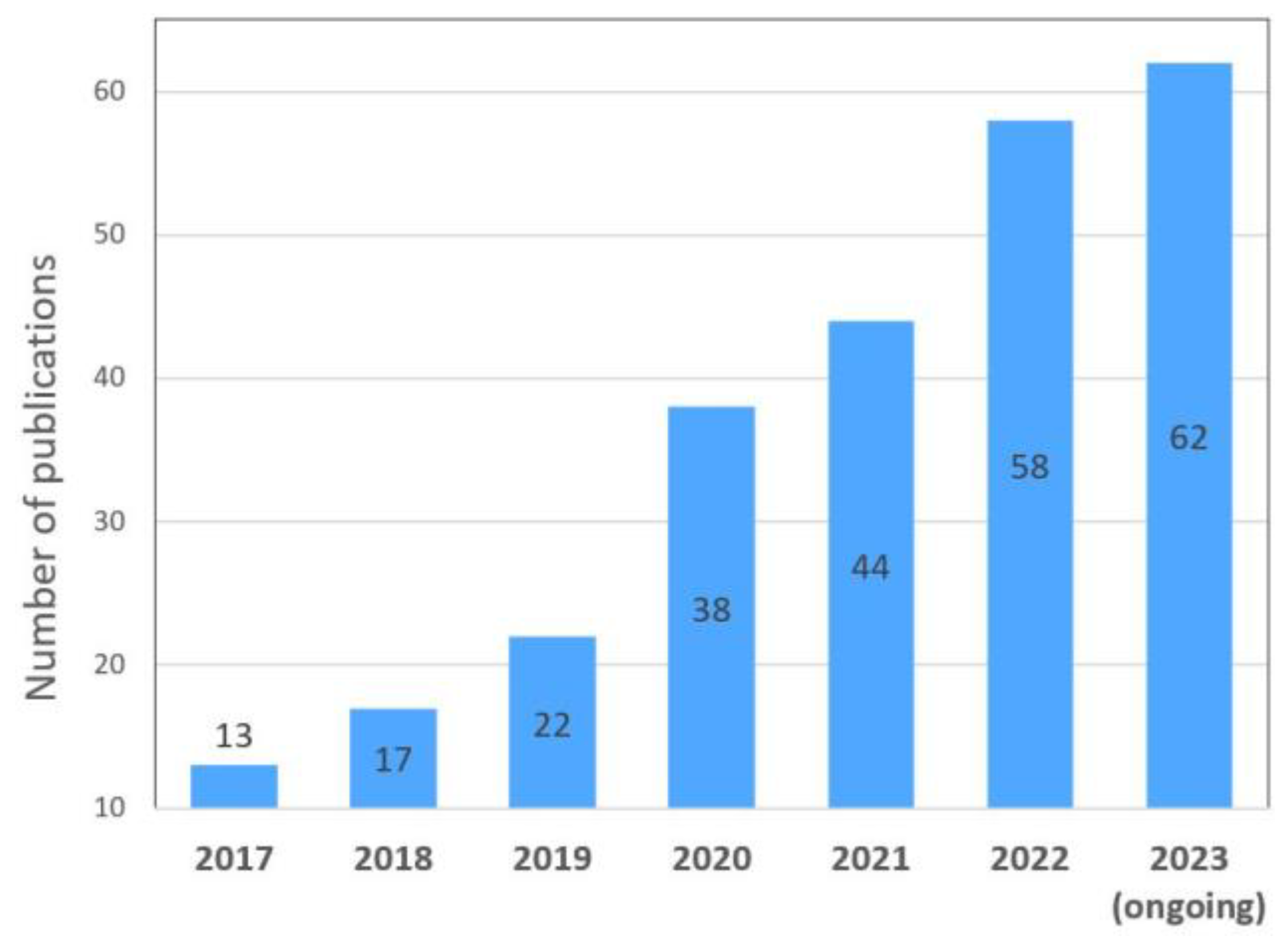
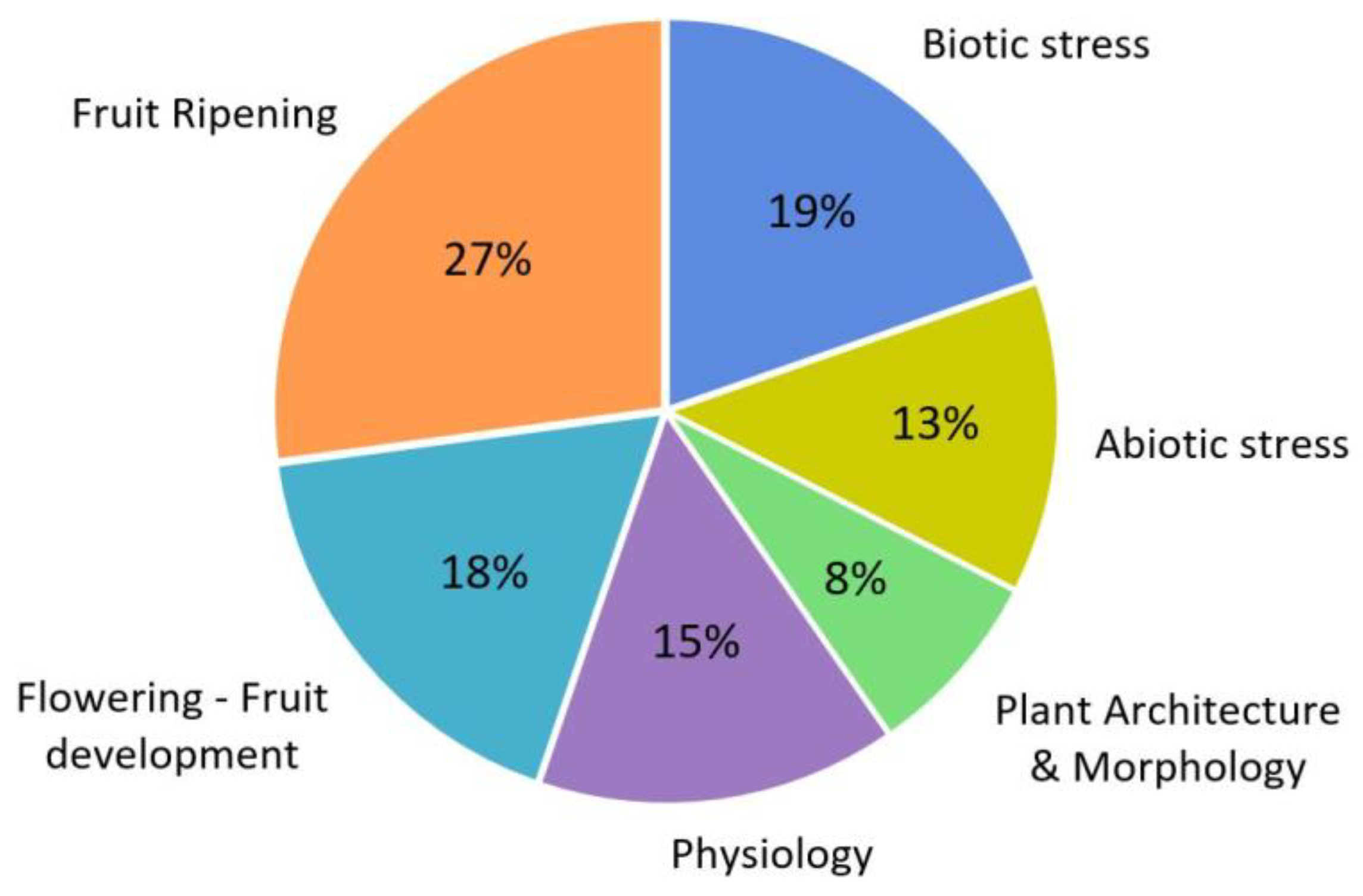
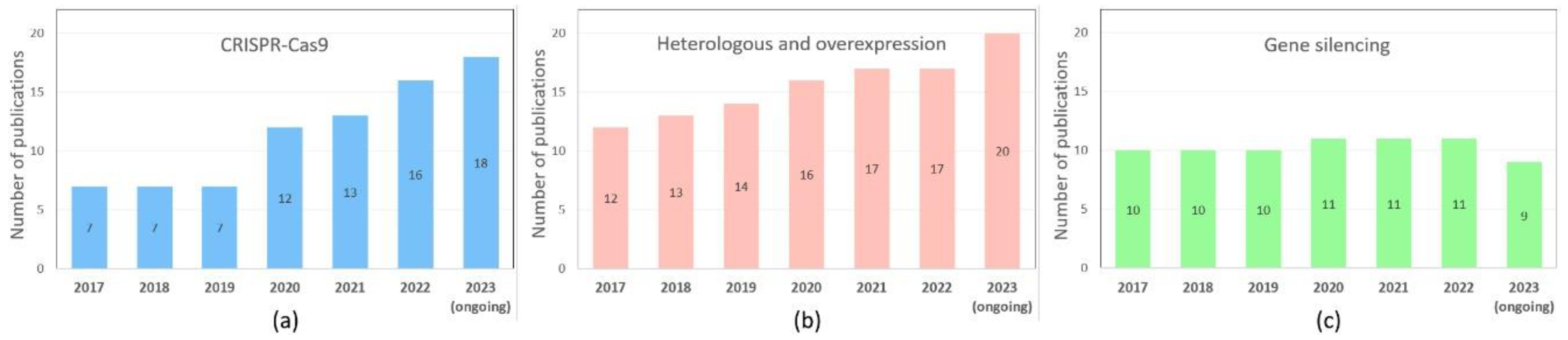
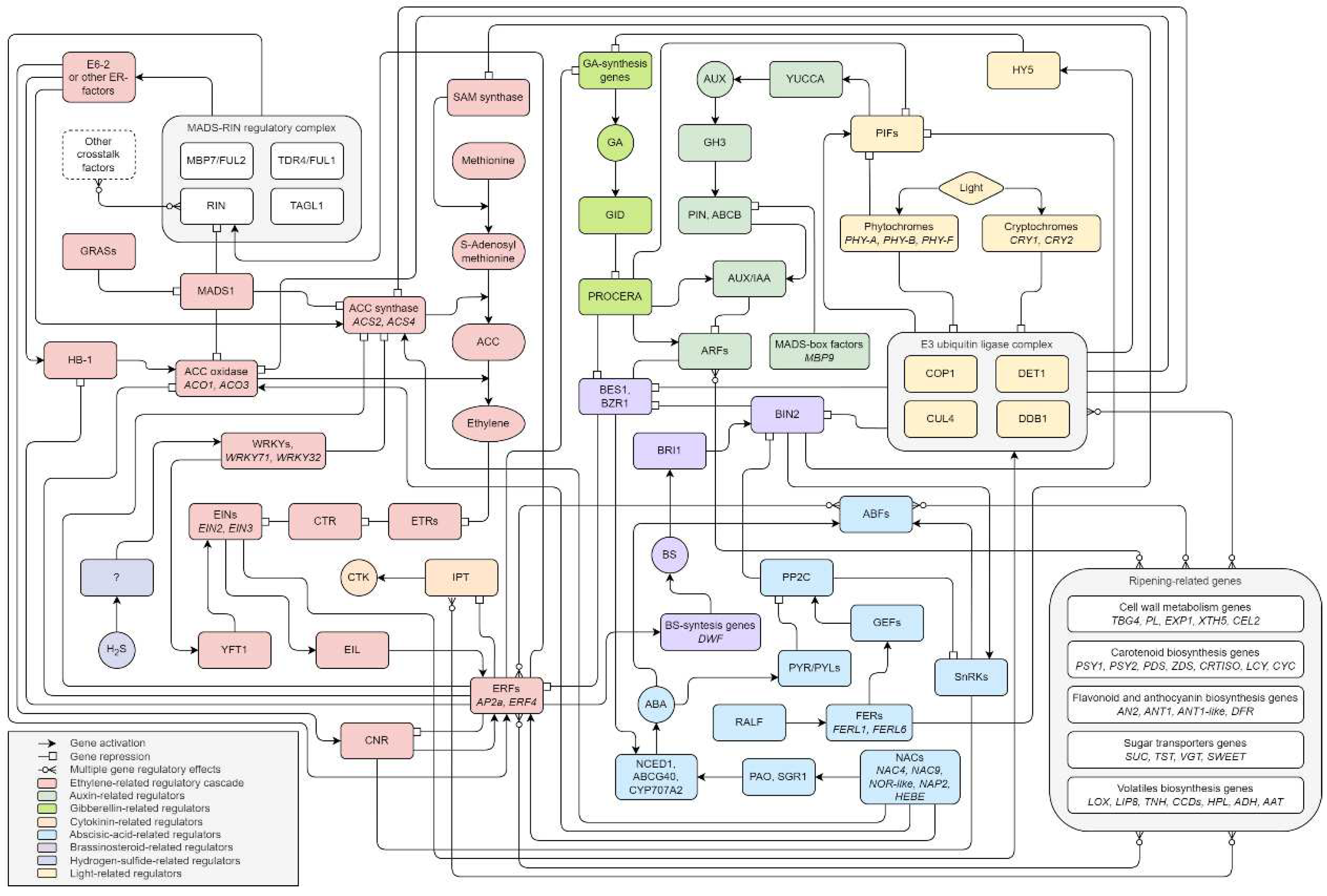
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




