Submitted:
11 December 2023
Posted:
11 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Tumor Samples
2.2. Cell Lines
2.3. RNA Isolation from FFPE Surgical Specimens
2.4. MicroRNA Expression Profiling
Array Card Expression Analysis for 754 miRNAs
2.5. Single microRNA Expression Assays
2.6. miR-221 and miR-483-3p Mimic Transfection
2.7. Bulk RNA-Sequencing (RNA-seq) in Transfected OE-19 Cells
2.8. Data Analysis
3. Results
3.1. Discovery Dataset: Identification of Deregulated miRNAs in EAC
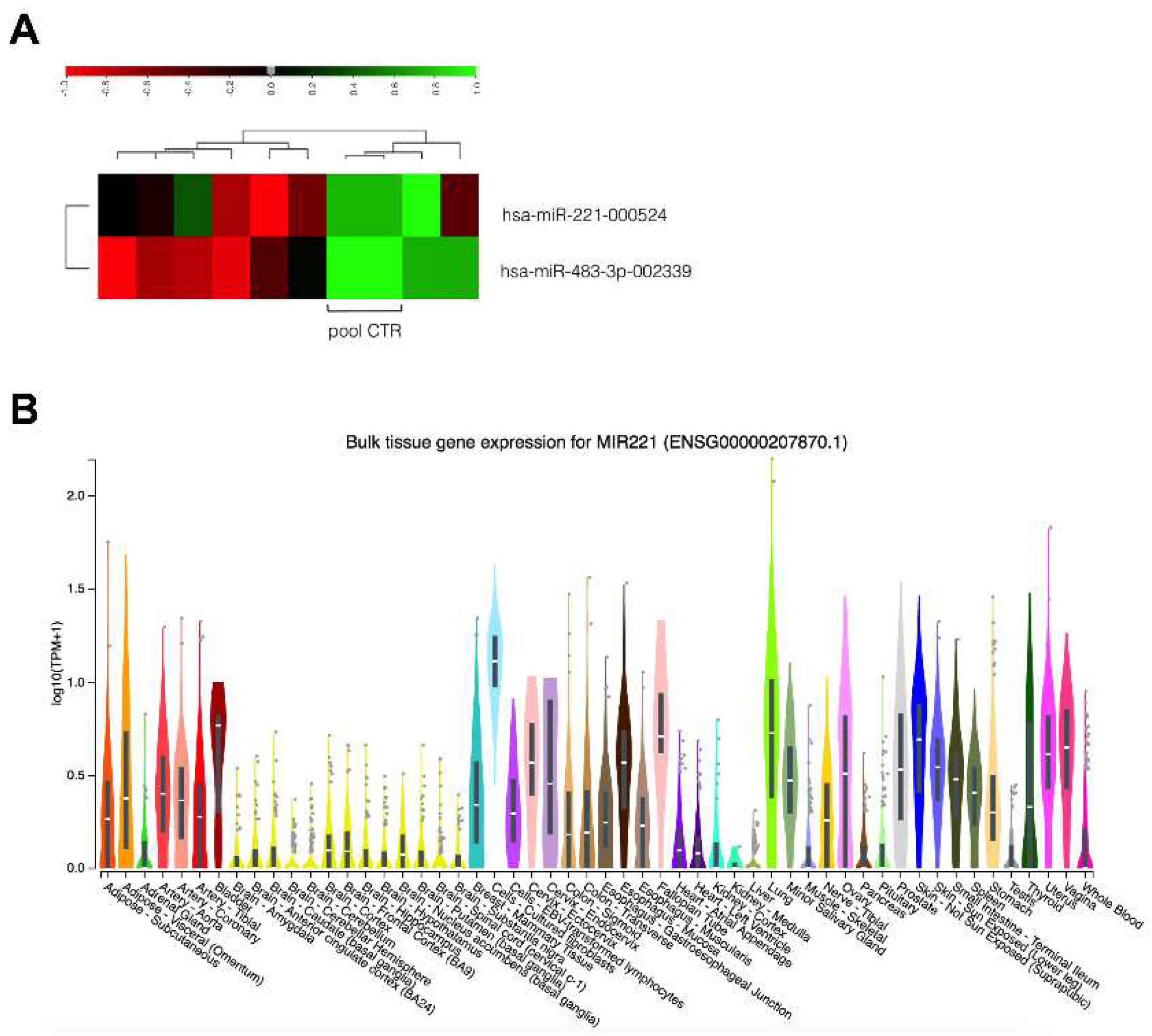
3.2. Replication Dataset (EACGSE Cohort): Single miRNA Analysis
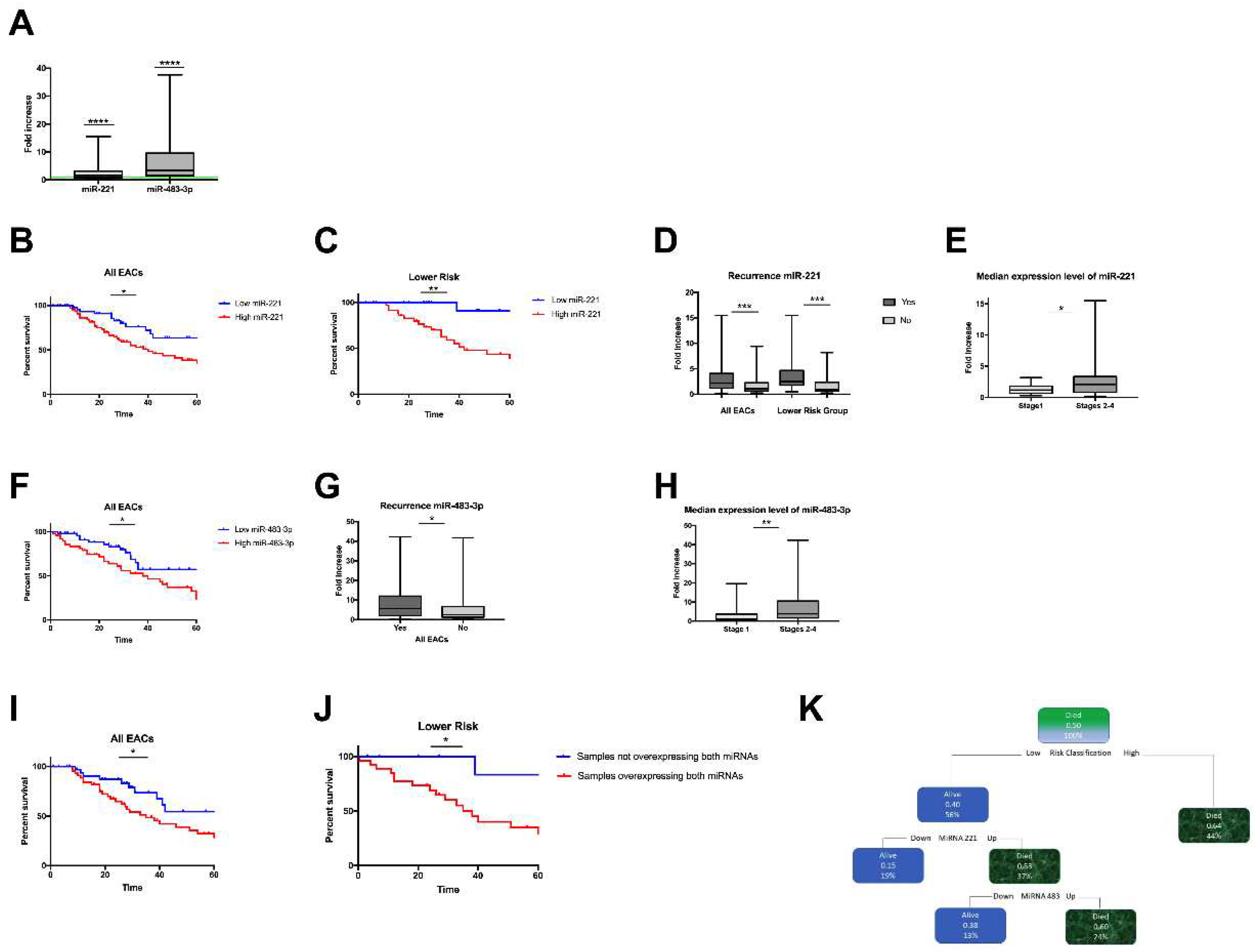
3.3. Correlation between miRNA-221 Expression and EAC Clinicopathological Features
3.4. Correlation between miRNA-483-3p Expression and EAC Clinicopathological Features
3.5. Concurrent miRNA-221 and 483-3p Overexpression is Correlated with Poor Survival
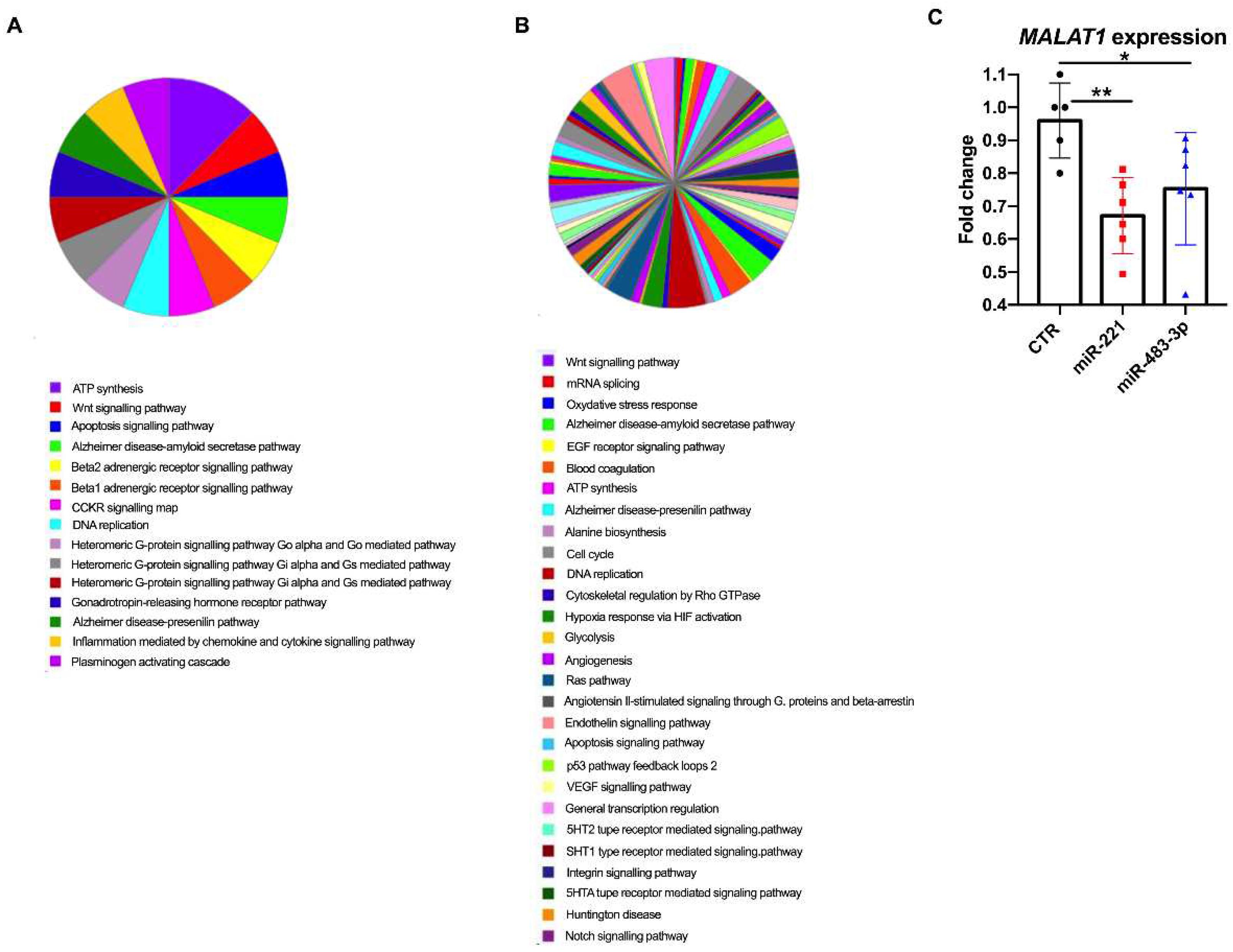
3.6. miRNA Overexpression and Transcriptome Analysis In Vitro
4. Discussion
Supplementary Materials
Acknowledgments
References
- Dubecz, A.; Gall, I.; Solymosi, N.; Schweigert, M.; Peters, J. H.; Feith, M.; Stein, H. J. Temporal Trends in Long-Term Survival and Cure Rates in Esophageal Cancer: A SEER Database Analysis. J Thorac Oncol 2012, 7 (2), 443–447. [CrossRef]
- Velanovich, V.; Hollingsworth, J.; Suresh, P.; Ben-Menachem, T. Relationship of Gastroesophageal Reflux Disease with Adenocarcinoma of the Distal Esophagus and Cardia. Dig Surg 2002, 19 (5), 349–353. [CrossRef]
- Curtius, K.; Rubenstein, J. H.; Chak, A.; Inadomi, J. M. Computational Modelling Suggests That Barrett’s Oesophagus May Be the Precursor of All Oesophageal Adenocarcinomas. Gut 2020, 70 (8), 1435–1440. [CrossRef]
- Rice, T. W.; Ishwaran, H.; Ferguson, M. K.; Blackstone, E. H.; Goldstraw, P. Cancer of the Esophagus and Esophagogastric Junction: An Eighth Edition Staging Primer. J Thorac Oncol 2017, 12 (1), 36–42. [CrossRef]
- Mattioli, S.; Ruffato, A.; Di Simone, M. P.; Corti, B.; D’Errico, A.; Lugaresi, M. L.; Mattioli, B.; D’Ovidio, F. Immunopathological Patterns of the Stomach in Adenocarcinoma of the Esophagus, Cardia, and Gastric Antrum: Gastric Profiles in Siewert Type I and II Tumors. Ann Thorac Surg 2007, 83 (5), 1814–1819. [CrossRef]
- Ruffato, A.; Mattioli, S.; Perrone, O.; Lugaresi, M.; Di Simone, M. P.; D’Errico, A.; Malvi, D.; Aprile, M. R.; Raulli, G.; Frassineti, L. Esophagogastric Metaplasia Relates to Nodal Metastases in Adenocarcinoma of Esophagus and Cardia. Ann Thorac Surg 2013, 95 (4), 1147–1153. [CrossRef]
- Al-Batran, S.-E.; Hofheinz, R. D.; Pauligk, C.; Kopp, H.-G.; Haag, G. M.; Luley, K. B.; Meiler, J.; Homann, N.; Lorenzen, S.; Schmalenberg, H.; Probst, S.; Koenigsmann, M.; Egger, M.; Prasnikar, N.; Caca, K.; Trojan, J.; Martens, U. M.; Block, A.; Fischbach, W.; Mahlberg, R.; Clemens, M.; Illerhaus, G.; Zirlik, K.; Behringer, D. M.; Schmiegel, W.; Pohl, M.; Heike, M.; Ronellenfitsch, U.; Schuler, M.; Bechstein, W. O.; Königsrainer, A.; Gaiser, T.; Schirmacher, P.; Hozaeel, W.; Reichart, A.; Goetze, T. O.; Sievert, M.; Jäger, E.; Mönig, S.; Tannapfel, A. Histopathological Regression after Neoadjuvant Docetaxel, Oxaliplatin, Fluorouracil, and Leucovorin versus Epirubicin, Cisplatin, and Fluorouracil or Capecitabine in Patients with Resectable Gastric or Gastro-Oesophageal Junction Adenocarcinoma (FLOT4-AIO): Results from the Phase 2 Part of a Multicentre, Open-Label, Randomized Phase 2/3 Trial. Lancet Oncol 2016, 17 (12), 1697–1708. [CrossRef]
- van der Kaaij, R. T.; Snaebjornsson, P.; Voncken, F. E. M.; van Dieren, J. M.; Jansen, E. P. M.; Sikorska, K.; Cats, A.; van Sandick, J. W. The Prognostic and Potentially Predictive Value of the Laurén Classification in Oesophageal Adenocarcinoma. Eur J Cancer 2017, 76, 27–35. [CrossRef]
- Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Oesophageal Carcinoma. Nature 2017, 541 (7636), 169–175. [CrossRef]
- Secrier, M.; Li, X.; de Silva, N.; Eldridge, M. D.; Contino, G.; Bornschein, J.; MacRae, S.; Grehan, N.; O’Donovan, M.; Miremadi, A.; Yang, T.-P.; Bower, L.; Chettouh, H.; Crawte, J.; Galeano-Dalmau, N.; Grabowska, A.; Saunders, J.; Underwood, T.; Waddell, N.; Barbour, A. P.; Nutzinger, B.; Achilleos, A.; Edwards, P. A. W.; Lynch, A. G.; Tavaré, S.; Fitzgerald, R. C.; Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) Consortium. Mutational Signatures in Esophageal Adenocarcinoma Define Etiologically Distinct Subgroups with Therapeutic Relevance. Nat Genet 2016, 48 (10), 1131–1141. [CrossRef]
- Isidori, F.; Bozzarelli, I.; Mastracci, L.; Malvi, D.; Lugaresi, M.; Molinari, C.; Söderström, H.; Räsänen, J.; D’Errico, A.; Fiocca, R.; Seri, M.; Krishnadath, K. K.; Bonora, E.; Mattioli, S. Targeted Sequencing of Sorted Esophageal Adenocarcinoma Cells Unveils Known and Novel Mutations in the Separated Subpopulations. Clin Transl Gastroenterol 2020, 11 (9), e00202. [CrossRef]
- Bornschein, J.; Wernisch, L.; Secrier, M.; Miremadi, A.; Perner, J.; MacRae, S.; O’Donovan, M.; Newton, R.; Menon, S.; Bower, L.; Eldridge, M. D.; Devonshire, G.; Cheah, C.; Turkington, R.; Hardwick, R. H.; Selgrad, M.; Venerito, M.; Malfertheiner, P.; OCCAMS Consortium; Fitzgerald, R. C. Transcriptomic Profiling Reveals Three Molecular Phenotypes of Adenocarcinoma at the Gastroesophageal Junction. Int J Cancer 2019, 145 (12), 3389–3401. [CrossRef]
- Jammula, S.; Katz-Summercorn, A. C.; Li, X.; Linossi, C.; Smyth, E.; Killcoyne, S.; Biasci, D.; Subash, V. V.; Abbas, S.; Blasko, A.; Devonshire, G.; Grantham, A.; Wronowski, F.; O’Donovan, M.; Grehan, N.; Eldridge, M. D.; Tavaré, S.; Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) consortium; Fitzgerald, R. C. Identification of Subtypes of Barrett’s Esophagus and Esophageal Adenocarcinoma Based on DNA Methylation Profiles and Integration of Transcriptome and Genome Data. Gastroenterology 2020, 158 (6), 1682-1697.e1. [CrossRef]
- Antonowicz, S.; Bodai, Z.; Wiggins, T.; Markar, S. R.; Boshier, P. R.; Goh, Y. M.; Adam, M. E.; Lu, H.; Kudo, H.; Rosini, F.; Goldin, R.; Moralli, D.; Green, C. M.; Peters, C. J.; Habib, N.; Gabra, H.; Fitzgerald, R. C.; Takats, Z.; Hanna, G. B. Endogenous Aldehyde Accumulation Generates Genotoxicity and Exhaled Biomarkers in Esophageal Adenocarcinoma. Nat Commun 2021, 12 (1), 1454. [CrossRef]
- Bartel, D. P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116 (2), 281–297. [CrossRef]
- Macfarlane, L.-A.; Murphy, P. R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics 2010, 11 (7), 537–561. [CrossRef]
- Di Leva, G.; Croce, C. M. Roles of Small RNAs in Tumor Formation. Trends Mol Med 2010, 16 (6), 257–267. [CrossRef]
- Shah, M. Y.; Calin, G. A. MicroRNAs as Therapeutic Targets in Human Cancers. Wiley Interdiscip Rev RNA 2014, 5 (4), 537–548. [CrossRef]
- Feber, A.; Xi, L.; Luketich, J. D.; Pennathur, A.; Landreneau, R. J.; Wu, M.; Swanson, S. J.; Godfrey, T. E.; Litle, V. R. MicroRNA Expression Profiles of Esophageal Cancer. J Thorac Cardiovasc Surg 2008, 135 (2), 255–260; discussion 260. [CrossRef]
- Gu, J.; Wang, Y.; Wu, X. MicroRNA in the Pathogenesis and Prognosis of Esophageal Cancer. Curr Pharm Des 2013, 19 (7), 1292–1300. [CrossRef]
- Gao, S.; Zhao, Z.-Y.; Zhang, Z.-Y.; Zhang, Y.; Wu, R. Prognostic Value of MicroRNAs in Esophageal Carcinoma: A Meta-Analysis. Clin Transl Gastroenterol 2018, 9 (11), 203. [CrossRef]
- Smith, C.-M.; Watson, D. I.; Michael, M. Z.; Hussey, D. J. MicroRNAs, Development of Barrett’s Esophagus, and Progression to Esophageal Adenocarcinoma. World J Gastroenterol 2010, 16 (5), 531–537. [CrossRef]
- Revilla-Nuin, B.; Parrilla, P.; Lozano, J. J.; de Haro, L. F. M.; Ortiz, A.; Martínez, C.; Munitiz, V.; de Angulo, D. R.; Bermejo, J.; Molina, J.; Cayuela, M. L.; Yélamos, J. Predictive Value of MicroRNAs in the Progression of Barrett Esophagus to Adenocarcinoma in a Long-Term Follow-up Study. Ann Surg 2013, 257 (5), 886–893. [CrossRef]
- 24. Orsini A, Mastracci L, Bozzarelli I, Ferrari A, Isidori F, Fiocca R, Lugaresi M, D'Errico A, Malvi D, Cataldi-Stagetti E, Spaggiari P, Tomezzoli A, Albarello L, Ristimäki A, Bottiglieri L, Krishnadath KK, Rosati R, Fumagalli Romario U, De Manzoni G, Räsänen J, Martinelli G, Mattioli S, Bonora E, On Behalf Of The Eacsge Consortium. Correlations between Molecular Alterations, Histopathological Characteristics, and Poor Prognosis in Esophageal Adenocarcinoma. Cancers (Basel). 2023, 15(5):1408. doi: 10.3390/cancers15051408.
- Fiocca, R.; Mastracci, L.; Lugaresi, M.; Grillo, F.; D’Errico, A.; Malvi, D.; Spaggiari, P.; Tomezzoli, A.; Albarello, L.; Ristimäki, A.; Bottiglieri, L.; Bonora, E.; Krishnadath, K. K.; Raulli, G. D.; Rosati, R.; Fumagalli Romario, U.; De Manzoni, G.; Räsänen, J.; Mattioli, S. The Prognostic Impact of Histology in Esophageal and Esophago-Gastric Junction Adenocarcinoma. Cancers (Basel) 2021, 13 (20), 5211. [CrossRef]
- Rockett, J. C.; Larkin, K.; Darnton, S. J.; Morris, A. G.; Matthews, H. R. Five Newly Established Oesophageal Carcinoma Cell Lines: Phenotypic and Immunological Characterization. Br J Cancer 1997, 75 (2), 258–263. [CrossRef]
- Babraham, B. FastQC A Quality Control Tool for High Throughput Sequence Data. Accessed July 4, 2023. Https://Www.Bioinformatics.Babraham.Ac.Uk/Projects/Fastqc/.
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32 (19), 3047–3048. [CrossRef]
- Dobin, A.; Davis, C. A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T. R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29 (1), 15–21. [CrossRef]
- Danecek, P.; Bonfield, J. K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M. O.; Whitwham, A.; Keane, T.; McCarthy, S. A.; Davies, R. M.; Li, H. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10 (2), giab008. [CrossRef]
- Putri, G. H.; Anders, S.; Pyl, P. T.; Pimanda, J. E.; Zanini, F. Analysing High-Throughput Sequencing Data in Python with HTSeq 2.0. Bioinformatics 2022, 38 (10), 2943–2945. [CrossRef]
- Ritchie, M. E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C. W.; Shi, W.; Smyth, G. K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res 2015, 43 (7), e47. [CrossRef]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P. D. PANTHER Version 11: Expanded Annotation Data from Gene Ontology and Reactome Pathways, and Data Analysis Tool Enhancements. Nucleic Acids Res 2017, 45 (D1), D183–D189. [CrossRef]
- Boonstra, J. J.; van Marion, R.; Beer, D. G.; Lin, L.; Chaves, P.; Ribeiro, C.; Pereira, A. D.; Roque, L.; Darnton, S. J.; Altorki, N. K.; Schrump, D. S.; Klimstra, D. S.; Tang, L. H.; Eshleman, J. R.; Alvarez, H.; Shimada, Y.; van Dekken, H.; Tilanus, H. W.; Dinjens, W. N. M. Verification and Unmasking of Widely Used Human Esophageal Adenocarcinoma Cell Lines. J Natl Cancer Inst 2010, 102 (4), 271–274. [CrossRef]
- Hutchinson, J. N.; Ensminger, A. W.; Clemson, C. M.; Lynch, C. R.; Lawrence, J. B.; Chess, A. A Screen for Nuclear Transcripts Identifies Two Linked Noncoding RNAs Associated with SC35 Splicing Domains. BMC Genomics 2007, 8, 39. [CrossRef]
- Njei, B.; McCarty, T. R.; Birk, J. W. Trends in Esophageal Cancer Survival in United States Adults from 1973 to 2009: A SEER Database Analysis. J Gastroenterol Hepatol 2016, 31 (6), 1141–1146. [CrossRef]
- Lagergren, J.; Smyth, E.; Cunningham, D.; Lagergren, P. Oesophageal Cancer. Lancet 2017, 390 (10110), 2383–2396. [CrossRef]
- Dulak, A. M.; Stojanov, P.; Peng, S.; Lawrence, M. S.; Fox, C.; Stewart, C.; Bandla, S.; Imamura, Y.; Schumacher, S. E.; Shefler, E.; McKenna, A.; Carter, S. L.; Cibulskis, K.; Sivachenko, A.; Saksena, G.; Voet, D.; Ramos, A. H.; Auclair, D.; Thompson, K.; Sougnez, C.; Onofrio, R. C.; Guiducci, C.; Beroukhim, R.; Zhou, Z.; Lin, L.; Lin, J.; Reddy, R.; Chang, A.; Landrenau, R.; Pennathur, A.; Ogino, S.; Luketich, J. D.; Golub, T. R.; Gabriel, S. B.; Lander, E. S.; Beer, D. G.; Godfrey, T. E.; Getz, G.; Bass, A. J. Exome and Whole-Genome Sequencing of Esophageal Adenocarcinoma Identifies Recurrent Driver Events and Mutational Complexity. Nat Genet 2013, 45 (5), 478–486. [CrossRef]
- Malumbres, M.; Carnero, A. Cell Cycle Deregulation: A Common Motif in Cancer. Prog Cell Cycle Res 2003, 5, 5–18.
- Acunzo, M.; Romano, G.; Wernicke, D.; Croce, C. M. MicroRNA and Cancer--a Brief Overview. Adv Biol Regul 2015, 57, 1–9. [CrossRef]
- Santiago, K.; Chen Wongworawat, Y.; Khan, S. Differential MicroRNA-Signatures in Thyroid Cancer Subtypes. J Oncol 2020, 2020, 2052396. [CrossRef]
- Garzon, R.; Croce, C. M. MicroRNAs in Normal and Malignant Hematopoiesis. Curr Opin Hematol 2008, 15 (4), 352–358. [CrossRef]
- Gramantieri, L.; Fornari, F.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G. A.; Grazi, G. L.; Croce, C. M.; Bolondi, L.; Negrini, M. MicroRNA-221 Targets Bmf in Hepatocellular Carcinoma and Correlates with Tumor Multifocality. Clin Cancer Res 2009, 15 (16), 5073–5081. [CrossRef]
- Matsuzaki, J.; Suzuki, H.; Tsugawa, H.; Watanabe, M.; Hossain, S.; Arai, E.; Saito, Y.; Sekine, S.; Akaike, T.; Kanai, Y.; Mukaisho, K.; Auwerx, J.; Hibi, T. Bile Acids Increase Levels of microRNAs 221 and 222, Leading to Degradation of CDX2 during Esophageal Carcinogenesis. Gastroenterology 2013, 145 (6), 1300–1311. [CrossRef]
- Wang, Y.; Zhao, Y.; Herbst, A.; Kalinski, T.; Qin, J.; Wang, X.; Jiang, Z.; Benedix, F.; Franke, S.; Wartman, T.; Camaj, P.; Halangk, W.; Kolligs, F. T.; Jauch, K. W.; Nelson, P. J.; Bruns, C. J. miR-221 Mediates Chemoresistance of Esophageal Adenocarcinoma by Direct Targeting of DKK2 Expression. Ann Surg 2016, 264 (5), 804–814. [CrossRef]
- Fu, H.; Tie, Y.; Xu, C.; Zhang, Z.; Zhu, J.; Shi, Y.; Jiang, H.; Sun, Z.; Zheng, X. Identification of Human Fetal Liver miRNAs by a Novel Method. FEBS Lett 2005, 579 (17), 3849–3854. [CrossRef]
- Lapunzina, P. Risk of Tumorigenesis in Overgrowth Syndromes: A Comprehensive Review. Am J Med Genet C Semin Med Genet 2005, 137C (1), 53–71. [CrossRef]
- Pepe, F.; Visone, R.; Veronese, A. The Glucose-Regulated MiR-483-3p Influences Key Signaling Pathways in Cancer. Cancers (Basel) 2018, 10 (6), 181. [CrossRef]
- Livingstone, C. IGF2 and Cancer. Endocr Relat Cancer 2013, 20 (6), R321-339. [CrossRef]
- Rainier, S.; Johnson, L. A.; Dobry, C. J.; Ping, A. J.; Grundy, P. E.; Feinberg, A. P. Relaxation of Imprinted Genes in Human Cancer. Nature 1993, 362 (6422), 747–749. [CrossRef]
- Veronese, A.; Lupini, L.; Consiglio, J.; Visone, R.; Ferracin, M.; Fornari, F.; Zanesi, N.; Alder, H.; D’Elia, G.; Gramantieri, L.; Bolondi, L.; Lanza, G.; Querzoli, P.; Angioni, A.; Croce, C. M.; Negrini, M. Oncogenic Role of miR-483-3p at the IGF2/483 Locus. Cancer Res 2010, 70 (8), 3140–3149. [CrossRef]
- Tang, W.; Pei, M.; Li, J.; Xu, N.; Xiao, W.; Yu, Z.; Zhang, J.; Hong, L.; Guo, Z.; Lin, J.; Dai, W.; Xiao, Y.; Wu, X.; Liu, G.; Zhi, F.; Li, G.; Xiong, J.; Chen, Y.; Zhang, H.; Xiang, L.; Li, A.; Liu, S.; Wang, J. The miR-3648/FRAT1-FRAT2/c-Myc Negative Feedback Loop Modulates the Metastasis and Invasion of Gastric Cancer Cells. Oncogene 2022, 41 (43), 4823–4838. [CrossRef]
- Saitoh, T.; Katoh, M. FRAT1 and FRAT2, Clustered in Human Chromosome 10q24.1 Region, Are up-Regulated in Gastric Cancer. Int J Oncol 2001, 19 (2), 311–315. [CrossRef]
- Sari, I. N.; Yang, Y.-G.; Wijaya, Y. T.; Jun, N.; Lee, S.; Kim, K. S.; Bajaj, J.; Oehler, V. G.; Kim, S.-H.; Choi, S.-Y.; Park, S.-H.; Kim, D.-W.; Reya, T.; Han, J.; Kwon, H. Y. AMD1 Is Required for the Maintenance of Leukemic Stem Cells and Promotes Chronic Myeloid Leukemic Growth. Oncogene 2021, 40 (3), 603–617. [CrossRef]
- Gao, H.; Li, H.; Wang, J.; Xu, C.; Zhu, Y.; Tuluhong, D.; Li, X.; Wang, S.; Li, J. Polyamine Synthesis Enzyme AMD1 Is Closely Related to the Tumorigenesis and Prognosis of Human Breast Cancer. Exp Cell Res 2022, 417 (2), 113235. [CrossRef]
- Xu, L.; You, X.; Cao, Q.; Huang, M.; Hong, L.-L.; Chen, X.-L.; Lei, L.; Ling, Z.-Q.; Chen, Y. Polyamine Synthesis Enzyme AMD1 Is Closely Associated with Tumorigenesis and Prognosis of Human Gastric Cancers. Carcinogenesis 2020, 41 (2), 214–222. [CrossRef]
- Agarwal, S.; Behring, M.; Hale, K.; Al Diffalha, S.; Wang, K.; Manne, U.; Varambally, S. MTHFD1L, A Folate Cycle Enzyme, Is Involved in Progression of Colorectal Cancer. Transl Oncol 2019, 12 (11), 1461–1467. [CrossRef]
- Lee, D.; Xu, I. M.-J.; Chiu, D. K.-C.; Lai, R. K.-H.; Tse, A. P.-W.; Lan Li, L.; Law, C.-T.; Tsang, F. H.-C.; Wei, L. L.; Chan, C. Y.-K.; Wong, C.-M.; Ng, I. O.-L.; Wong, C. C.-L. Folate Cycle Enzyme MTHFD1L Confers Metabolic Advantages in Hepatocellular Carcinoma. J Clin Invest 2017, 127 (5), 1856–1872. [CrossRef]
- He, Z.; Wang, X.; Zhang, H.; Liang, B.; Zhang, J.; Zhang, Z.; Yang, Y. High Expression of Folate Cycle Enzyme MTHFD1L Correlates with Poor Prognosis and Increased Proliferation and Migration in Colorectal Cancer. J Cancer 2020, 11 (14), 4213–4221. [CrossRef]
- Yang, Y.-S.; Yuan, Y.; Hu, W.-P.; Shang, Q.-X.; Chen, L.-Q. The Role of Mitochondrial Folate Enzyme MTHFD1L in Esophageal Squamous Cell Carcinoma. Scand J Gastroenterol 2018, 53 (5), 533–540. [CrossRef]
- Tada, Y.; Yokomizo, A.; Shiota, M.; Song, Y.; Kashiwagi, E.; Kuroiwa, K.; Oda, Y.; Naito, S. Ectonucleoside Triphosphate Diphosphohydrolase 6 Expression in Testis and Testicular Cancer and Its Implication in Cisplatin Resistance. Oncol Rep 2011, 26 (1), 161–167. [CrossRef]
- Sun, Y.; Ma, L. New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers (Basel) 2019, 11 (2), 216. [CrossRef]
- Li, Z.; Zhang, Q.; Wu, Y.; Hu, F.; Gu, L.; Chen, T.; Wang, W. lncRNA Malat1 Modulates the Maturation Process, Cytokine Secretion and Apoptosis in Airway Epithelial Cell-Conditioned Dendritic Cells. Exp Ther Med 2018, 16 (5), 3951–3958. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




