Submitted:
11 December 2023
Posted:
12 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. The Characteristics of the Stationary Trial Plots
2.2. Molecular Genetic Identification of Fungi
2.2.1. Sampling and Molecular Analysis
2.2.2. Bioinformatics
2.2.3. Assessment of Biodiversity Indices
2.2.4. Statistics
3. Results
3.1. Analysis of Fungal Sequences
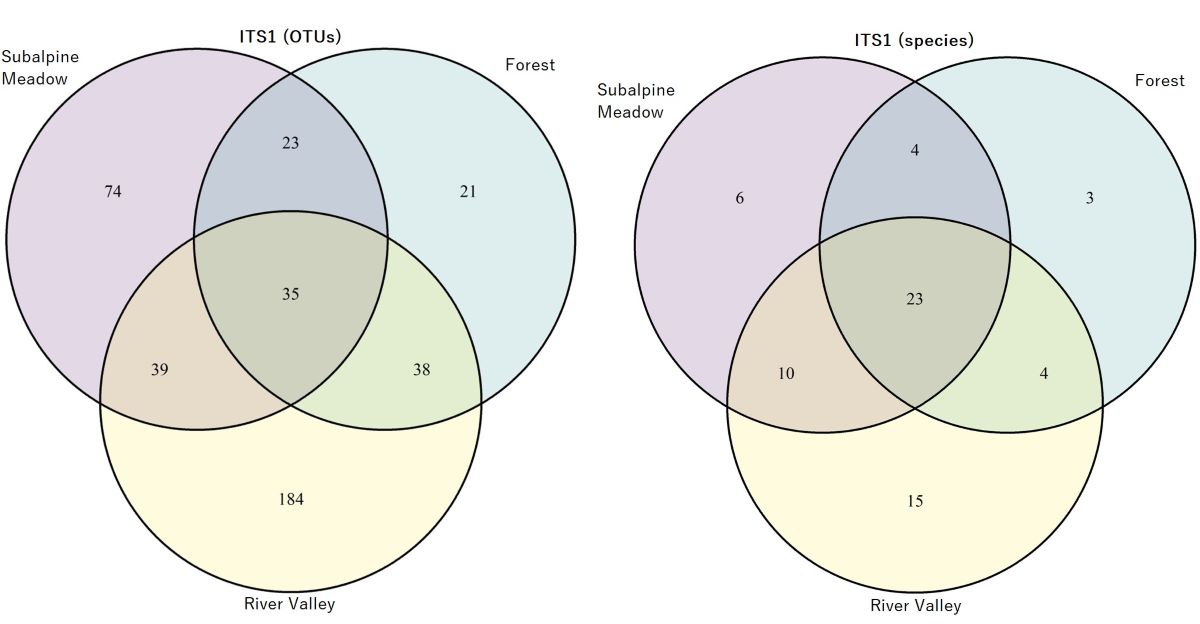
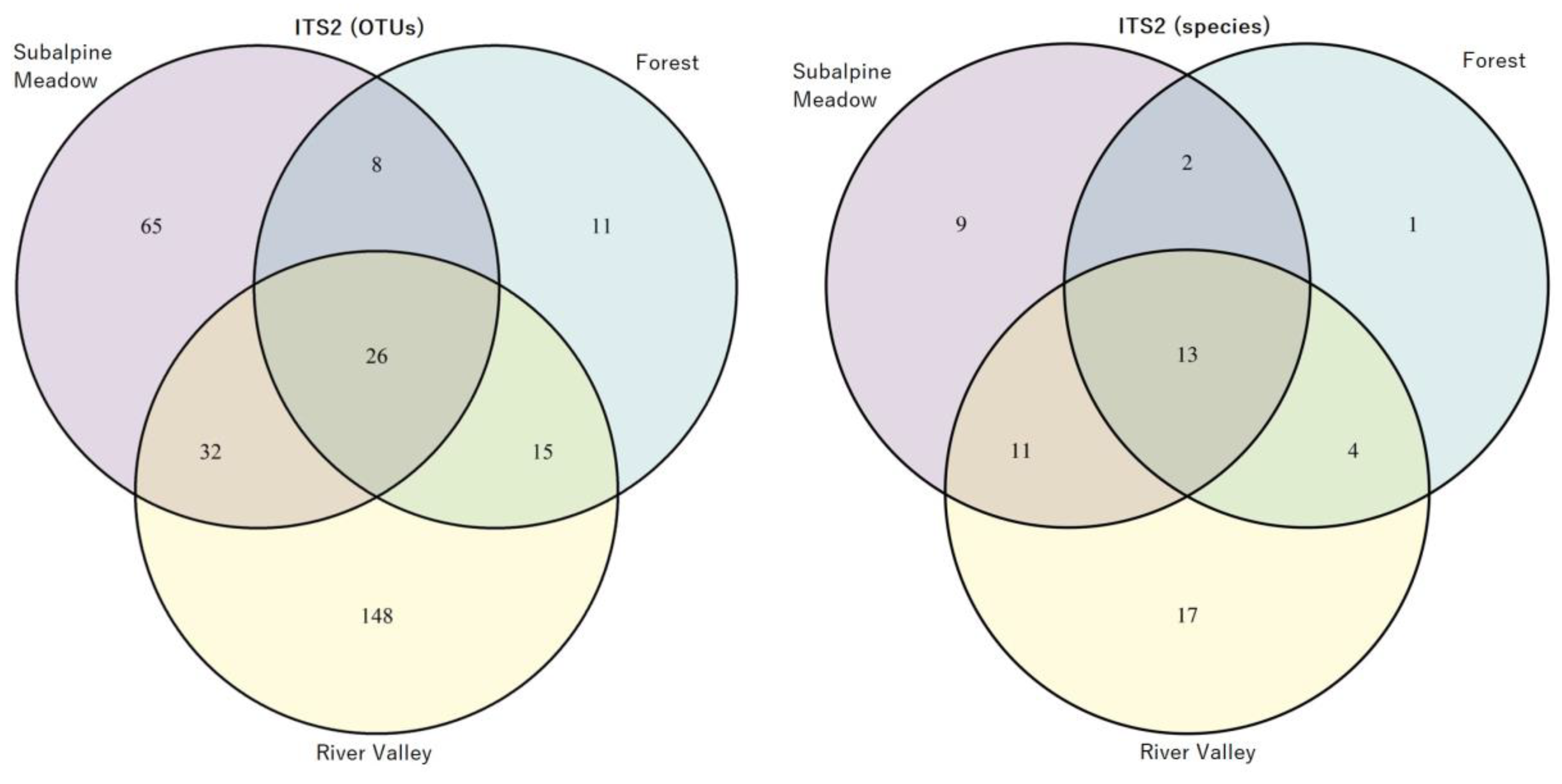
3.2. Taxonomic Composition of Fungal Phyla and Classes in Soil Samples from the River Valley, Subalpine Meadow and Forest
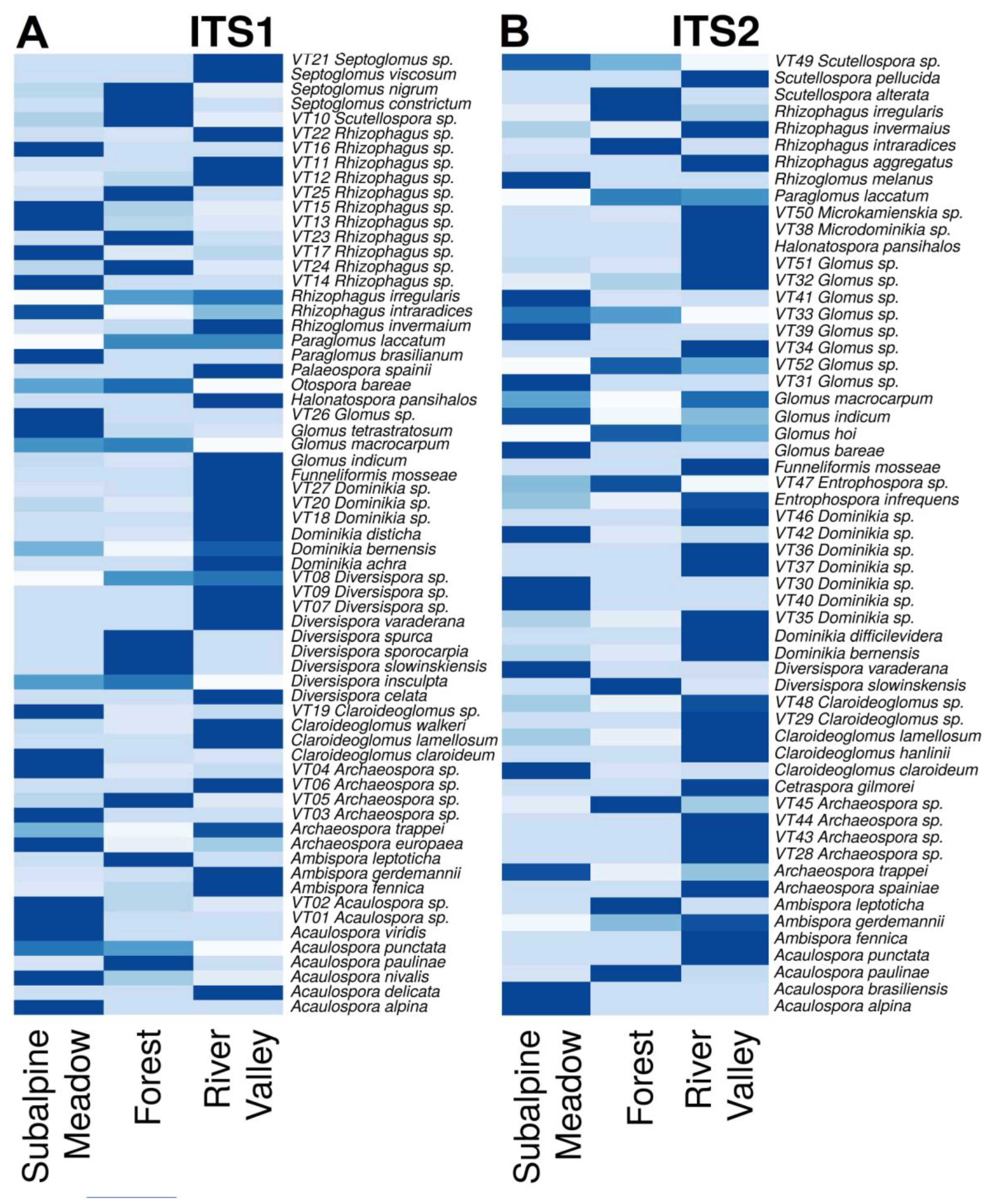
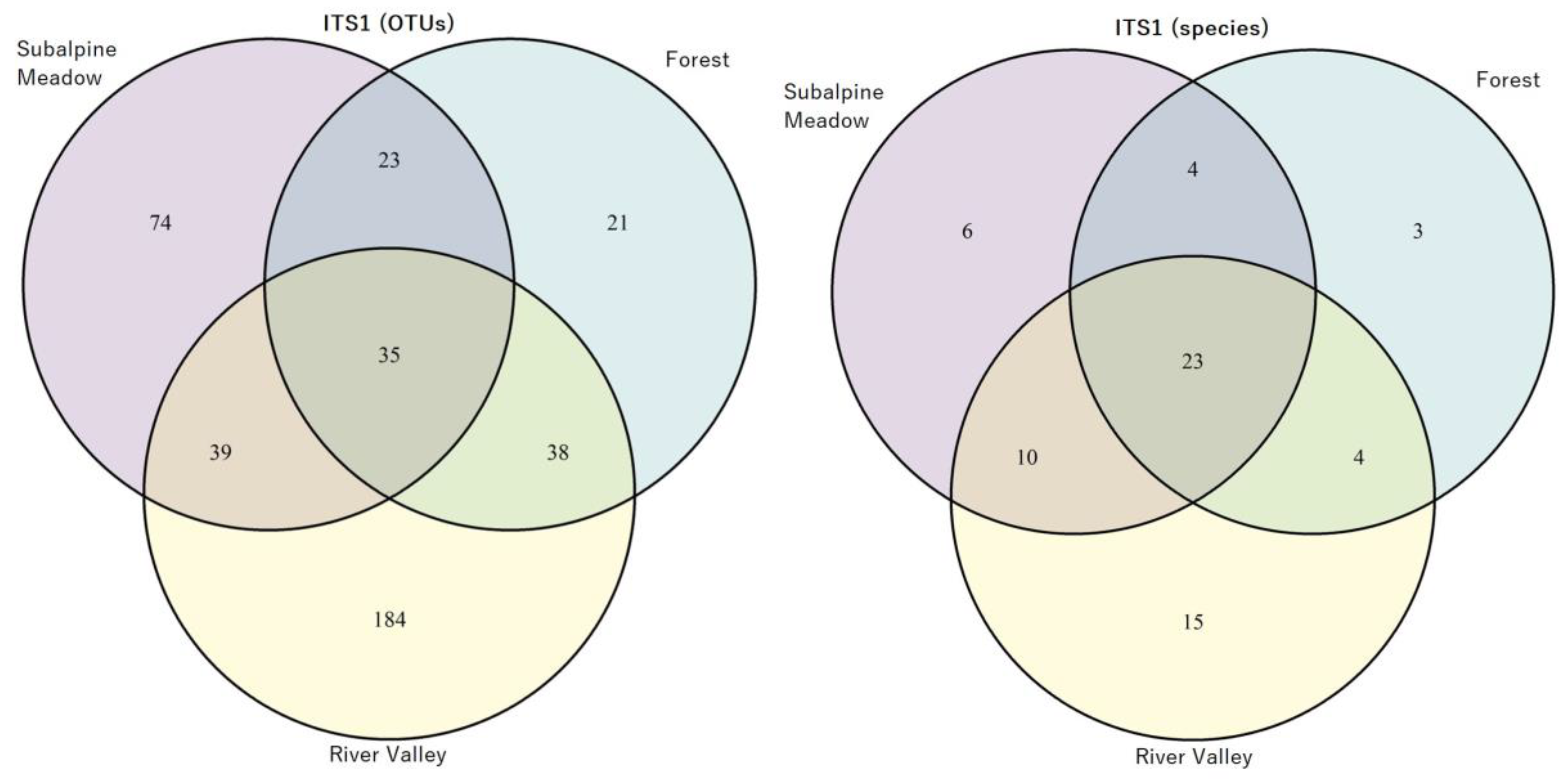
3.3. Identification of AMF Species in the River Valley, Subalpine Meadow and Forest Biotopes
- -
- subalpine meadow (Acaulospora alpina, Aс. nivalis, Aс. nivalis, Aс. paulinae, Aс. punctata, Aс. viridis, Ambispora gerdemannii, Am. leptoticha, Archaeospora europaea, Ar. trappei, Claroideoglomus claroideum, Cl. walkeri, Diversispora insculpta, Dominikia bernensis, Do. disticha, Glomus indicum, G. macrocarpum, G. tetrastratosum, Otospora bareae, Paraglomus brasilianum, Paraglomus laccatum, Rhizophagus intraradices, Rhizophagus irregularis, Septoglomus constrictum, S. nigrum), or 24 species from 12 genera, as well as 19 virtual taxa at the species level;
- -
- forest (Acaulospora nivalis, Ac. paulinae, Ac. punctata, Ambispora fennica, Am. gerdemannii, Am. leptoticha, Claroideoglomus claroideum, Diversispora insculpta, Di. slowinskiensis, Di. sporocarpia, Di. spurca, Dominikia bernensis, Glomus indicum, G. macrocarpum, G. tetrastratosum, Otospora bareae, Paraglomus laccatum, Rhizoglomus invermaium, Rhizophagus intraradices, Rhizophagus irregularis, Septoglomus constrictum, S. nigrum), or 22 species from 11 genera, as well as 12 virtual taxa at the species level;
- -
- river valley (Acaulospora delicata, Ac. paulinae, Ambispora fennica, Am. gerdemannii, Archaeospora europaea, Ar. trappei, Claroideoglomus claroideum, Cl. lamellosum, Cl. walkeri, Diversispora celata, Di. varaderana, Dominikia achra, Do. bernensis, Do. disticha, Funneliformis mosseae, Glomus indicum, G. macrocarpum, G. tetrastratosum, Halonatospora pansihalos, Otospora bareae, Palaeospora spainii, Paraglomus laccatum, Rhizoglomus invermaium, Rhizophagus intraradices, Rhizophagus irregularis, Septoglomus constrictum, S. nigrum, S. viscosum), or 28 species from 15 genera, as well as 24 virtual taxa at the species level.
- -
- subalpine meadow (Acaulospora alpina, Ac. brasiliensis, Ac. paulinae, Ambispora gerdemannii, Am. leptoticha, Archaeospora trappei, Claroideoglomus claroideum, C. lamellosum, Diversispora varaderana, Dominikia bernensis, Entrophospora infrequens, Glomus bareae, G. indicum, G. macrocarpum, Paraglomus laccatum, Rhizoglomus melanus, Rhizophagus intraradices, Rhizophagus invermaius, Rhizophagus irregularis), or 19 species from 11 genera, as well as 15 virtual taxa at the species level;
- -
- forest (Acaulospora paulinae, Ambispora gerdemannii, Am. leptoticha, Claroideoglomus claroideum, C. lamellosum, Diversispora slowinskensis, Entrophospora infrequens, Glomus hoi, G. indicum, Paraglomus laccatum, Rhizophagus intraradices, Rhizophagus irregularis, Scutellospora alterata), or 13 species from 9 genera, as well as 7 virtual taxa at the species level;
- -
- river valley (Acaulospora paulinae, Ac. punctata, Ambispora fennica, Am. gerdemannii, Archaeospora spainiae, Ar. trappei, Cetraspora gilmorei, Claroideoglomus claroideum, Cl. hanlinii, Cl. lamellosum, Dominikia bernensis, Do. difficilevidera, Entrophospora infrequens, Funneliformis mosseae, Glomus hoi, G. indicum, G. macrocarpum, Halonatospora pansihalos, Paraglomus laccatum, Rhizophagus aggregatus, Rhizophagus intraradices, Rhizophagus invermaius, Rhizophagus irregularis, Scutellospora pellucida), or 24 species from 13 genera, as well as 20 virtual taxa at the species level.
3.4. Correlation Analysis of the Interlinks between AMF Species Richness with STP Height and Agrochemical Parameters of Sampled Rhizosphere Soils
4. Discussion
4.1. Applicability of Molecular Genetic Identification of AMF in the Study of Their Biodiversity
4.2. Comparative Analysis of AMF Diversity in Different Biotopes
4.3. Reasons for Higher AMF Diversity in River Valley Biotopes
- (1)
- Human activity (livestock grazing, etc.) is much more developed in river valleys than in mountain forests and subalpine meadows [12]. The invasion of AM fungi by transferring spores on human shoes and hooves of livestock (cows, horses, etc.) is quite common [48,103]. However, some evidances indicate that ungulate grazing may be associated with the decrease of AM fungal abundance in soil [21]. Along with that AMF spores can migrate with water flows from the mountains to the river valley during erosion [103–105] and after sedimentation it actively enter into nonspecific symbiotic relationship. It is developed despite the fact that the organic reserves in these ecosystems are much higher because of livestock grazing, moreover the ecosystems themselves may have signs of soil degradation and are considered to be disturbed. Nevertheless, AM fungi spores, as a rule, are significantly larger (>40 microns) than the spores of many other fungi, so their distribution distance is relatively short [106]. According to Guo et al. [107], terrain slope can also affect AMF diversity. The biotopes of the river valley analyzed in our study were characterized by much more gentle slope in contrast to the biotopes of subalpine meadows and forests (Table S2). Therefore, it can be assumed that a flat slope will have a positive correlation with AMF biodiversity.
- (2)
- In conditions of intensive percolation water regime and good drainage, there is no stagnation of water and oxygen deficiency in soil, negatively affecting the development of AM fungi [108,109]. The percolation water regime can reduce the content of available phosphate (Pi) in the soil, which makes root mycorrhization an important adaptation for P uptake. It is the water regime is typical for river valleys in the North Caucasus. The correlation between the parameters of AMF biodiversity and “Pi, mg/kg” (inorganic P available for plant nutrition) in many cases was significantly (P<0.05) positive (Table S10). Our data is consistent with results of Guo et al. in terms of that the diversity of AMF had a positive correlation with available P content [107]. Nevertheless, large-scale studies have shown that AMF diversity and abundance decrease with phosphorus available in the soil [Ma et al., 2023]. This issue requires further consideration.
- (3)
- STPs altitude may be a factor influencing AM development. The analyzed biotopes of the forest and subalpine meadow located above the biotopes of the river valley were characterized by a reduced number of identified AMF at the species level (Figure 5 and Figure 6), although linear reliable correlations for the North Caucasus STPs were not found (Table S10). Our results are consistent with literature data [10,113–116]. Thus, AMF biodiversity was practically independent of altitude [50,54–57], or the correlations were negative [10,113–116]. However, this rule is not applicable for the Zackenberg valley in the High Arctic [43]: with an increase in altitude from 33 to 479 m (a small altitude above the sea level), an increase in AM fungi occurrence was observed. Perhaps the reason for the lack of correlation between altitude and AMF biodiversity in the North Caucasus is that there are no biotopes of alpine meadows in the analyzed STPs, which are characterized by lower temperatures. However temperature is considered an important factor for the development of AM [109,117,118]. The optimal average air temperature of the warmest month is +19 °C, but mycorrhizal colonization can be intensively increased with the frosty period not less than 2 months [119].
- (4)
- The biodiversity of AMF and the development of AM are affected by such factors as pollution, salinity, drought, extreme temperatures, CO2, liming, acidity, etc. [120], as well as soil composition, altitude, species composition of plant communities, climatic factors [46–49]. But it is pH, along with temperature, that are the main factors determining AMF biodiversity [59]. pH can have an important direct effect on the growth and productivity of the AM fungus [Wang et al., 1993; Coughlan et al., 2000]. The positive correlation between AMF diversity and pH is mentioned in [107,121]. This is consistent with our data on a significant positive correlation of the pHKCl with a number of biodiversity indicators (“Glomeromycota total reads”, “OTUs number for total species”, “total species number”, both for ITS1 and ITS2; Table S10; Figures S6 and S7).
- (5)
- The phenotypic diversity of OTU AMF is supposed to be under the effect of the phenotypic diversity of plants, which decreases with altitude in the mountains [122]. The colonization of root by AM fungi is not species–specific. The roots of one plant can be colonized by several species of AM fungi, and one AM fungus can colonize different plant species [123]. However, despite the absence of direct correlations between the diversity of AMF and the total number of herbaceous plants (Table S10), it might be expected a decrease in the spectrum of potential partners in the mutualistic symbiotic system, and thus affect AMF species diversity [124]. At the same time, annual plant species have a higher diversity of AMF than perennial plant species, and half of the currently identified AMF species may be more specific to one plant species [53]. Moreover, S. Horn et al. [50] demonstrated that the influence of biotic factors (interaction of AMF with plants) is more significant in comparison with the effect of abiotic factors on the composition of AMF genera. It is known that in the process of succession with an increase in the proportion of woody plants the density of AM fungi spores decreased [51,52]. Thus, higher abundance of annual plants (see “Percentage of annual plants” in Table S2) in river valleys in comparison with the biotopes of the subalpine meadow may be a key factor positively affecting AMF taxonomic diversity. Meanwhile, our studies confirm the relationship between the proportion of annual plant forms and the diversity of AMF. For instance, it was shown that the linear correlation coefficients were reliable (P<0.05; Table S10). The correlation coefficient between “Percentage of annual plants” and “Glomeromycota total reads” r = 0.76 and 0.81 (for ITS1 and ITS2, respectively), and the correlation coefficient between “Percentage of annual plants” and “OTUs number (for total species)” r = 0.67 и 0.77 (for ITS1 and ITS2, respectively). A weak correlation between the proportion of annual plants and AMF species diversity was also shown (r = 0.40 and 0.56 for ITS1 and ITS2, respectively; see Table S10). Similar results were obtained in [42]: the “natural grassland” ecosystem had the highest AMF species diversity among 20 ecosystems of interest. The opposite is also possible. It is shown that in the Teberdinsky National Park, experimental suppression of AM symbiosis always is followed by a decrease in the species richness and number of plants [125]. The observations of OTU diversity in the studied territories in a different season may provide new information, since the species composition and numerical ratios of different OTUs may alter due to the season [126,127]. Changes in the mycorrhization of plants by AM fungi thrughout a year in the Teberdinsky National Park have already been studied earlier [81], but their biodiversity has not been assessed.
4.4. Practical Application of the Results of AMF Biodiversity Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.G.; da Fonseca, A.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Marchese, C. Biodiversity hotspots: A shortcut for a more complicated concept. Glob. Ecol. Conserv. 2015, 3, 297–309. [Google Scholar] [CrossRef]
- Myers, N. Threatened biotas: “Hot spots” in tropical forests. Environ. 1988, 83(8), 187–208. [Google Scholar] [CrossRef]
- CEPF (Critical Ecosystem Parthnership Fund). Explore the Biodiversity Hotspots. 2023. Available online: https://www.cepf.net/our-work/biodiversity-hotspots (accessed on 30 October 2023).
- Stork, N.E.; Habel, J.C. Can biodiversity hotspots protect more than tropical forest plants and vertebrates? J. Biogeogr. 2014, 41, 421–428. [Google Scholar] [CrossRef]
- Brundrett, M.C.; Tedersoo, L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018, 220, 1108–1115. [Google Scholar] [CrossRef]
- Newshan, K.K.; Fitter, A.H.; Watkinson, A.R. Multi-functionality and biodiversity in arbuscular mycorrhizas. Review. Trends Ecol. Evol. 1995, 10(10), 407–411. [Google Scholar] [CrossRef]
- De Carvalho, F.; de Souza, F.A.; Carrenho, R.; Escobar, I.E.C.; Oehl, F.; da Silva, G.A. The mosaic of habitats in the high-altitude Brazilian rupestrian fields is a hotspot for arbuscular mycorrhizal fungi. Appl. Soil Ecol. 2012, 52, 9–19. [Google Scholar]
- Bonfim, J.A.; Vasconcellos, R.L.F.; Gumiere, T.; Mescolotti, D.L.C.; Oehl, F.; Cardoso, E.J.B.N. Diversity of arbuscular mycorrhizal fungi in a brazilian atlantic forest toposequence. Microb. Ecol. 2016, 71, 164–177. [Google Scholar] [CrossRef]
- Vieira, L. C. , da Silva, D. K. A., de Melo, M. A. C., Escobar, I. E. C., Oehl, F., da Silva, G. A. Edaphic factors influence the distribution of arbuscular mycorrhizal fungi along an altitudinal gradient of a Tropical Mountain. Microb. Ecol. 2019, 78, 904–913. [Google Scholar] [CrossRef]
- Zazanashvili, N. The Caucasus hotspot. In Status and Protection of Globally Threatened Species in the Caucasus; Zazanashvili, N., Mallon, D., Eds.; Contour Ltd.: Tbilisi, Georgia, 2004; pp. 15–25. [Google Scholar]
- Zernov, A.S.; Alekseev, Yu.E.; Onipchenko, V.G. The determinant of vascular plants of the Karachay-Cherkessia Republic; Association of Scientific Publications “KMK”: Moscow, Russia, 2015. [Google Scholar]
- Vasar, M.; Davison, J.; Sepp, S.-K.; Oja, J.; Al-Quraishy, S.; Bueno, C.G.; Cantero, J.J.; Fabiano, E.C.; Decocq, G.; Fraser, L.; Hiiesalu, I.; Hozzein, W.N.; Koorem, K.; Moora, M.; Mucina, L.; Onipchenko, V.; Öpik, M.; Pärtel, M.; Phosri, C.; Vahter, T.; Tedersoo, L.; Zobel, M. Global taxonomic and phylogenetic assembly of AM fungi. Mycorrhiza 2022, 32, 135–144. [Google Scholar] [CrossRef]
- Tedersoo, L.; Mikryukov, V.; Anslan, S.; Bahram, M.; Khalid, A.N.; Corrales, A.; Agan, A.; Vasco-Palacios, A.-M.; Saitta, A.; Antonelli, A.; et al. The Global Soil Mycobiome consortium dataset for boosting fungal diversity research. Fungal Divers. 2021, 111, 573–588. [Google Scholar] [CrossRef]
- Rożek, K.; Błaszkowski, J.; Nowak, A.; Zalewska-Gałosz, J.; Nobis, M.; Mleczko, P.; Zubek, S. Arbuscular mycorrhizal fungi in Georgia, the Caucasus region: The first report of species diversity and root colonization. Nov. Hedwigia. 2018, 106, 473–483. [Google Scholar] [CrossRef]
- Bidondo, L.F.; Colombo, R.P.; Recchi, R.; Silvani, V.A.; Pérgola, M.; Martínez, A.; Godeas, A.M. Detection of arbuscular mycorrhizal fungi associated with pecan (Carya illinoinensis) trees by molecular and morphological approaches. MycoKeys 2018, 42, 73–88. [Google Scholar] [CrossRef]
- Lara-Pérez, L.A.; Oros-Ortega, I.; Córdova-Lara, I.; Estrada-Medina, H.; O'Connor-Sánchez, A.; Góngora-Castillo, E.; Sáenz-Carbonell, L. Seasonal shifts of arbuscular mycorrhizal fungi in Cocos nucifera roots in Yucatan, Mexico. Mycorrhiza 2020, 30, 269–283. [Google Scholar] [CrossRef]
- Nam, Y.J.; Kim, H.; Lee, J.H.; Yoon, H.; Kim, J.-G. Metagenomic analysis of soil fungal communities on Ulleungdo and Dokdo Islands. J. Gen. Appl. Microbiol. 2015, 61, 67–74. [Google Scholar] [CrossRef]
- Botnen, S.; Kauserud, H.; Carlsen, T.; Blaalid, R.; Høiland, K. Mycorrhizal fungal communities in coastal sand dunes and heaths investigated by pyrosequencing analyses. Mycorrhiza 2015, 25, 447–456. [Google Scholar] [CrossRef]
- Peña-Venegas, C.P.; Kuyper, T.W.; Davison, J.; Jairus, T.; Vasar, M.; Stomph, T.J.; Struik, P.C.; Öpik, M. Distinct arbuscular mycorrhizal fungal communities associate with different manioc landraces and Amazonian soils. Mycorrhiza 2019, 29, 263–275. [Google Scholar] [CrossRef]
- Stevens, B.M.; Propster, J.R.; Öpik, O.; Wilson, G.W.T.; Alloway, S.L.; Mayemba, E.; Johnson, N.C. Arbuscular mycorrhizal fungi in roots and soil respond differently to biotic and abiotic factors in the Serengeti. Mycorrhiza 2020, 30, 79–95. [Google Scholar] [CrossRef]
- Ji, L.; Zhang, Y.; Yang, Y.; Yang, L.; Yang, N.; Zhang, D. Long-term effects of mixed planting on arbuscular mycorrhizal fungal communities in the roots and soils of Juglans mandshurica plantations. BMC Microbiol. 2020, 20(1), 304. [Google Scholar] [CrossRef]
- Xu, Z.; Lv, Y.; Fang, M.; Lui, J.; Zeng, H.; Ban, Y. Diverse and abundant arbuscular mycorrhizal fungi in ecological floating beds used to treat eutrophic water. Appl. Microbiol. Biotechnol. 2021, 105, 6959–6975. [Google Scholar] [CrossRef]
- Kolaříková, Z.; Slavíková, R.; Krüger, C.; Krüger, M.; Kohout, P. PacBio sequencing of Glomeromycota rDNA: a novel amplicon covering all widely used ribosomal barcoding regions and its applicability in taxonomy and ecology of arbuscular mycorrhizal fungi. New Phytol. 2021, 231, 490–499. [Google Scholar] [CrossRef]
- Baldrian, P.; Větrovský, T.; Lepinay, C.; Kohout, P. High-throughput sequencing view on the magnitude of global fungal diversity. Fungal Divers. 2022, 114, 539–547. [Google Scholar] [CrossRef]
- Alaux, P.-L.; Mison, C.; Senés-Guerrero, C.; Moreau, V.; Manssens, G.; Foucart, G.; Cranenbrouck, S.; Declerck, S. Diversity and species composition of arbuscular mycorrhizal fungi across maize fields in the southern part of Belgium. Mycorrhiza 2021, 31, 265–272. [Google Scholar] [CrossRef]
- Vieira, C.K.; Marascalchi, M.N.; Rodrigues, A.V.; de Armas, R.D.; Stürmer, S.L. Morphological and molecular diversity of arbuscular mycorrhizal fungi in revegetated iron-mining site has the same magnitude of adjacent pristine ecosystems. J. Environ. Sci. (China) 2018, 67, 330–343. [Google Scholar] [CrossRef]
- Morgan, B.S.T.; Egerton-Warburton, L.M. Barcoded NS31/AML2 primers for sequencing of arbuscular mycorrhizal communities in environmental samples. Appl. Plant Sci. 2017, 5, 1700017. [Google Scholar] [CrossRef]
- Magurno, F.; Malicka, M.; Posta, K.; Wozniak, G.; Lumini, E.; Piotrowska-Seget, Z. Glomalin gene as molecular marker for functional diversity of arbuscular mycorrhizal fungi in soil. Biol. Fertil. Soils. 2019, 554(55), 411–417. [Google Scholar] [CrossRef]
- Schüßler, A. Glomeromycota species list. Available online: http://www.amf-phylogeny.com and https://docs.google.com/spreadsheets/d/1vHHj9XtfIYj9B60L7UP4rUci8bJeolUP/edit#gid=626676380, last updated 14 Jan 2023 (accessed on 30 October 2023).
- GenBank NCBI. 2023. Available online: https://www.ncbi.nlm.nih.gov/genbank (accessed on 30 October 2023).
- Stockinger, H.; Krüger, M.; Schüssler, A. DNA barcoding of arbuscular mycorrhizal fungi. New Phytol. 2010, 187(2), 461–474. [Google Scholar] [CrossRef]
- Borriello, R.; Bianciotto, V.; Orgiazzi, A.; Lumini, E.; Bergero, R. Sequencing and comparison of the mitochondrial COI gene from isolates of Arbuscular Mycorrhizal Fungi belonging to Gigasporaceae and Glomeraceae families. Mol. Phylogenet. Evol. 2014, 75, 1–10. [Google Scholar] [CrossRef]
- Börstler, B.; Thiéry, O.; Sýkorová, Z.; Berner, A.; Redecker, D. Diversity of mitochondrial large subunit rDNA haplotypes of Glomus intraradices in two agricultural field experiments and two semi-natural grasslands. Mol. Ecol. 2010, 19(7), 1497–1511. [Google Scholar] [CrossRef]
- James, T.Y.; Kauff, F.; Schoch, C.L.; Matheny, P.B.; Hofstetter, V.; Cox, C.J.; Celio, G.; Gueidan, C.; Fraker, E., Miadlikowska J.; et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 2006, 443(7113), 818–822. [Google Scholar] [CrossRef]
- Sokolski, S.; Dalpé, Y.; Séguin, S.; Khasa, D.; Lévesque, C.A.; Piché, Y. Conspecificity of DAOM 197198, the model arbuscular mycorrhizal fungus, with Glomus irregulare: molecular evidence with three protein-encoding genes. Botany 2010, 88(9), 829–838. [Google Scholar] [CrossRef]
- Msiska, Z.; Morton, J. Phylogenetic analysis of the Glomeromycota by a partial beta-tubulin gene. Mycorrhiza 2009, 19(4), 247–254. [Google Scholar] [CrossRef]
- Öpik, M.; Davison, J.; Moora, M.; Zobel, M. DNA-based detection and identification of Glomeromycota: the virtual taxonomy of environmental sequences. Botany 2014, 92(2), 135–147. [Google Scholar] [CrossRef]
- Błaszkowski, J.; Sánchez-García, M.; Niezgoda, P.; Zubek, S.; Fernández, F.; Vila, A.; Al-Yahya'ei, M.N.; Symanczik, S.; Milczarski, P.; Malinowski, R.; Cabello, M.; Goto, B.T.; Casieri, L.; Malicka, M.; Bierza, W.; Magurno, F. A new order, Entrophosporales, and three new Entrophospora species in Glomeromycota. Front. Microbiol. 2022, 13, 962856. [Google Scholar] [CrossRef]
- Błaszkowski, J.; Jobim, K.; Niezgoda, P.; Meller, E.; Malinowski, R.; Milczarski, P.; Zubek, S.; Magurno, F.; Casieri, L.; Bierza, W.; Błaszkowski, T.; Crossay, T.; Goto, B.T. New glomeromycotan taxa, Dominikia glomerocarpica sp. nov. and Epigeocarpum crypticum gen. nov. et sp. nov. from Brazil, and Silvaspora gen. nov. from New Caledonia. Front. Microbiol. 2021, 12, 655–910. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; James, T.Y.; O'Donnell, K.; Roberson, R.W.; Taylor, T.N.; Uehling, J.; Vilgalys, R.; White, M.M.; Stajich, J.E. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2016, 108(5), 1028–1046. [Google Scholar]
- Castillo, C.G.; Borie, F.; Oehl, F.; Sieverding, E. Arbuscular mycorrhizal fungi biodiversity: prospecting in Southern-Central zone of Chile. A review. J. Soil Sci. Plant Nutr. 2016, 16(2), 400–422. [Google Scholar] [CrossRef]
- Rasmussen, P.U.; Abrego, N.; Roslin, T.; Opik, M.; Sepp, S.-K.; Blanchet, F.G.; Huotari, T.; Hugerth, L.W.; Tack, A.J.M. Elevation and plant species identity jointly shape a diverse arbuscular mycorrhizal fungal community in the High Arctic. New Phytol. 2022, 236, 671–683. [Google Scholar] [CrossRef]
- Öpik, M.; Vanatoa, A.; Vanatoa, E.; Moora, M.; Davison, J.; Kalwij, J.M.; Reier, U.; Zobel, M. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol. 2010, 188, 223–241. [Google Scholar] [CrossRef]
- MaarjAM Database. 2023. Available online: http://maarjam.botany.ut.ee (accessed on 30 October 2023).
- Kruger, C.; Kohout, P.; Janouskova, M.; Puschel, D.; Frouz, J.; Rydlova, J. Plant communities rather than soil properties structure arbuscular mycorrhizal fungal communities along primary succession on a mine spoil. Front. Microbiol. 2017, 8, 719. [Google Scholar] [CrossRef]
- Ezeokoli, O.T.; Nwangburuka, C.C.; Adeleke, R.A.; Roopnarain, A.; Paterson, D.G.; Maboeta, M.S.; Bezuidenhout, C.C. Assessment of arbuscular mycorrhizal fungal spore density and viability in soil stockpiles of south African opencast coal mines. South African J. Plant Soil 2019, 36, 91–99. [Google Scholar] [CrossRef]
- Clavel, J.; Lembrechts, J.; Alexander, J.; Haider, S.; Lenoir, J.; Milbau, A.; Nuñez, M.A.; Pauchard, A.; Nijs, I.; Verbruggen, E. The role of arbuscular mycorrhizal fungi in nonnative plant invasion along mountain roads. New Phytol. 2021, 230(3), 1156–1168. [Google Scholar] [CrossRef]
- Yan, P.; Hou, H.; Lv, Y.; Zhang, H.; Li, J.; Shao, L.; Xie, Q.; Liang, Y.; Li, J.; Ni, X. Diversity characteristics of arbuscular mycorrhizal fungi communities in the soil along successional altitudes of Helan Mountain, arid, and semi-arid regions of China. Front. Microbiol. 2023, 14, 1099131. [Google Scholar] [CrossRef]
- Horn, S.; Caruso, T.; Verbruggen, E.; Rillig, M.C.; Hempel, S. Arbuscular mycorrhizal fungal communities are phylogenetically clustered at small scales. ISME J. 2014, 8(11), 2231–2242. [Google Scholar] [CrossRef]
- Zangaro, W.; Assis, R.; Rostirola, L.; Souza, P.; Gonçalves, M.; Andrade, G.; Nogueira, M. Changes in arbuscular mycorrhizal associations and fine root traits in sites under different plant successional phases in southern Brazil. Mycorrhiza 2008, 19, 37–45. [Google Scholar] [CrossRef]
- Zangaro, W.; Rostirola, L.; Souza, P.; Almeida Alves, R.; Lescano, L.; Rondina, A.; Nogueira, M.; Carrenho, R. Root colonization and spore abundance of arbuscular mycorrhizal fungi in distinct successional stages from an Atlantic rainforest biome in southern Brazil. Mycorrhiza 2013, 23, 221–233. [Google Scholar] [CrossRef]
- Torrecillas, E.; Alguacil, M.M.; Roldan, A. Differences in the AMF diversity in soil and roots between two annual and perennial gramineous plants co-occurring in a Mediterranean, semiarid degraded area. Plant Soil 2012, 354, 97–106. [Google Scholar] [CrossRef]
- Gai, J.P.; Tian, H.; Yang, F.Y.; Christie, P.; Li, X.L.; Klironomos, J.N. Arbuscular mycorrhizal fungal diversity along a Tibetan elevation gradient. Pedobiol. 2012, 55, 145–151. [Google Scholar] [CrossRef]
- Coutinho, E.S.; Fernandes, G.W.; Berbara, R.L.L.; Valerio, H.M.; Goto, B.T. Variation of arbuscular mycorrhizal fungal communities along an altitudinal gradient in rupestrian grasslands in Brazil. Mycorrhiza 2015, 25, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.C.; Ni, Y.Y.; Liang, W.J.; Wang, J.J.; Chu, H.Y. Distinct soil bacterial communities along a small-scale elevational gradient in alpine tundra. Front. Microbiol. 2015, 6, 582. [Google Scholar] [CrossRef]
- Senés-Guerrero, C.; Schüßler, A. A conserved arbuscular mycorrhizal fungal core-species community colonizes potato roots in the Andes. Fungal Divers. 2015, 77(1), 317–333. [Google Scholar] [CrossRef]
- Zernov, A.S. Flora of the North-West Caucasus; Association of Scientific Publications “KMK”: Moscow, Russia, 2006. [Google Scholar]
- Davison, J.; Moora, M.; Semchenko, M.; Adenan, S.B.; Ahmed, T.; Akhmetzhanova, A.A.; Alatalo, J.M.; Al-Quraishy, S.; Andriyanova, E.; Anslan, S.; et al. Temperature and pH define the realised niche space of arbuscular mycorrhizal fungi. New Phytol. 2021, 231(2), 763–776. [Google Scholar] [CrossRef]
- Sokolov, A.V. Agrochemical Methods of Soil Studies; Nauka: Moscow, Russia, 1975. [Google Scholar]
- Maniatis, T.; Fritsch, E.F.; Sambrook, J. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor Laboratory: NY, USA, 1982.
- Kryukov, A.A.; Gorbunova, A.O.; Machs, E.M.; Mikhaylova, Y.V.; Rodionov, A.V.; Zhurbenko, P.M.; Yurkov, A.P. Perspectives of using Illumina MiSeq for identification of arbuscular mycorrhizal fungi. Vavilovskii Zhurnal Genet. Selektsii 2020, 24(2), 158–167. [Google Scholar]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26(19), 2460–2461. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Meth. 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Bogusz, M.; Whelan, S. Phylogenetic tree estimation with and without alignment: new distance methods and benchmarking. Syst. Biol. 2017, 66(2), 218–231. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Tedersoo, L.; Ryberg, M.; Kristiansson, E.; Hartmann, M.; Unterseher, M.; Porter, T.M.; Bengtsson-Palme, J.; Walker, D.M.; de Sousa, F.; Gamper, H.A.; Larsson, E.; Larsson, K.H.; Kõljalg, U.; Edgar, R.C.; Abarenkov, K. A comprehensive, automatically updated fungal ITS sequence dataset for reference-based chimera control in environmental sequencing efforts. Microbes Environ. 2015, 30(2), 140–150. [Google Scholar] [CrossRef]
- MEGA, Molecular Evolutionary Genetics Analysis. 2023. Available online: https://www.megasoftware.net (accessed on 30 October 2023).
- Yan, L.; Kenchanmane Raju, S.K.; Lai, X.; Zhang, Y.; Dai, X.; Rodriguez, O.; Mahboub, S.; Roston, R.L.; Schnable, J.C. Parallels between natural selection in the cold-adapted crop-wild relative Tripsacum dactyloides and artificial selection in temperate adapted maize. Plant J. 2019, 99(5), 965–977. [Google Scholar] [CrossRef]
- Hsieh, T.C.; Ma, K.H.; Chao, A. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 2016, 7(12), 1451–1456. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2023. Available online: https://www.R-project.org (accessed on 30 October 2023).
- Stacklies, W.; Redestig, H.; Scholz, M.; Walther, D.; Selbig, J. pca Methods – a Bioconductor package providing PCA methods for incomplete data. Bioinformatics 2007, 23, 1164–1167. [Google Scholar] [CrossRef]
- Galili, T. dendextend: an R package for visualizing, adjusting, and comparing trees of ierarchical clustering. Bioinformatics 2015, 31(22), 3718–3720. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer-Verlag: NY, USA, 2016. [Google Scholar]
- Chen, H.; Boutros, P.C. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 2011, 12, 35. [Google Scholar] [CrossRef]
- Alberdi, A.; Aizpurua, O.; Gilbert, M.; Bohmann, K. Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods Ecol. Evol. 2018, 9, 134–147. [Google Scholar] [CrossRef]
- Delavaux, C.S.; Ramos, R.J.; Sturmer, S.L.; Bever, J.D. Environmental identification of arbuscular mycorrhizal fungi using the LSU rDNA gene region: an expanded database and improved pipeline. Mycorrhiza 2022, 32(2), 145–153. [Google Scholar] [CrossRef]
- Krüger, M.; Stockinger, H.; Krüger, C.; Schüßler, A. DNA-based species level detection of Glomeromycota: one PCR primer set for all arbuscular mycorrhizal fungi. New Phytol. 2009, 183, 212–223. [Google Scholar] [CrossRef]
- Bruns, T.D.; Taylor, J.W. Comment on “Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism”. Science 2016, 351, 826–826. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Bender, S.F.; Mascher, F.; Russo, G.; Patrignani, A.; Camenzind, T.; Hempel, S.; Rillig, M.C.; Heijden, M.G. High-resolution community profiling of arbuscular mycorrhizal fungi. New Phytol. 2016, 212, 780–791. [Google Scholar] [CrossRef]
- Baikalova, A.S.; Onipchenko, V.G. Mycosymbiotrophism of alpine plants in the Teberda Nature Reserve. In Opyt issledovaniya rastitel’nykh soobshchestv v zapovednikakh (Study of Communities in Nature Reserves), Tr. Tsentr. Nauchno-Issled. Lab. Okhotn. Khoz. Zapoved., Moscow, Russia, 1988; 93–107.
- Onipchenko, V.G.; Guzhova, G.A.; Semenova, G.V.; Rabotnova, M.V. Population strategy of Alpine plants of the northwestern Caucasus. In Ekologiya populyatsii (Ecology of Populations); Nauka: Moscow, Russia, 1991; pp. 165–180. [Google Scholar]
- Onipchenko, V.G.; Zobel, M. Mycorrhiza, vegetative mobility and responses to disturbance of alpine plants in the Northwestern Caucasus. Folia Geobot. 2000, 35(1), 1–11. [Google Scholar] [CrossRef]
- Wipf, D.; Krajinski, F.; van Tuinen, D.; Recorbet, G.; Courty, P.E. Trading on the arbuscular mycorrhiza market: from arbuscules to common mycorrhizal networks. New Phytol. 2019, 223(3), 1127–1142. [Google Scholar] [CrossRef]
- Rosendahl, S. Communities, populations and individuals of arbuscular mycorrhizal fungi. New Phytol. 2008, 178(2), 253–266. [Google Scholar] [CrossRef]
- Mathieu, S.; Cusant, L.; Roux, C.; Corradi, N. Arbuscular mycorrhizal fungi: intraspecific diversity and pangenomes. New Phytol. 2018, 220(4), 1129–1134. [Google Scholar] [CrossRef]
- Schoen, C.; Montibeler, M.; Costa, M.D.; Antunes, P.M.; Stürmer, S.L. Inter and intra-specific variability in arbuscular mycorrhizal fungi affects hosts and soil health. Symbiosis 2021, 1–17. [Google Scholar] [CrossRef]
- Daubois, L.; Beaudet, D.; Hijri, M.; de la Providencia, I. Independent mitochondrial and nuclear exchanges arising in Rhizophagus irregularis crossed-isolates support the presence of a mitochondrial segregation mechanism. BMC Microbiol. 2016, 16(1), 1–12. [Google Scholar] [CrossRef]
- Yurkov, A.P.; Kryukov, A.A.; Gorbunova, A.O.; Kojemyakov, A.P.; Stepanova, G.V.; Machs, E.M.; Rodionov, A.V.; Shishova, M.F. Molecular genetic identification of arbuscular mycorrhizal fungi. Ecol. Genet. 2018, 16(2), 11–23. [Google Scholar] [CrossRef]
- Stockinger, H.; Walker, C.; Schüßler, A. 'Glomus intraradices DAOM197198', a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol. 2009, 183(4), 1176–1187. [Google Scholar] [CrossRef]
- Hosny, M.; Gianinazzi-Pearson, V.; Dulieu, H. Nuclear DNA content of 11 fungal species in Glomales. Genome 1998, 41(3), 422–428. [Google Scholar] [CrossRef]
- Marleau, J.; Dalpé, Y.; St-Arnaud, M.; Hijri, M. Spore development and nuclear inheritance in arbuscular mycorrhizal fungi. BMC Evol. Biol. 2011, 11, 51. [Google Scholar] [CrossRef]
- Rodionov, A.V.; Amosova, A.V.; Krainova, L.M.; Machs, E.M.; Mikhailova, Yu.V.; Gnutikov, A.A.; Muravenko, O.V.; Loskutov, I.G. Phenomenon of multiple mutations in the 35S rRNA genes of the C subgenome of polyploid Avena L. species. Russ. J. Genet. 2020, 56(6), 674–683. [Google Scholar] [CrossRef]
- Borowska-Zuchowska, N.; Kovarik, A.; Robaszkiewicz, E.; Tuna, M.; Tuna, G.S.; Gordon, S.; Vogel, J.P.; Hasterok, R. The fate of 35S rRNA genes in the allotetraploid grass Brachypodium hybridum. Plant J. 2020, 103(5), 1810–1825. [Google Scholar] [CrossRef]
- Lunerova, J.; Renny-Byfield, S.; Matyášek, R.; Leitch, A.; Kovařík, A. Concerted evolution rapidly eliminates sequence variation in rDNA coding regions but not in intergenic spacers in Nicotiana tabacum allotetraploid. Plant Sys. Evol. 2017, 303(8), 1043–1060. [Google Scholar] [CrossRef]
- Robbins, C.; Corella, J.C.; Aletti, C.; Seiler, R.; Mateus, I.D.; Lee, S.J.; Masclaux, F.G.; Sanders, I.R. Generation of unequal nuclear genotype proportions in Rhizophagus irregularis progeny causes allelic imbalance in gene transcription. New Phytol. 2021, 231(5), 1984. [Google Scholar] [CrossRef]
- Chen, E.C.H.; Mathieu, S.; Hoffrichter, A.; Sedzielewska-Toro, K.; Peart, M.; Pelin, A.; Ndikumana, S.; Ropars, J.; Dreissig, S.; Fuchs, J.; Brachmann, A.; Corradi, N. Single nucleus sequencing reveals evidence of inter-nucleus recombination in arbuscular mycorrhizal fungi. eLife 2018, 7, e39813. [Google Scholar] [CrossRef]
- Tisserant, E.; Kohler, A.; Dozolme-Seddas, P.; Balestrini, R.; Benabdellah, K.; Colard, A.; Croll, D.; Da Silva, C.; Gomez, S.K.; Koul, R.; et al. The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol. 2012, 193(3), 755–769. [Google Scholar] [CrossRef]
- Corradi, N.; Brachmann, A. Fungal mating in the most widespread plant symbionts? Trends Plant Sci. 2017, 22, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Formey, D.; Molès, M.; Haouy, A.; Savelli, B.; Bouchez, O.; Bécard, G.; Roux, C. Comparative analysis of mitochondrial genomes of Rhizophagus irregularis – syn. Glomus irregulare – reveals a polymorphism induced by variability generating elements. New Phytol. 2012, 196(4), 1217–1227. [Google Scholar] [CrossRef]
- Sieverding, E.; Oehl, F. Revision of Entrophospora and description of Kuklospora and Intraspora, two new genera in the arbuscular mycorrhizal Glomeromycetes. J. Appl. Bot. Food Qual. 2006, 80(1), 69–81. [Google Scholar]
- Zhang, M.; Shi, Z.; Yang, M.; Lu, S.; Cao, L.; Wang, X. Molecular diversity and distribution of arbuscular mycorrhizal fungi at different elevations in Mt. Taibai of Qinling mountain. Front. Microbiol. 2021, 12, 609386. [Google Scholar] [CrossRef]
- Anderson, R.S.; Homola, R.L.; Davis, R.B.; Jacobson, Jr.G.L. Fossil remains of the mycorrhizal fungal Glomus fasciculatum complex in postglacial lake sediments from Maine. Rev. Can. Bot. 1984, 62, 2325–2328. [Google Scholar] [CrossRef]
- Miehe, G.; Miehe, S.; Kaiser, K.; Reudenbach, C.; Behrendes, L.; La, D.; Schlütz, F. How old is pastoralism in Tibet? An ecological approach to the making of a Tibetan landscape. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2009, 276(1-4), 130–147. [CrossRef]
- Ghosh, R.; Paruya, D.K.; Acharya, K.; Ghorai, N.; Bera, S. How reliable are non-pollen palynomorphs in tracing vegetation changes and grazing activities? Study from the Darjeeling Himalaya, India. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2017, 475, 23–40. [Google Scholar] [CrossRef]
- Gou, H.; Wei, H.; Duan, R.; Tianyuan, C. Spatial distribution of modern pollen and fungal spores and their ecological indication in Qinghai Lake on northeastern Tibetan Plateau, China. Ecol. Indic. 2022, 144(19), 109474. [Google Scholar] [CrossRef]
- Guo, Y.N.; Zhang, H.D.; Bao, Y.Y.; Tan, H.Z.; Liu, X.H.; Rahman, Z.U. Distribution characteristics of soil AM fungi community in soft sandstone area. J. Environ. Manag. 2022, 316, 115193. [Google Scholar] [CrossRef]
- Selivanov, I.A. Mikosimbiotrofizm kak forma konsortivnykh svyazei v rastitel’nom pokrove Sovetskogo Soyuza (Mycosymbiotrophism as a Form of Consortive Relations in Vegetation Cover of Soviet Union); Nauka: Moscow, Russia, 1981. [Google Scholar]
- Lavrenov, N.G.; Zernov, A.S.; Kipkeev, A.M.; Tekeev, D.K.; Semenova, R.B.; Akhmetzhanova, A.A.; Perevedentseva, L.G.; Sudzilovskaya, N.A.; Korneecheva, M.Yu.; Onipchenko, V.G. Plant mycorrhiza under extreme conditions of snow beds Alpine communities in Armenia. Biol. Bull. Rev. 2018, 8(5), 401–405. [Google Scholar] [CrossRef]
- Lugo, M.A.; Ferrero, M.; Menoyo, E.; Estévez, M.C.; Siñeriz, F.; Anton, A. Arbuscular mycorrhizal fungi and rhizospheric bacteria diversity along an altitudinal gradient in South American Puna grassland. Microb. Ecol. 2008, 55, 705–713. [Google Scholar] [CrossRef]
- Lugo, M.A.; Negritto, M.A.; Jofré, M.; Anton, A.; Galetto, L. Colonization of native Andean grasses by arbuscular mycorrhizal fungi in puna: a matter of altitude, host photosynthetic pathway and host life cycles. FEMS Microbiol. Ecol. 2012, 81, 455–466. [Google Scholar] [CrossRef]
- Kernaghan, G.; Harper, K.A. Community structure of ectomycorrhizal fungi across an alpine/subalpine ecotone Ecography 2001, 24(2), 181–188. [CrossRef]
- De Mesquita, C.P.B.; Sartwell, S.A.; Ordemann, E.V.; Porazinska, D.L.; Farrer, E.C.; King, A.J.; Spasojevic, M.J.; Smith, J.G.; Suding, K.N.; Schmidt, S.K. Patterns of root colonization by arbuscular mycorrhizal fungi and dark septate endophytes across a mostly-unvegetated, high-elevation landscape. Fungal Ecol. 2018, 36, 63–74. [Google Scholar] [CrossRef]
- Nozadze, L.M. Mycosymbiotrophism of herbaceous plants in some plant communities of the Inguri River basin related to vertical zonality. In Mikoriza i drugie formy konsortativnykh otnoshenii v prirode (Mycorrhiza and Other Forms of Consortive Relations in Nature); Permsk. Gos. Pedagog. Inst.: Perm, Russia, 1981; pp. 50–52. [Google Scholar]
- Hempel, S.; Gotzenberger, L.; Kuhn, I.; Michalski, S.G.; Rillig, M.C.; Zobel, M.; Moora, M. Mycorrhizas in the Central European flora: relationships with plant life history traits and ecology. Ecology 2013, 94(6), 1389–1399. [Google Scholar] [CrossRef]
- Soudzilovskaia, N.A.; Douma, J.C.; Akhmetzhanova, A.A.; van Bodegom, P.M.; Cornwell, W.C.; Moens, E.; Treseder, K.; Tibbett, M.; Wang, Y.; Cornelissen, J. Global patterns of plant root colonization intensity by mycorrhizal fungi explained by climate and soil chemistry. Global Ecol. Biogeogr. 2015, 24(3), 371–382. [Google Scholar] [CrossRef]
- Lenoir, I.; Fontaine, J.; Sahraoui, A.L. Arbuscular mycorrhizal fungal responses to abiotic stresses: a review. Phytochemistry 2016, 1(123), 4–15. [Google Scholar] [CrossRef]
- Dumbrell, A.J.; Nelson, M.; Helgason, T.; Dytham, C.; Fitter, A.H. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 2010, 4, 337–345. [Google Scholar] [CrossRef]
- López-Angulo, J.; Pescador, D.S.; Sánchez, A.M.; Luzuriaga, A.L.; Cavieres, L.A.; Escudero, A. Impacts of climate, soil and biotic interactions on the interplay of the different facets of alpine plant diversity. Sci. Total Environ. 2020, 698, 133960. [Google Scholar] [CrossRef]
- Makarov, M.I. The role of mycorrhiza in transformation of nitrogene compounds in soil and nitrogene nutrition of plant: a review. Eurasian Soil Sci. 2019, 52(2), 193–205. [Google Scholar] [CrossRef]
- Soteras, F.; Menoyo, E.; Grilli, G.; Becerra, A.G. Arbuscular mycorrhizal fungal communities of high mountain ecosystems of South America: relationship with microscale and macroscale factors. In Mycorrhizal Fungi in South America; Springer: Cham, Switzerland, 2019; pp. 257–275. [Google Scholar]
- Farish, N.R.; Lavrenov, N.G. Statistical analysis of the mycorrhizae impact on communities of alpine maedows. Reports of Int. Conf. “Mathematical Biology AND BioInformatics”; V.D. Lahno; IMPB RAN: Puschino, Russia, 2018; 7, e102. [CrossRef]
- Sigüenza, C.; Espejel, I.; Allen, E. Seasonality of mycorrhizae in coastal sand dunes of Baja California. Mycorrhiza 1996, 6, 151–157. [Google Scholar] [CrossRef]
- Liu, M.; Yue, Y.J.; Wang, Z.H.; Li, L.; Duan, G.Z.; Bai, S.L.; Li, T. Composition of the arbuscular mycorrhizal fungal community and changes in diversity of the rhizosphere of Clematis fruticosa over three seasons across different elevations. Eur. J. Soil Sci. 2020, 71, 511–523. [Google Scholar] [CrossRef]
- Berruti, A.; Lumini, E.; Balestrini, R.; Bianciotto, V. Arbuscular mycorrhizal fungi as natural biofertilizers: let's benefit from past successes. Front. Microbiol. 2016, 6, 1559. [Google Scholar] [CrossRef]
- Pellegrino, E.; Piazza, G.; Arduini, I.; Ercoli, L. Field inoculation of bread wheat with Rhizophagus irregularis under organic farming: variability in growth response and nutritional uptake of eleven old genotypes and a modern variety. Agronomy 2020, 10(3), 333. [Google Scholar] [CrossRef]
- Wang, J.; Fu, W.; Sun, C.; Cai, S.; Tang, C. Funneliformis mosseae inoculation enhances Cucurbita pepo L. plant growth and fruit yield by reshaping rhizosphere microbial community structure. Diversity 2022, 14(11), 932. [Google Scholar] [CrossRef]
- Veresoglou, S.D.; Shaw, L.J.; Sen, R. Glomus intraradices and Gigaspora margarita arbuscular mycorrhizal associations differentially affect nitrogen and potassium nutrition of Plantago lanceolata in a low fertility dune soil. Plant Soil 2011, 340, 481–490. [Google Scholar] [CrossRef]
- Kaur, S.; Campbell, B.J.; Suseela, V. Root metabolome of plant-arbuscular mycorrhizal symbiosis mirrors the mutualistic or parasitic mycorrhizal phenotype. New Phytol. 2022, 234, 672–687. [Google Scholar] [CrossRef]
- Edlinger, A.; Garland, G.; Hartman, K.; Banerjee, S.; Degrune, F.; García-Palacios, P.; Hallin, S.; Valzano-Held, A.; Herzog, C.; Jansa, J.; Kost, E.; Maestre, F.T.; Pescador, D.S.; Philippot, L.; Rillig, M.C.; Romdhane, S.; Saghaï, A.; Spor, A.; Frossard, E.; van der Heijden, M.G.A. Agricultural management and pesticide use reduce the functioning of beneficial plant symbionts. Nat Ecol Evol. 2022, 6(8), 1145–1154. [Google Scholar] [CrossRef]
- Kakouridis, A.; Hagen, J.A.; Kan, M.P.; Mambelli, S.; Feldman, L.J.; Herman, D.J.; Weber, P.K.; Pett-Ridge, J.; Firestone, M.K. Routes to roots: direct evidence of water transport by arbuscular mycorrhizal fungi to host plants. New Phytol. 2022, 236(1), 210–221. [Google Scholar] [CrossRef]
- Bioproject PRJNA646244 in GenBank NCBI. 2023. Available online: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA646244 (accessed on 30 October 2023).
- Thiéry, O.; Moora, M.; Vasar, M.; Zobel, M.; Öpik, M. Inter- and intrasporal nuclear ribosomal gene sequence variation within one isolate of arbuscular mycorrhizal fungus, Diversispora sp. Symbiosis. 2012, 58(1-3), 135-147. [CrossRef]
- Warton, D.I.; Wright, T.W.; Wang, Y. Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol Evol. 2012, 3, 89–101. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.; Blanchet, F.; Kindt, R.; Legendre, P.; Minchin, P.; O'Hara, R.; Solymos, P.; Stevens, M.; Szoecs, E.; et al. _vegan: Community Ecology Package_. R package version 2.6-4. Available online: https://CRAN.R-project.org/package=vegan, last updated 11 Oct 2022 (accessed on 30 November 2023).
- Ma, X.; Xu, X.; Geng, Q.; Luo, Y.; Ju, C.; Li, Q.; Zhou, Y. Global arbuscular mycorrhizal fungal diversity and abundance decreases with soil available phosphorus. Glob Ecol Biogeogr. 2023, 32(8), 1423–1434. [Google Scholar] [CrossRef]
- Wang, G.M. Effects of pH on arbuscular mycorrhiza I. Field observations on the long-term liming experiments at Rothamsted and Woburn. New Phytol. 1993. 124, 465–472. [CrossRef]
- Coughlan, A.P. , Dalpé, Y., Lapointe, L., Piché, Y. Soil pH-induced changes in root colonization, diversity, and reproduction of symbiotic arbuscular mycorrhizal fungi from healthy and declining maple forests. CJFR 2000. 30, 1543–1554. [CrossRef]
| STP Number | Stationary Trial Plot | Coordinates | Altitude, m | Type of Soil | Soil Profile |
|---|---|---|---|---|---|
| 1 | Subalpine Meadow-4, Malaya Hatipara ridge | 43°25'50.0"N 41°42'20.0"E | 2437 | ID 199 Mountain-meadow sod-peatyWRB, 2006. Umbric LeptosolsFAO, 1988. Umbric Leptosols | О1/А1v-А1Вр-ВСр-Ср |
| 3 | Subalpine Meadow-3, Malaya Hatipara ridge | 43°25'48.0"N 41°42'31.0"E | 2401 | ID 200 Mountain-meadow soddyWRB, 2006. Umbric LeptosolsFAO, 1988. Umbric Leptosols | A1-A2-B |
| 4 | Subalpine Meadow-2, Malaya Hatipara ridge | 43°25'51.0"N 41°42'55.0"E | 2186 | –//– | –//– |
| 7 | Fir Forest-3, Malaya Hatipara mountain | 43°26'07.3"N 41°43'14.1"E | 1900 | ID 68 Brownzems raw-humic illuvial-humicWRB, 2006. Haplic CambisolsFAO, 1988. Dystric Cambisols | О(АО)-А1-А1А2-Вm,f,h(Bh,m)-С |
| 8 | Pine Forest-3, Malaya Hatipara mountain | 43°26'07.3"N 41°43'14.1"E | 1890 | –//– | –//– |
| 9 | Mixed forest near the Bolshaya Hatipara river, Bolshaya Hatipara mountain | 43°24'56.0"N 41°42'49.0"E | 1507 | –//– | –//– |
| 11 | Grassland in the valley of the Teberda river, Teberda town | 43°25'12.0"N 41°43'45.0"E | 1342 | ID 191 Alluvials compactWRB, 2006. Gleyic VertisolsFAO, 1988. Eutric Vertisols | A1v-A1-Bve-BC-C |
| 12 | Grassland in the valley of the Teberda river, the border of the New Teberda village | 43°39'37.0"N 41°53'12.0"E | 1026 | ID 188 Alluvials saturatedWRB, 2006. Haplic FluvisolsFAO, 1988. Eutric Fluvisols | A1-AB-B-BC-D |
| 13 | Grassland in the valley of the Kuban river, Ordzhonikidzevsky village | 43°51'38.0"N 41°54'22.0"E | 795 | –//– | –//– |
| Analyzed | Subalpine Meadow | Forest | River Valley | Total for 9 STPs | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Parameters | STP 1 | STP 3 | STP 4 | STP 7 | STP 8 | STP 9 | STP 11 | STP 12 | STP 13 | |
| Total reads | 1494700 | 619855 | 1648642 | 652998 | 1469158 | 970340 | 1108444 | 1271980 | 862545 | 10 098 662 |
| Merged reads after length trim | 401850 | 255057 | 626988 | 102472 | 586935 | 539334 | 363547 | 160449 | 347435 | 3 384 067 |
| Glomeromycota ITS1 total reads | 1243 | 1011 | 1116 | 183 | 76 | 697 | 6670 | 335 | 4638 | 15 969 |
| Glomeromycota ITS1 OTU number | 100 | 59 | 86 | 29 | 26 | 80 | 197 | 66 | 109 | 414 |
| 171 | 117 | 296 | ||||||||
| Glomeromycota ITS2 total reads | 546 | 493 | 580 | 7 | 24 | 583 | 1624 | 885 | 1758 | 6 500 |
| Glomeromycota ITS2 OTU number | 72 | 49 | 74 | 3 | 10 | 58 | 128 | 93 | 76 | 305 |
| 131 | 60 | 221 | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





