Submitted:
11 December 2023
Posted:
12 December 2023
You are already at the latest version
Abstract
Keywords:
Introduction
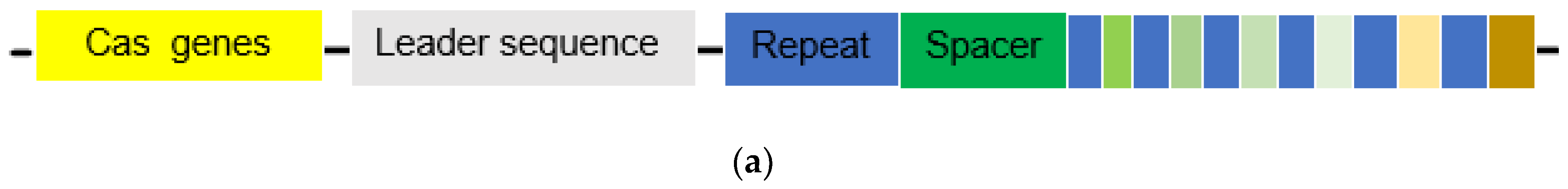
Genetic Structure and Function of the CRISPR System
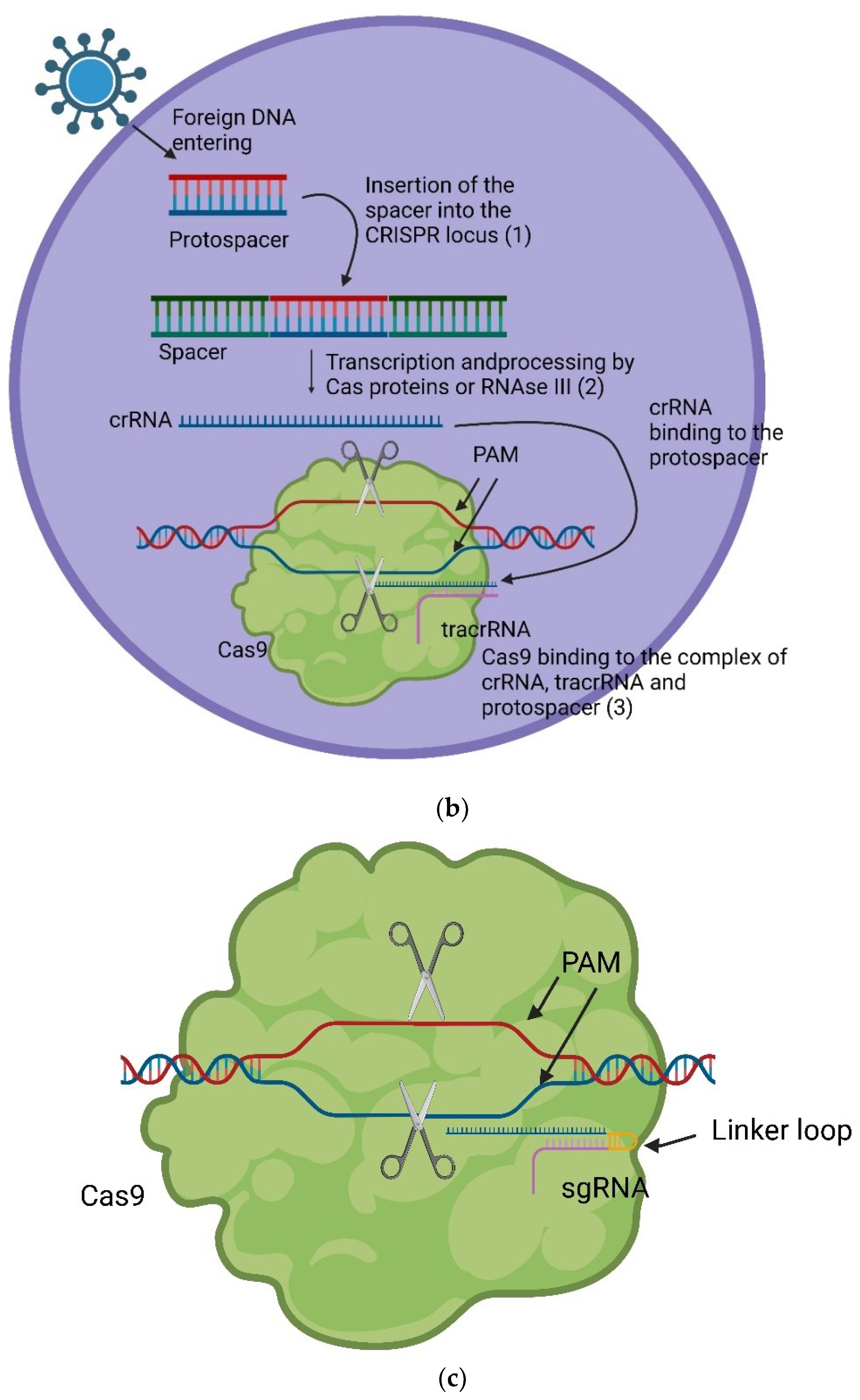
CRISPR/Cas9-mediated Genome Editing
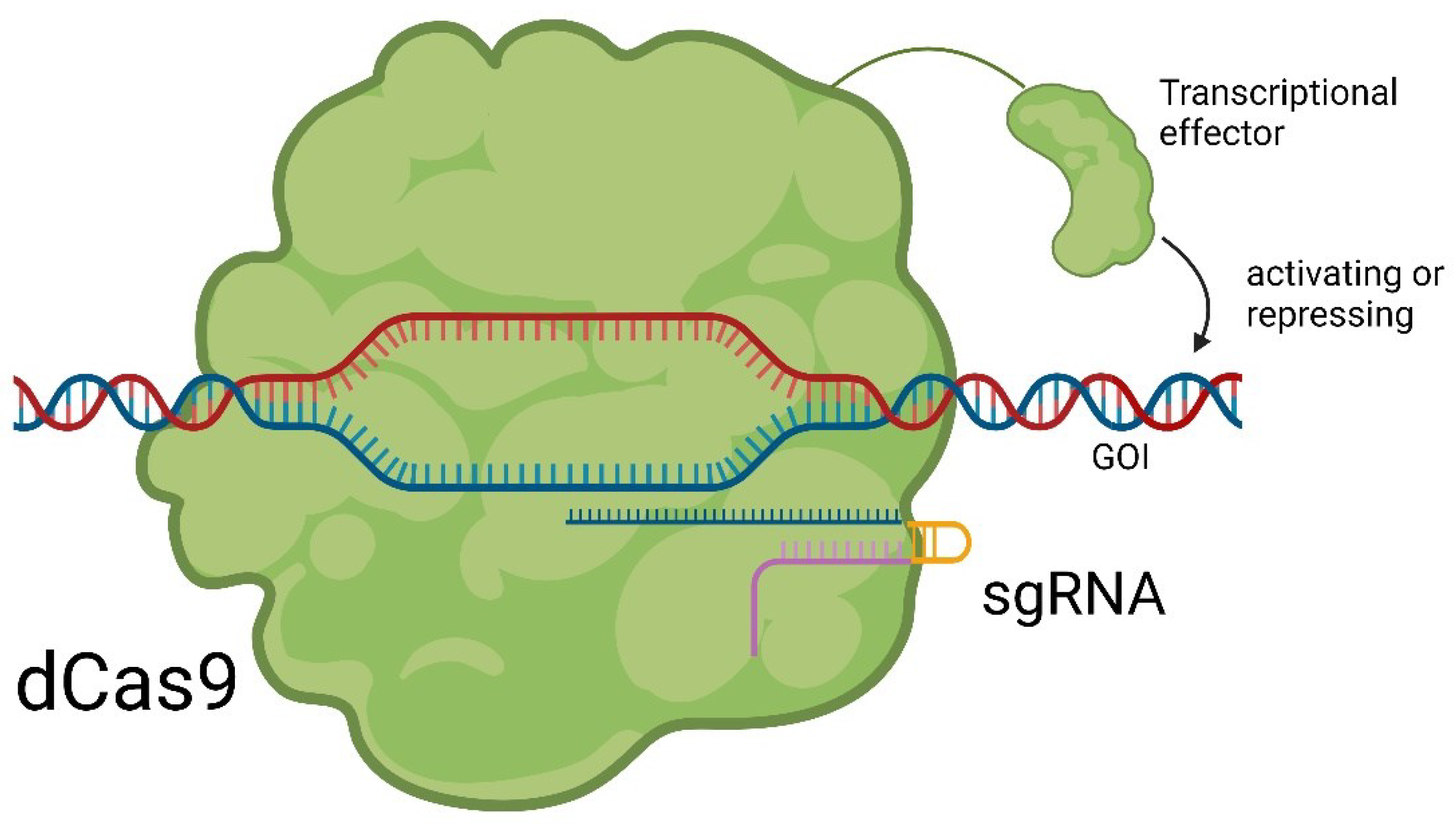
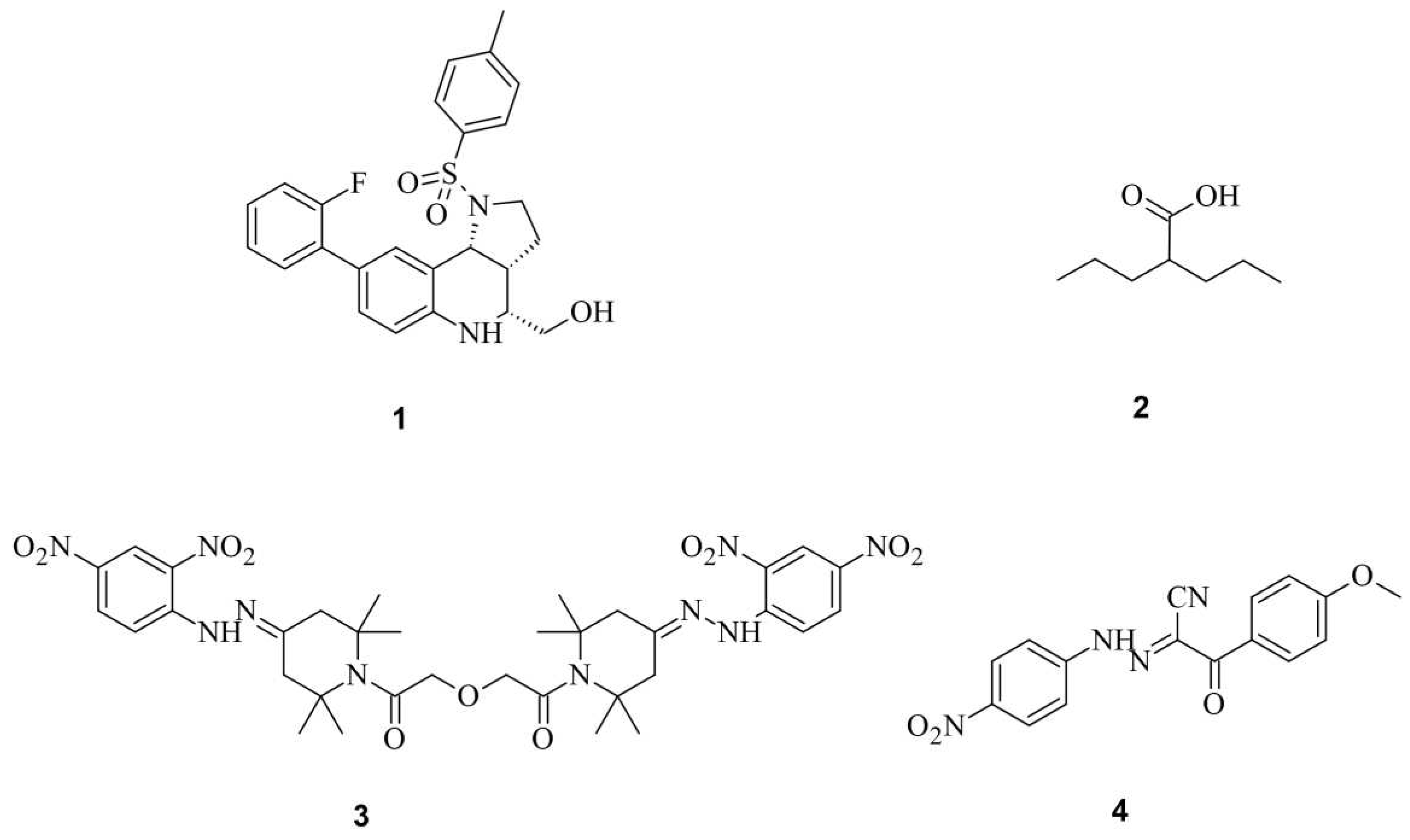
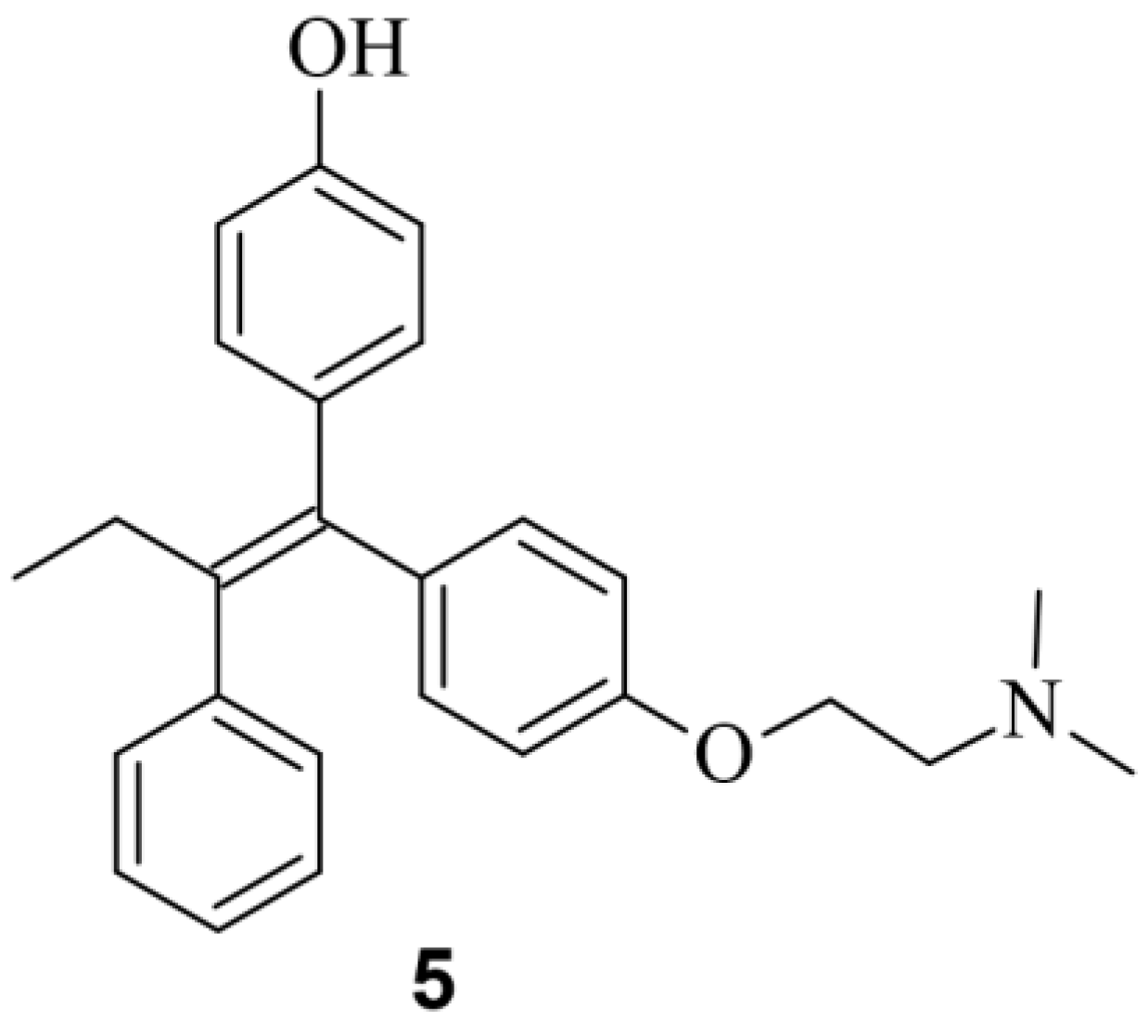
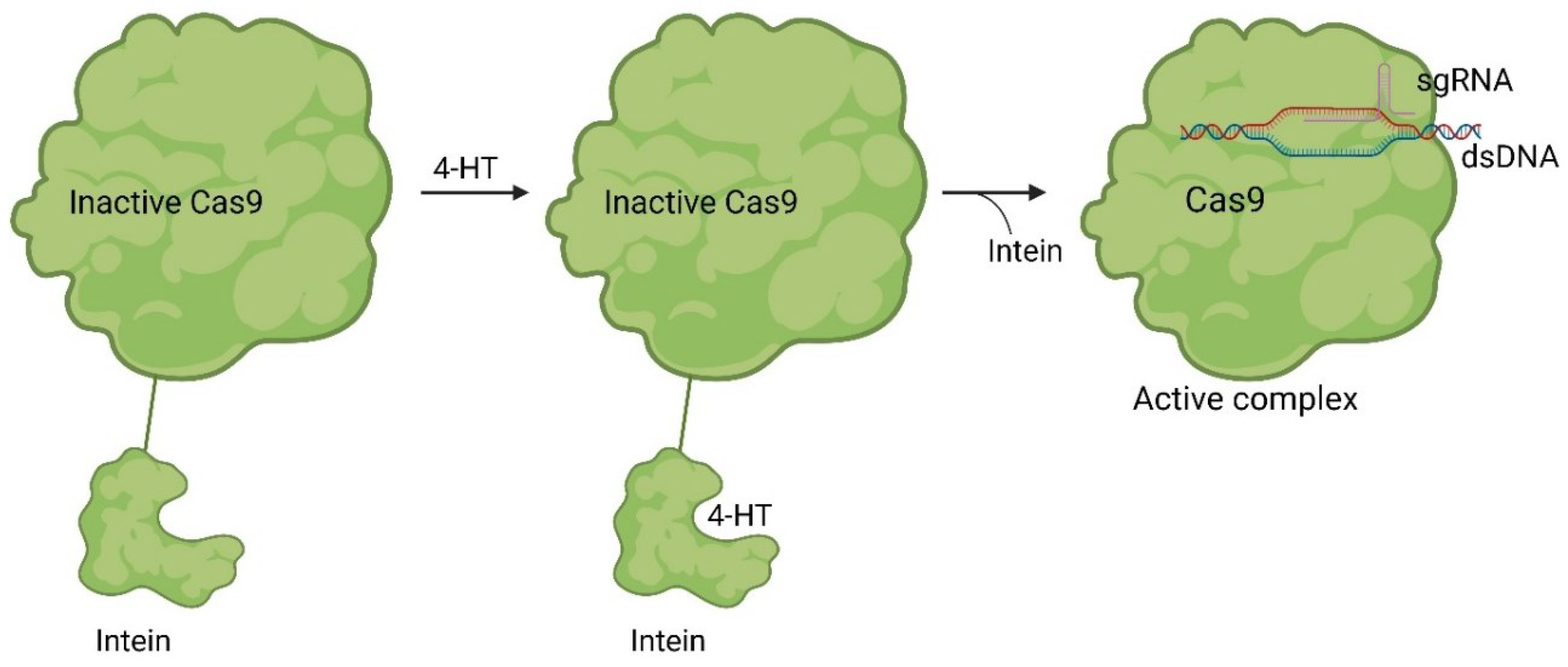
Small Molecules Modulate Wild-type Cas9 Protein
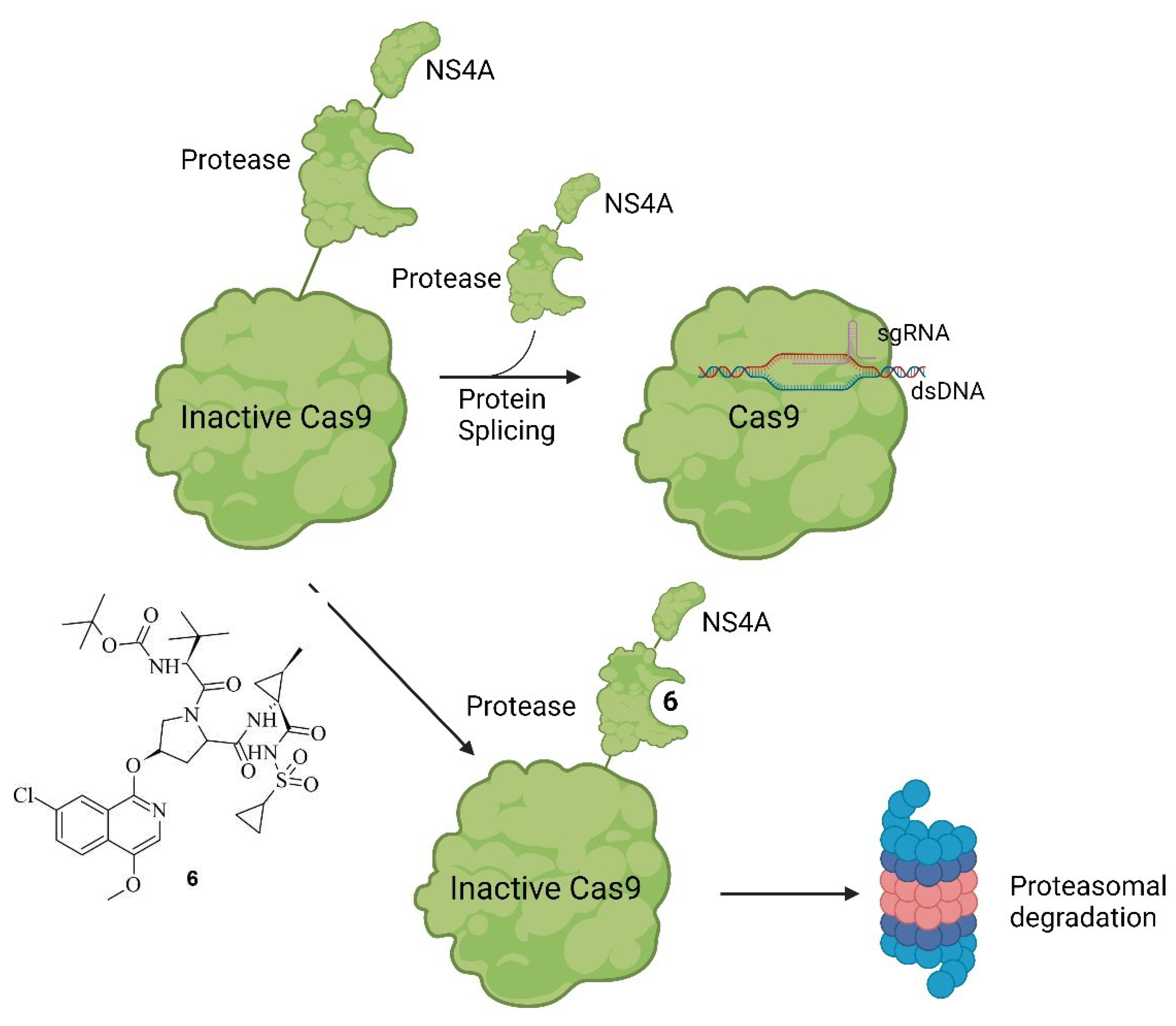
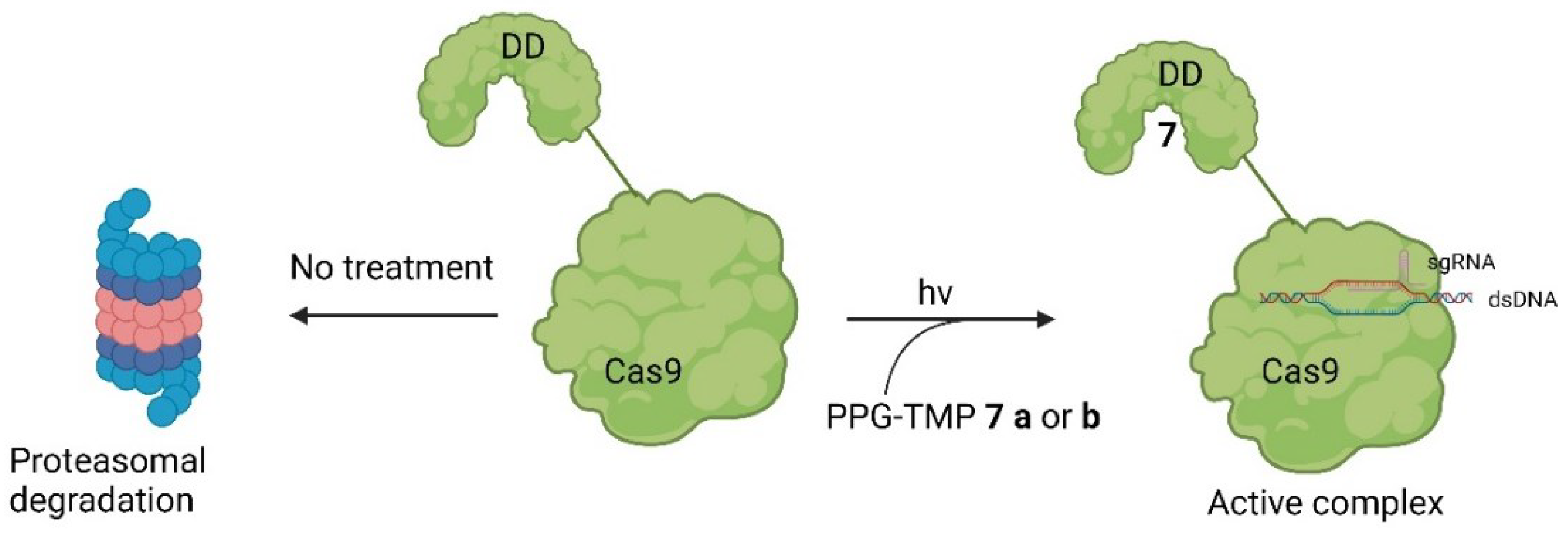
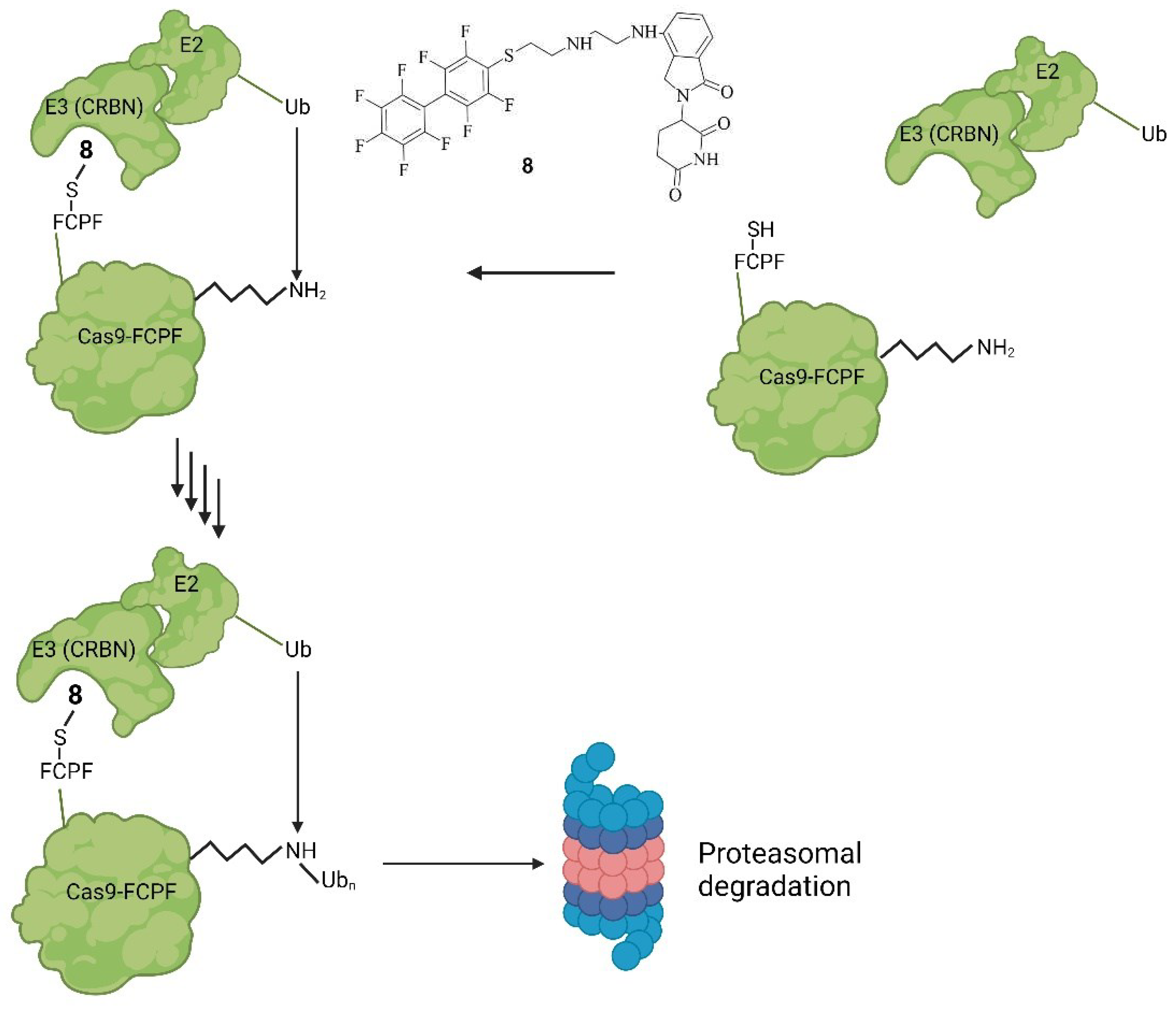
Small Molecules Modulate Engineered Cas9 Protein
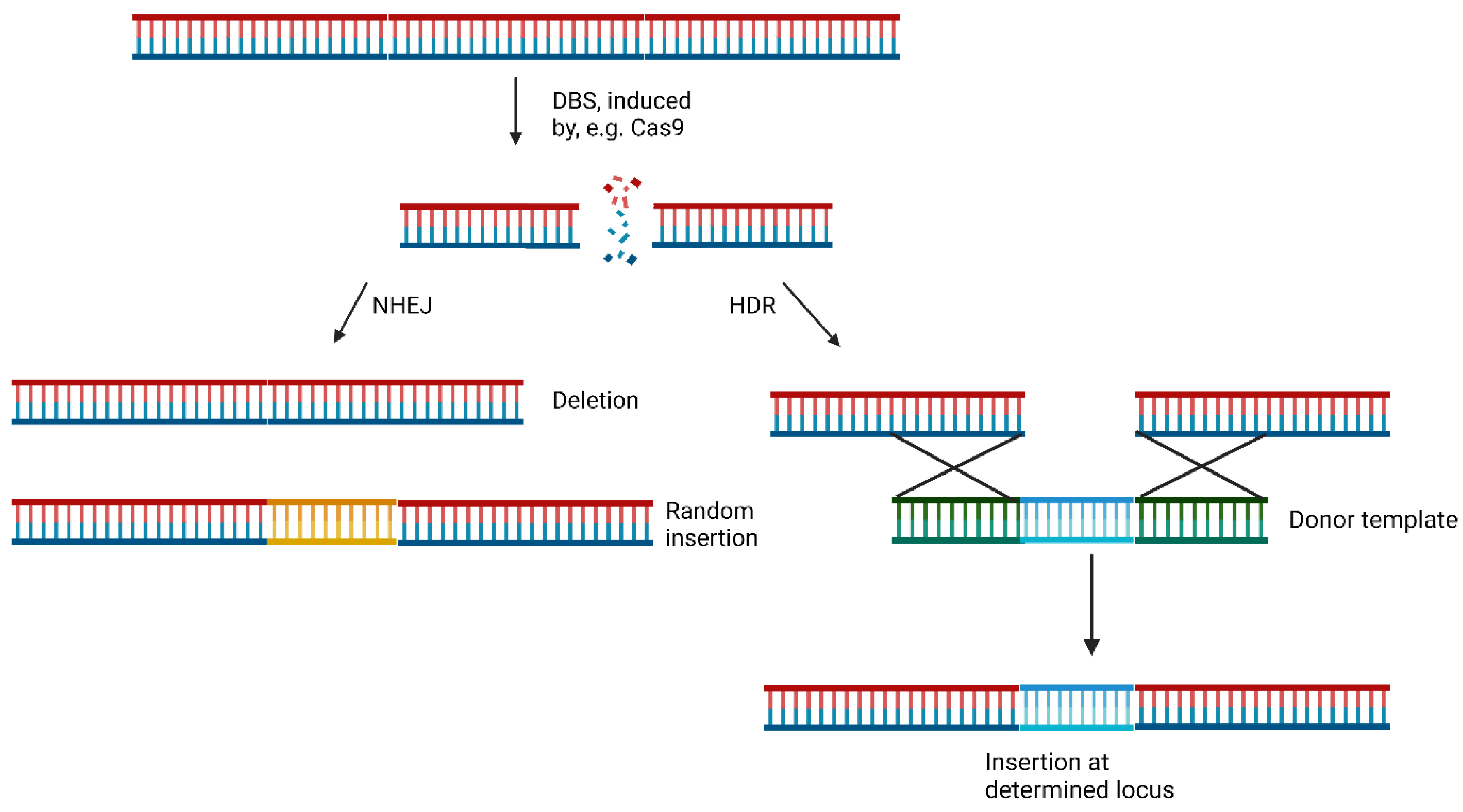
Small molecules regulate DSB Repair Mechanisms
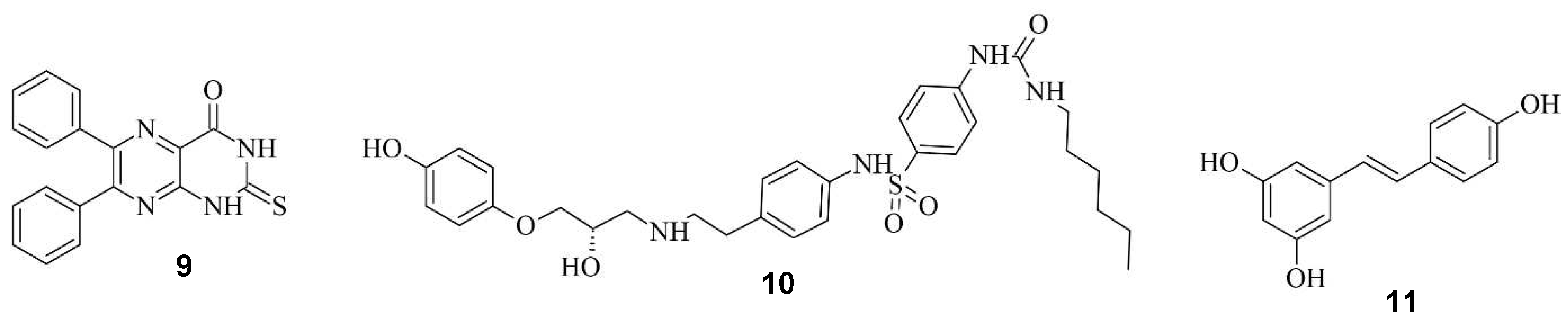
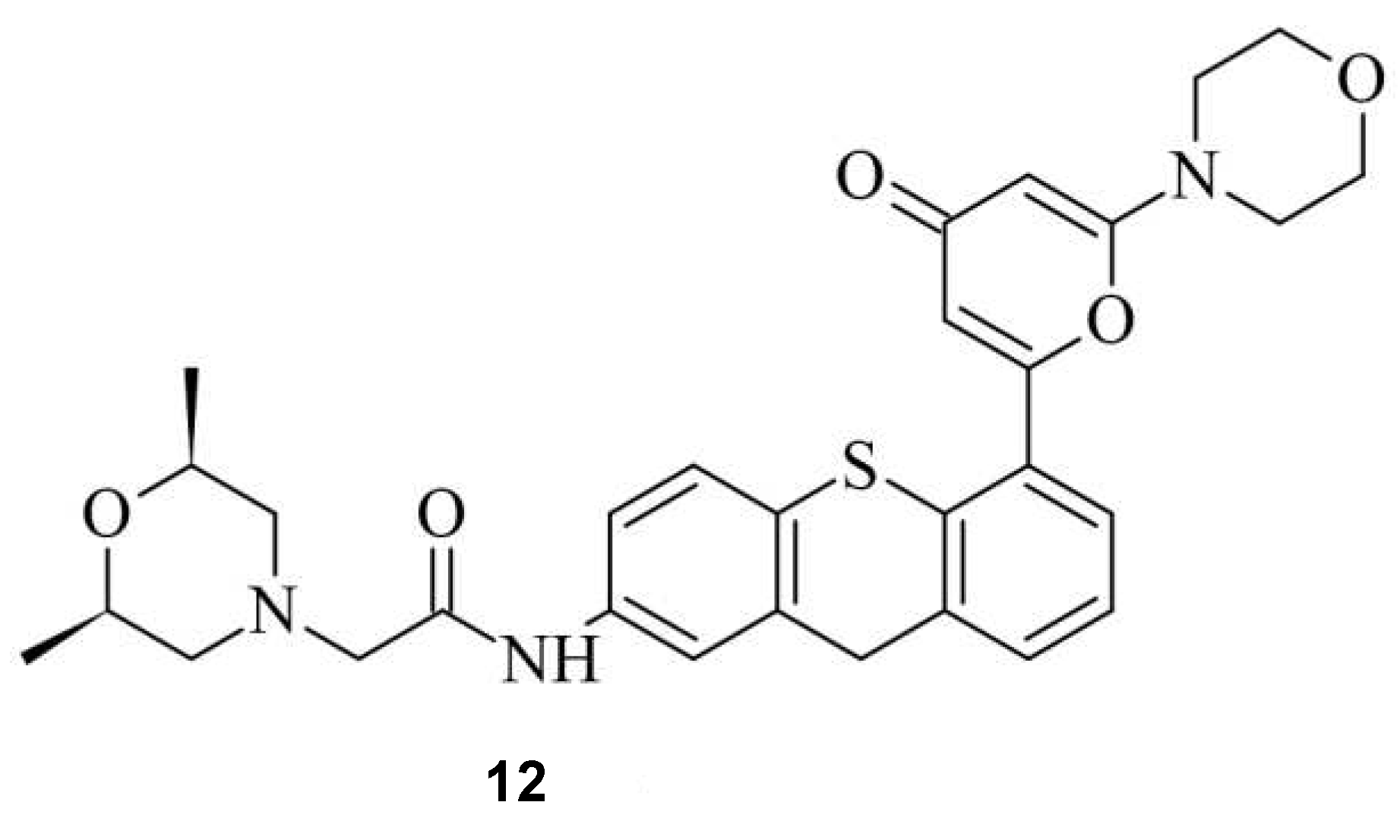
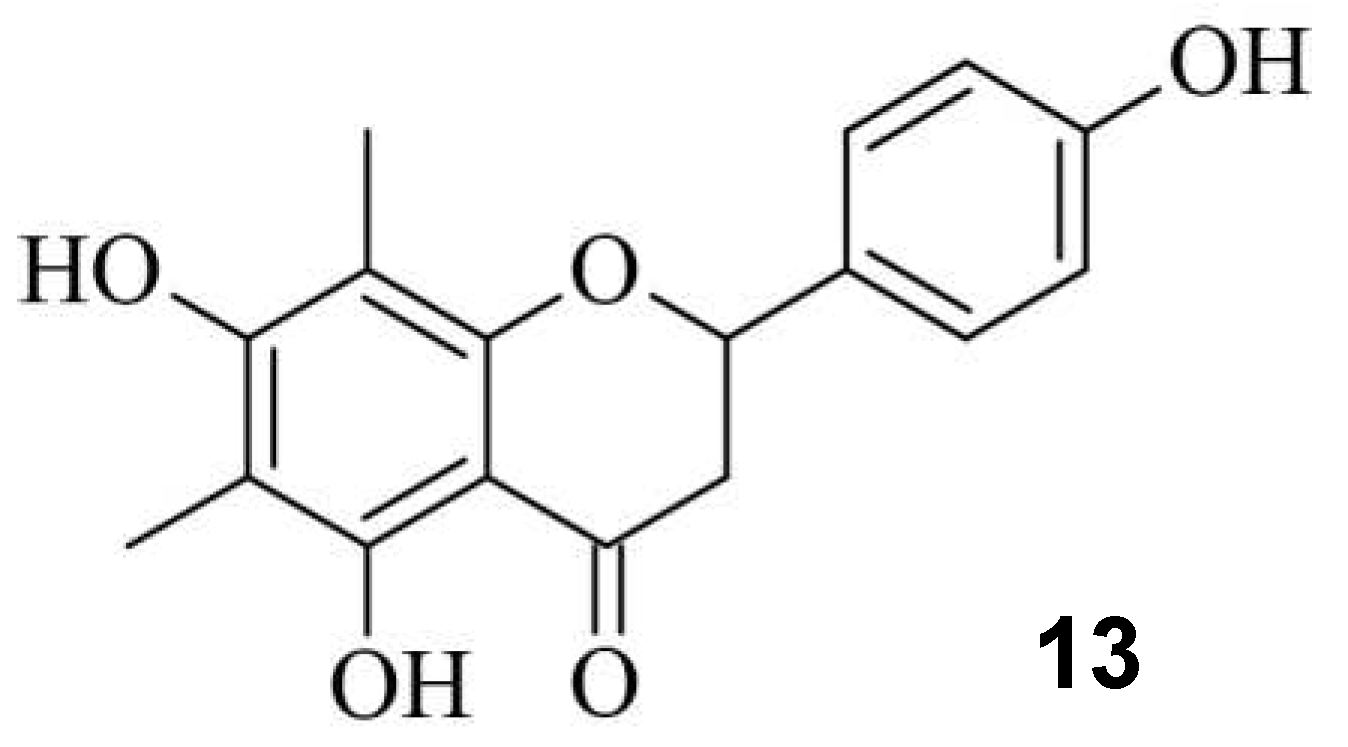
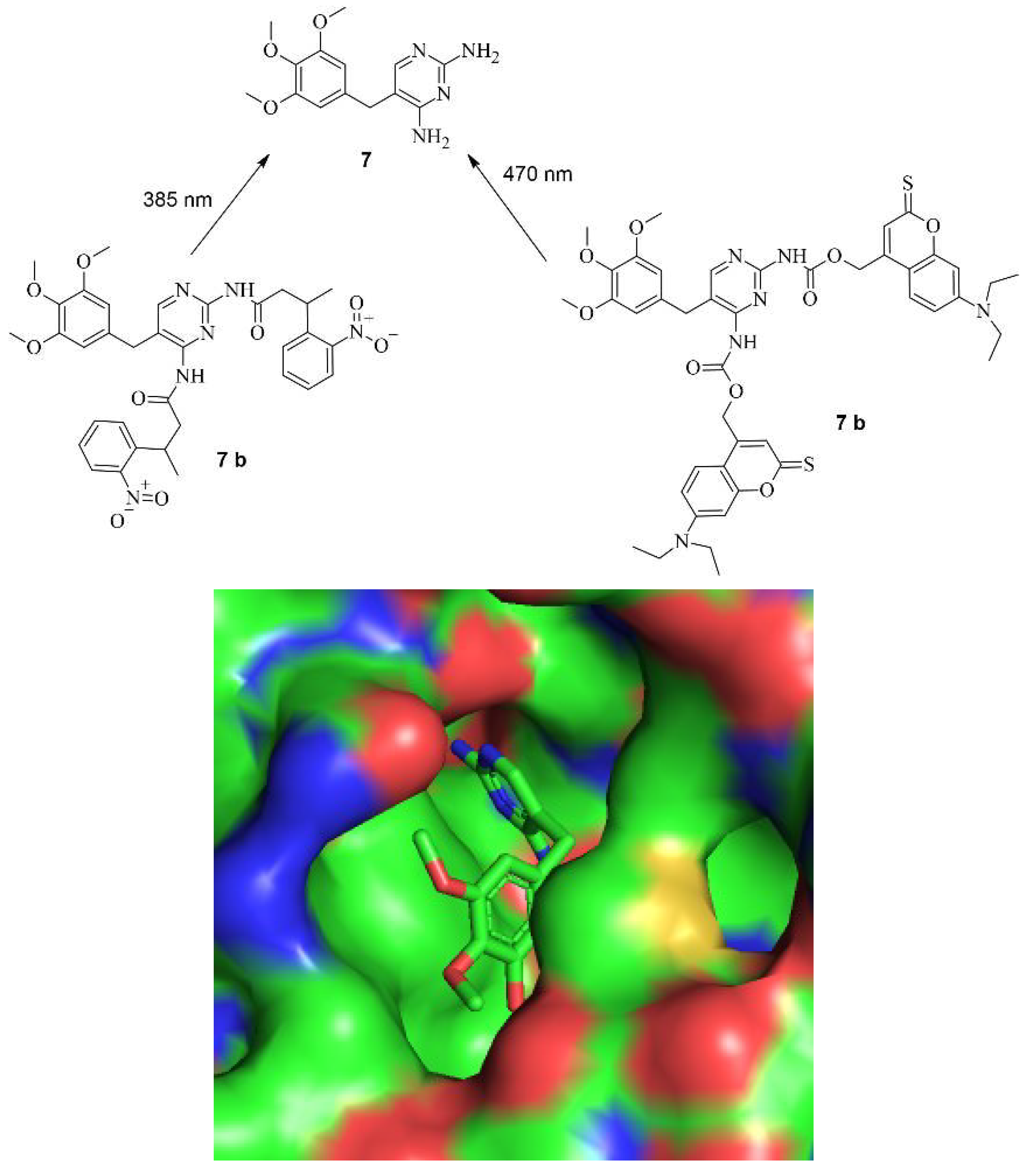
Small Molecules regulate sgRNAs
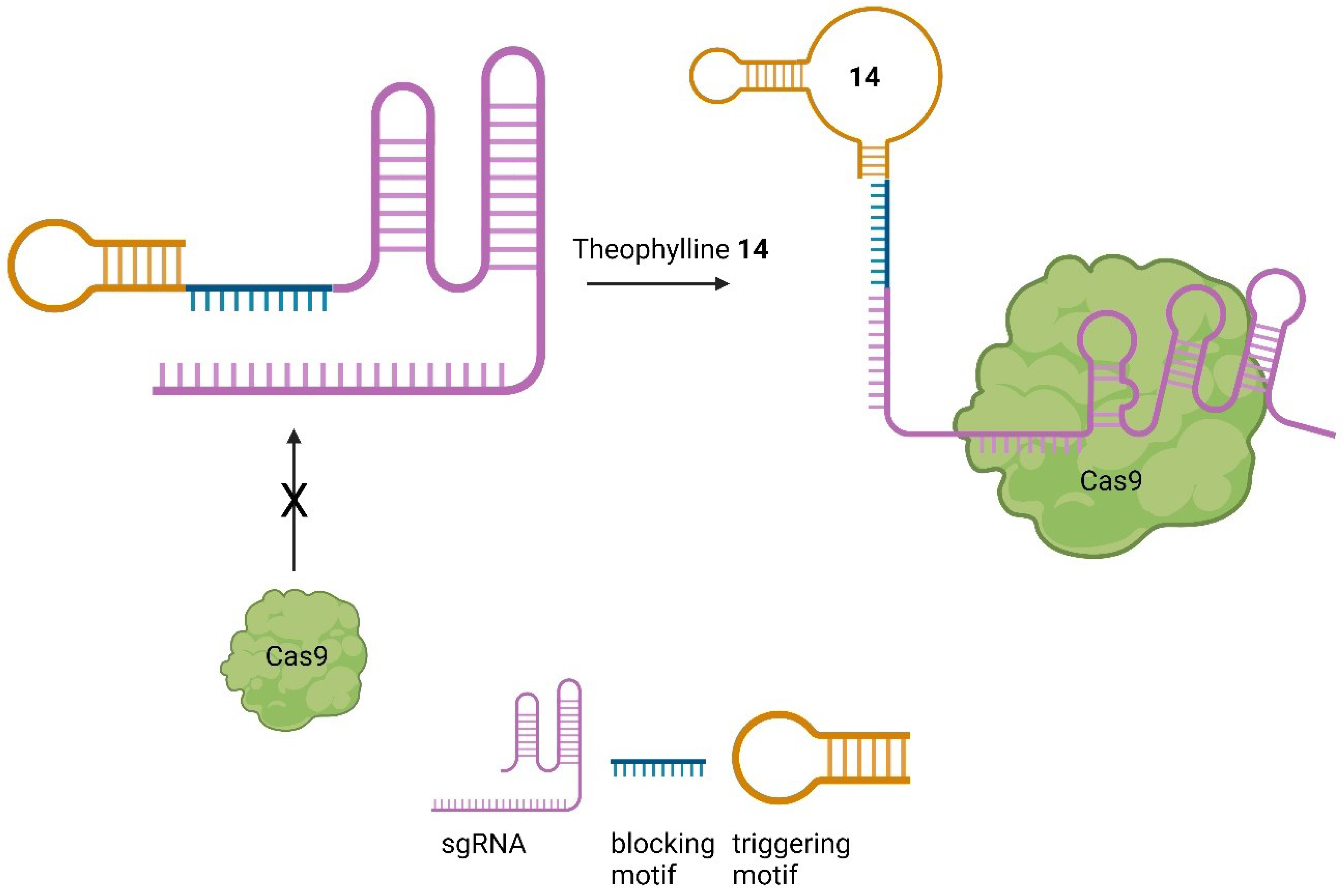
Conclusion
References
- B. Wiedenheft, S. H. Sternberg, and J. A. Doudna, ‘RNA-guided genetic silencing systems in bacteria and archaea’, Nature 2012 482:7385, vol. 482, no. 7385, pp. 331–338, Feb. 2012. [CrossRef]
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna, and E. Charpentier, ‘A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity’, Science (1979), vol. 337, no. 6096, 2012. [CrossRef]
- Y. Xu and Z. Li, ‘CRISPR-Cas systems: Overview, innovations and applications in human disease research and gene therapy’, Comput Struct Biotechnol J, vol. 18, pp. 2401–2415, Jan. 2020. [CrossRef]
- E. Shrock and M. Güell, ‘CRISPR in Animals and Animal Models’, Prog Mol Biol Transl Sci, vol. 152, pp. 95–114, 2017. [CrossRef]
- M. Adli, ‘The CRISPR tool kit for genome editing and beyond’, Nat Commun, vol. 9, pp. 1–13, 2018. [CrossRef]
- S. W. Lee et al., ‘Identification and Optimization of Novel Small-Molecule Cas9 Inhibitors by Cell-Based High-Throughput Screening’, J Med Chem, vol. 65, no. 4, pp. 3266–3305, Feb. 2022. [CrossRef]
- F. V. Karginov and G. J. Hannon, ‘The CRISPR System: Small RNA-Guided Defense in Bacteria and Archaea’, Mol Cell, vol. 37, no. 1, pp. 7–19, Jan. 2010. [CrossRef]
- F. Safari, K. Zare, M. Negahdaripour, M. Barekati-Mowahed, and Y. Ghasemi, ‘CRISPR Cpf1 proteins: structure, function and implications for genome editing’, Cell & Bioscience 2019 9:1, vol. 9, no. 1, pp. 1–21, May 2019. [CrossRef]
- M. A. Mengstie and B. Z. Wondimu, ‘Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing’, Biologics, vol. 15, p. 353, 2021. [CrossRef]
- K. S. Makarova and E. V. Koonin, ‘Annotation and classification of CRISPR-Cas systems’, Methods in Molecular Biology, vol. 1311, pp. 47–75, 2015. [CrossRef]
- F. Jiang and J. A. Doudna, ‘CRISPR–Cas9 Structures and Mechanisms’, https://doi.org/10.1146/annurevbiophys-062215-010822, vol. 46, pp. 505–529, May 2017. [CrossRef]
- E. Weterings and D. J. Chen, ‘The endless tale of non-homologous end-joining’, Cell Research 2008 18:1, vol. 18, no. 1, pp. 114–124, Jan. 2008. [CrossRef]
- M. R. Lieber, ‘The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway’, Annu Rev Biochem, vol. 79, pp. 181–211, Jul. 2010. [CrossRef]
- J. San Filippo, P. Sung, and H. Klein, ‘Mechanism of Eukaryotic Homologous Recombination’. https://doi.org/10.1146/annurev.biochem.77.061306.125255, vol. 77, pp. 229–257, Jun. 2008. [CrossRef]
- G. Li et al., ‘Small molecules enhance CRISPR/ Cas9-mediated homology-directed genome editing in primary cells OPEN’, Scientific Reports , vol. 7, pp. 1–11, 2017. [CrossRef]
- H. Wang and X. Xu, ‘Microhomology-mediated end joining: New players join the team’, Cell Biosci, vol. 7, no. 1, pp. 1–6, Jan. 2017. [CrossRef]
- C. A. Lino, J. C. Harper, J. P. Carney, and J. A. Timlin, ‘Delivering CRISPR: a review of the challenges and approaches’. https://doi.org/10.1080/10717544.2018.1474964, vol. 25, no. 1, pp. 1234–1257, 2018. [CrossRef]
- M. Lee and H. Kim, ‘Therapeutic application of the CRISPR system: current issues and new prospects’, Hum Genet, vol. 138, no. 6, pp. 563–590, Jun. 2019. [CrossRef]
- A. Mahas, C. Neal Stewart, and M. M. Mahfouz, ‘Harnessing CRISPR/Cas systems for programmable transcriptional and post-transcriptional regulation’, Biotechnol Adv, vol. 36, no. 1, pp. 295–310, Jan. 2018. [CrossRef]
- A. A. Dominguez, W. A. Lim, and L. S. Qi, ‘Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation’, Nat Rev Mol Cell Biol, vol. 17, 2015. [CrossRef]
- R. Urrutia, ‘KRAB-containing zinc-finger repressor proteins’, Genome Biol, vol. 4, no. 10, pp. 1–8, Sep. 2003. [CrossRef]
- J. Armando Casas-Mollano, M. H. Zinselmeier, S. E. Erickson, and M. J. Smanski, ‘CRISPR-Cas Activators for Engineering Gene Expression in Higher Eukaryotes’, CRISPR J, vol. 3, no. 5, pp. 350–364, Oct. 2020. [CrossRef]
- C. K. S. Karlson, S. N. Mohd-noor, N. Nolte, and B. C. Tan, ‘CRISPR/dCas9-Based Systems: Mechanisms and Applications in Plant Sciences’, Plants, vol. 10, no. 10, Oct. 2021. [CrossRef]
- S. A. Gangopadhyay et al., ‘Precision Control of CRISPR-Cas9 Using Small Molecules and Light’, Biochemistry, vol. 58, no. 4, p. 234, Jan. 2019. [CrossRef]
- D. Manna et al., ‘A Singular System with Precise Dosing and Spatiotemporal Control of CRISPR-Cas9’, Angewandte Chemie International Edition, vol. 58, no. 19, pp. 6285–6289, May 2019. [CrossRef]
- J. Bondy-Denomy, ‘Protein inhibitors of CRISPR-Cas9’, ACS Chem Biol, vol. 13, no. 2, pp. 417–423, 2018. [CrossRef]
- P. Vyas and Harish, ‘Anti-CRISPR proteins as a therapeutic agent against drug-resistant bacteria’, Microbiol Res, vol. 257, p. 126963, Apr. 2022. [CrossRef]
- S. E. Franklin and S. P. Mayfield, ‘Expert Opinion on Biological Therapy Recent developments in the production of human therapeutic proteins in eukaryotic algae Recent developments in the production of human therapeutic proteins in eukaryotic algae’, Expert Opin. Biol. Ther, vol. 5, no. 2, pp. 225–235, 2005. [CrossRef]
- H. Beck, M. Härter, B. Haß, C. Schmeck, and L. Baerfacker, ‘Small molecules and their impact in drug discovery: A perspective on the occasion of the 125th anniversary of the Bayer Chemical Research Laboratory’, Drug Discov Today, vol. 27, no. 6, pp. 1560–1574, Jun. 2022. [CrossRef]
- B. Maji et al., ‘A High-Throughput Platform to Identify Small-Molecule Inhibitors of CRISPR-Cas9’, Cell, vol. 177, no. 4, pp. 1067-1079.e19, May 2019. [CrossRef]
- Y. Yang et al., ‘Identification and Analysis of Small Molecule Inhibitors of CRISPR-Cas9 in Human Cells’, Cells, vol. 11, no. 22, p. 3574, Nov. 2022. [CrossRef]
- X. Cheng, ‘Valproic Acid Thermally Destabilizes and Inhibits SpyCas9 Activity’, Molecular Therapy, vol. 28, no. 12, pp. 2635–2641, Dec. 2020. [CrossRef]
- K. M. Davis, V. Pattanayak, D. B. Thompson, J. A. Zuris, and D. R. Liu, ‘Small molecule–triggered Cas9 protein with improved genome-editing specificity’, 2015. [CrossRef]
- R. Buskirk, Y. C. Ong, Z. J. Gartner, and D. R. Liu, ‘Directed evolution of ligand dependence: Small-molecule-activated protein splicing’, Proc Natl Acad Sci U S A, vol. 101, no. 29, pp. 10505–10510, Jul. 2004. [CrossRef]
- Y. Wu, L. Yang, T. Chang, F. Kandeel, and J. K. Yee, ‘A Small Molecule-Controlled Cas9 Repressible System’, Mol Ther Nucleic Acids, vol. 19, pp. 922–932, Mar. 2020. [CrossRef]
- J. C. P. N. A. , R. C. D. Rose, ‘Suppression of unwanted CRISPR-Cas9 editing by co-administration of catalytically inactivating truncated guide RNAs.’, Nat Commun, vol. 11, no. 2697, pp. 1–11, 2020.
- R. Sekine, T. Kawata, and T. Muramoto, ‘CRISPR/Cas9 mediated targeting of multiple genes in Dictyostelium’, Sci Rep, vol. 8, no. 1, 2018. [CrossRef]
- R. Weinstain, T. T. Slanina, D. Kand, and P. K. Klán, ‘Visible-to-NIR-Light Activated Release: From Small Molecules to Nanomaterials’, Chemicals Reviews, vol. 120, no. 24, pp. 13135–13272, 2020. [CrossRef]
- B. Maji et al., ‘Multidimensional chemical control of CRISPR–Cas9’, Nature Chemical Biology 2016 13:1, vol. 13, no. 1, pp. 9–11, Oct. 2016. [CrossRef]
- J. Krucinska et al., ‘Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens’, Communications Biology 2022 5:1, vol. 5, no. 1, pp. 1–14, May 2022. [CrossRef]
- R. A. Gama-Brambila et al., ‘A Chemical Toolbox for Labeling and Degrading Engineered Cas Proteins’, vol. 1, pp. 777–785, 2021. [CrossRef]
- C. Zhang et al., ‘π-Clamp-mediated cysteine conjugation’, Nat Chem, vol. 8, pp. 120–128, 2016. [CrossRef]
- M. Békés, D. R. Langley, and C. M. Crews, ‘PROTAC targeted protein degraders: the past is prologue’, Nat Rev Drug Discov, vol. 21, pp. 181–200, 2022. [CrossRef]
- S. Kim, D. Kim, S. W. Cho, J. Kim, and J. S. Kim, ‘Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins’, Genome Res, vol. 24, no. 6, pp. 1012–1019, Jun. 2014. [CrossRef]
- Bubeck et al., ‘Engineered anti-CRISPR proteins for optogenetic control of CRISPR–Cas9’, Nature Methods 2018 15:11, vol. 15, no. 11, pp. 924–927, Oct. 2018. [CrossRef]
- M. Srivastava et al., ‘An Inhibitor of Nonhomologous End-Joining Abrogates Double-Strand Break Repair and Impedes Cancer Progression’, Cell, vol. 151, no. 7, pp. 1474–1487, Dec. 2012. [CrossRef]
- H. C. Bermudez-Cabrera et al., ‘Small molecule inhibition of ATM kinase increases CRISPR-Cas9 1-bp insertion frequency’, Nat Commun, vol. 12, pp. 1–10, 2021. [CrossRef]
- S. Ueno, T. Sudo, and A. Hirasawa, ‘ATM: Functions of ATM Kinase and Its Relevance to Hereditary Tumors’, International Journal of Molecular Sciences 2022, Vol. 23, Page 523, vol. 23, no. 1, p. 523, Jan. 2022. [CrossRef]
- D. B. Lombard, K. F. Chua, R. Mostoslavsky, S. Franco, M. Gostissa, and F. W. Alt, ‘DNA Repair, Genome Stability, and Aging’, Cell, vol. 120, no. 4, pp. 497–512, Feb. 2005. [CrossRef]
- Z. Mao, M. Bozzella, A. Seluanov, and V. Gorbunova, ‘Comparison of nonhomologous end joining and homologous recombination in human cells’, DNA Repair (Amst), vol. 7, no. 10, pp. 1765–1771, Oct. 2008. [CrossRef]
- Y. Chen et al., ‘A PARP1-BRG1-SIRT1 axis promotes HR repair by reducing nucleosome density at DNA damage sites’, Nucleic Acids Res, vol. 47, no. 16, pp. 8563–8580, Sep. 2019. [CrossRef]
- I. E. Wassing et al., ‘The RAD51 recombinase protects mitotic chromatin in human cells’, Nature Communications 2021 12:1, vol. 12, no. 1, pp. 1–17, Sep. 2021. [CrossRef]
- W. Zhang et al., ‘A high-throughput small molecule screen identifies farrerol as a potentiator of CRISPR/Cas9-mediated genome editing’, Elife, vol. 9, pp. 1–25, Jul. 2020. [CrossRef]
- K. N. Meek, A. E. Rangel, and J. M. Heemstra, ‘Enhancing aptamer function and stability via in vitro selection using modified nucleic acids’, Methods, vol. 106, pp. 29–36, Aug. 2016. [CrossRef]
- R. S. Iwasaki, B. A. Ozdilek, A. D. Garst, A. Choudhury, and R. T. Batey, ‘Small molecule regulated sgRNAs enable control of genome editing in E. coli by Cas9’, Nature Communications 2020 11:1, vol. 11, no. 1, pp. 1–9, Mar. 2020. [CrossRef]
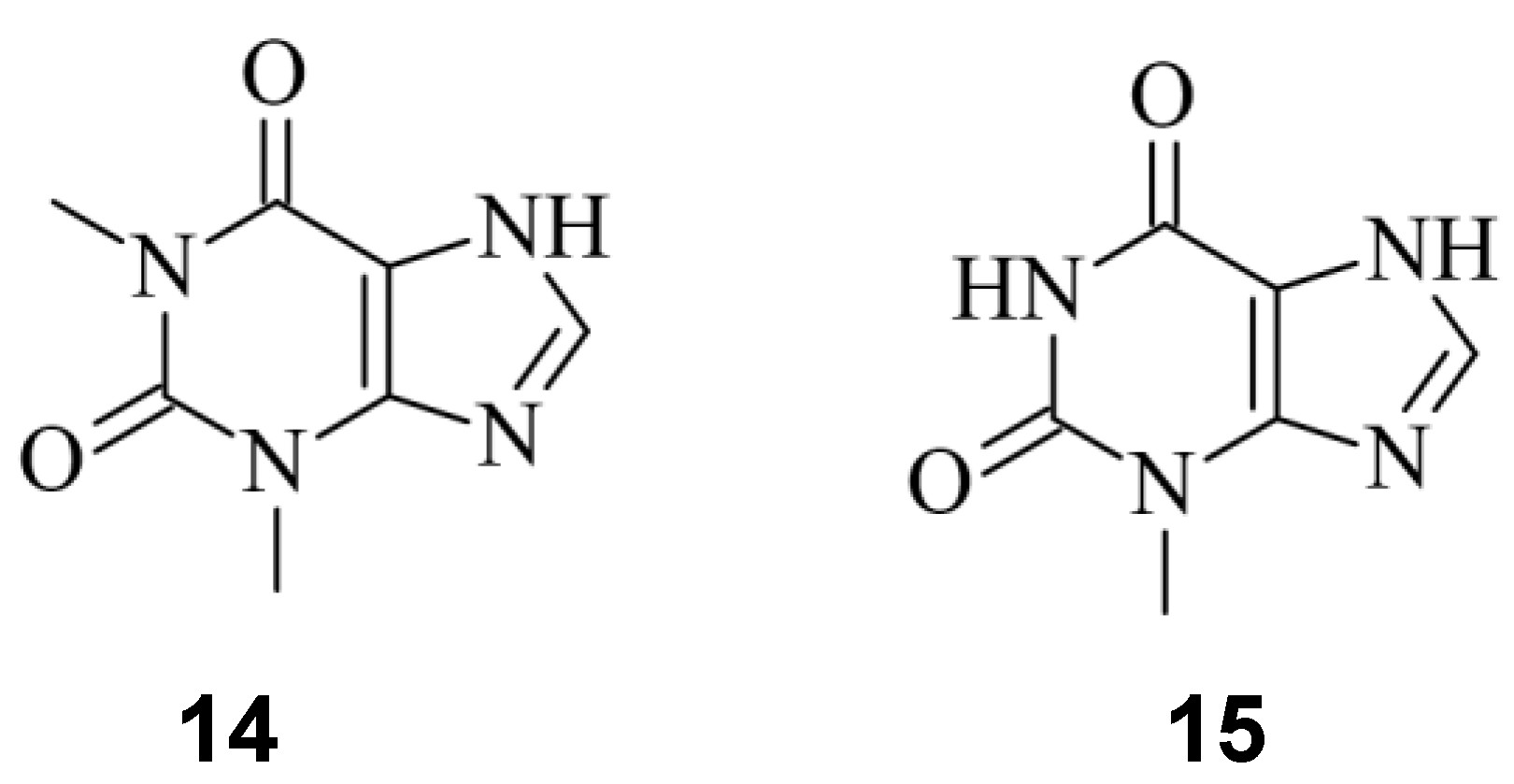
- A. Wrist, W. Sun, and R. M. Summers, ‘The Theophylline Aptamer: 25 Years as an Important Tool in Cellular Engineering Research’, ACS Synth Biol, vol. 9, no. 4, pp. 682–697, Apr. 2020. [CrossRef]
- S. W. Lee, L. Zhao, A. Pardi, and T. Xia, ‘Ultrafast dynamics show that the theophylline and 3-methylxanthine aptamers employ a conformational capture mechanism for binding their ligands’, Biochemistry, vol. 49, no. 13, pp. 2943–2951, Apr. 2010. [CrossRef]
- B. Lin et al., ‘Control of CRISPR-Cas9 with small molecule-activated allosteric aptamer regulating sgRNAs †’, Chem. Commun, vol. 55, p. 12223, 2019. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).