Submitted:
02 July 2024
Posted:
03 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
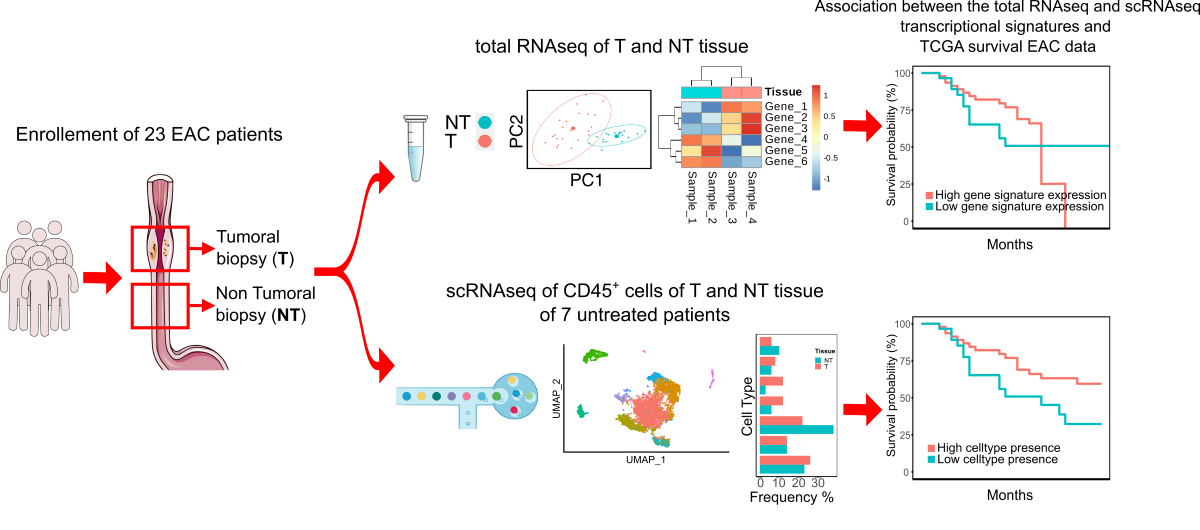
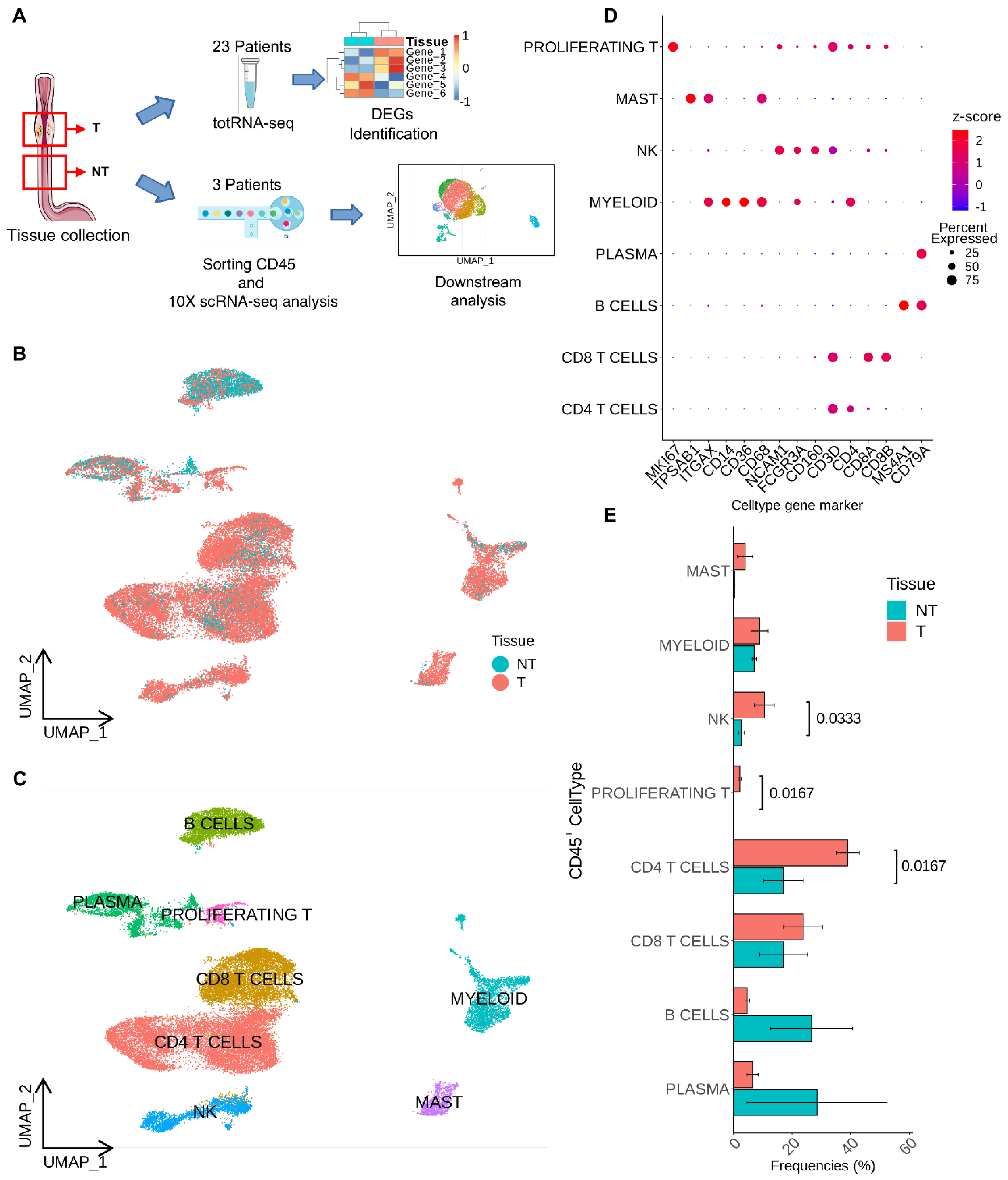
2.1. Patients’ Recruitment, Tissue Collection and Experimental Workflow
2.2. Single-Cell Sequencing: Cells’ Preparation, Library Preparation and Sequencing
2.3. Analysis of Single-cell RNA Sequencing Data
2.4. Identification of TF regulons
2.5. Polychromatic Flow Cytometry
2.6. Computational analysis of flow cytometry data
2.7. Analysis of Bulk RNA Sequencing Data
2.8. SODEGIR Analysis
2.9. Survival Analysis
3. Results
3.1. Single-Cell Level Analysis of Esophageal Adenocarcinoma Immune Infiltrate
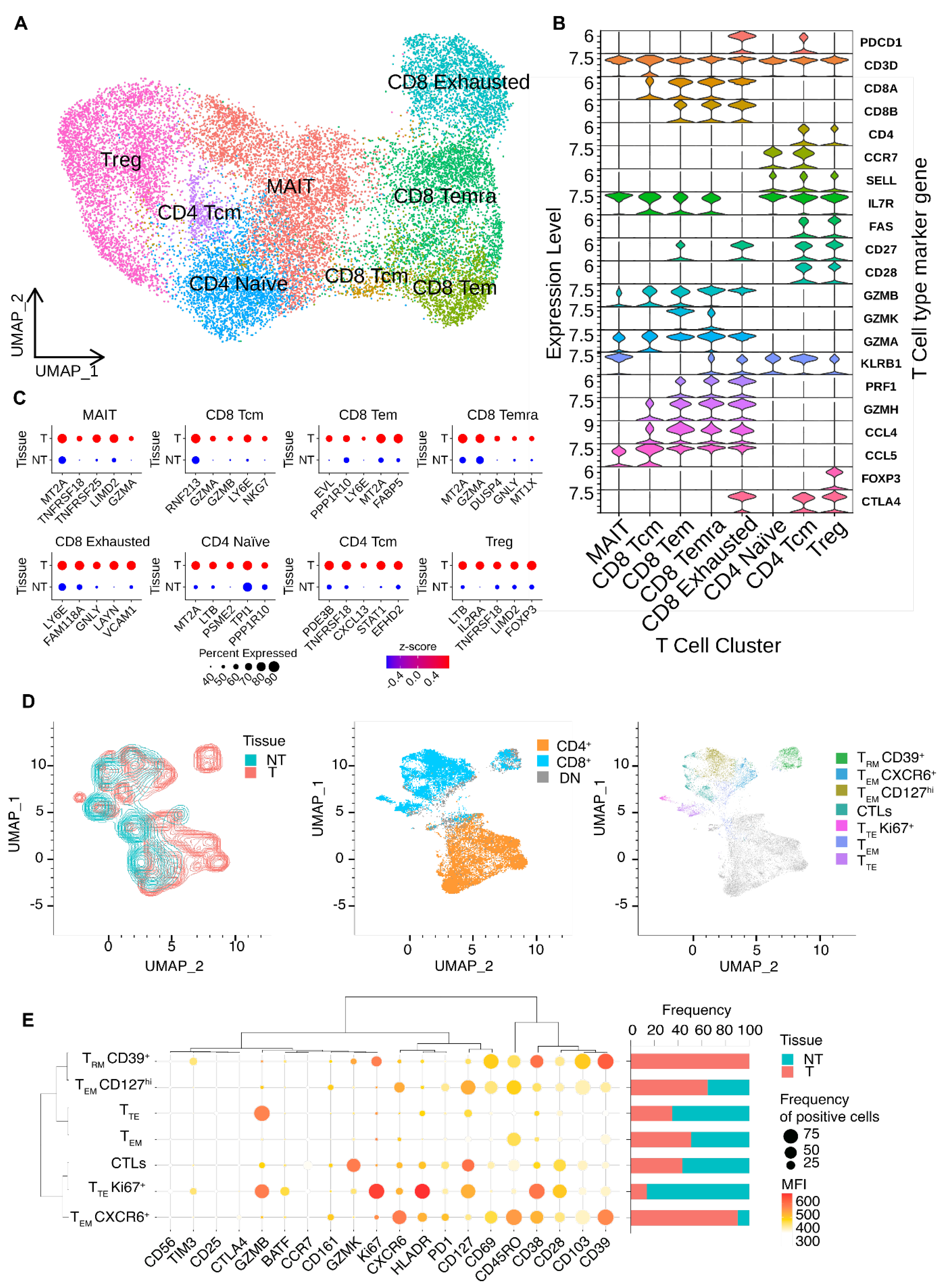
3.2. Dissection of T-cells Heterogeneity in Esophageal Adenocarcinoma
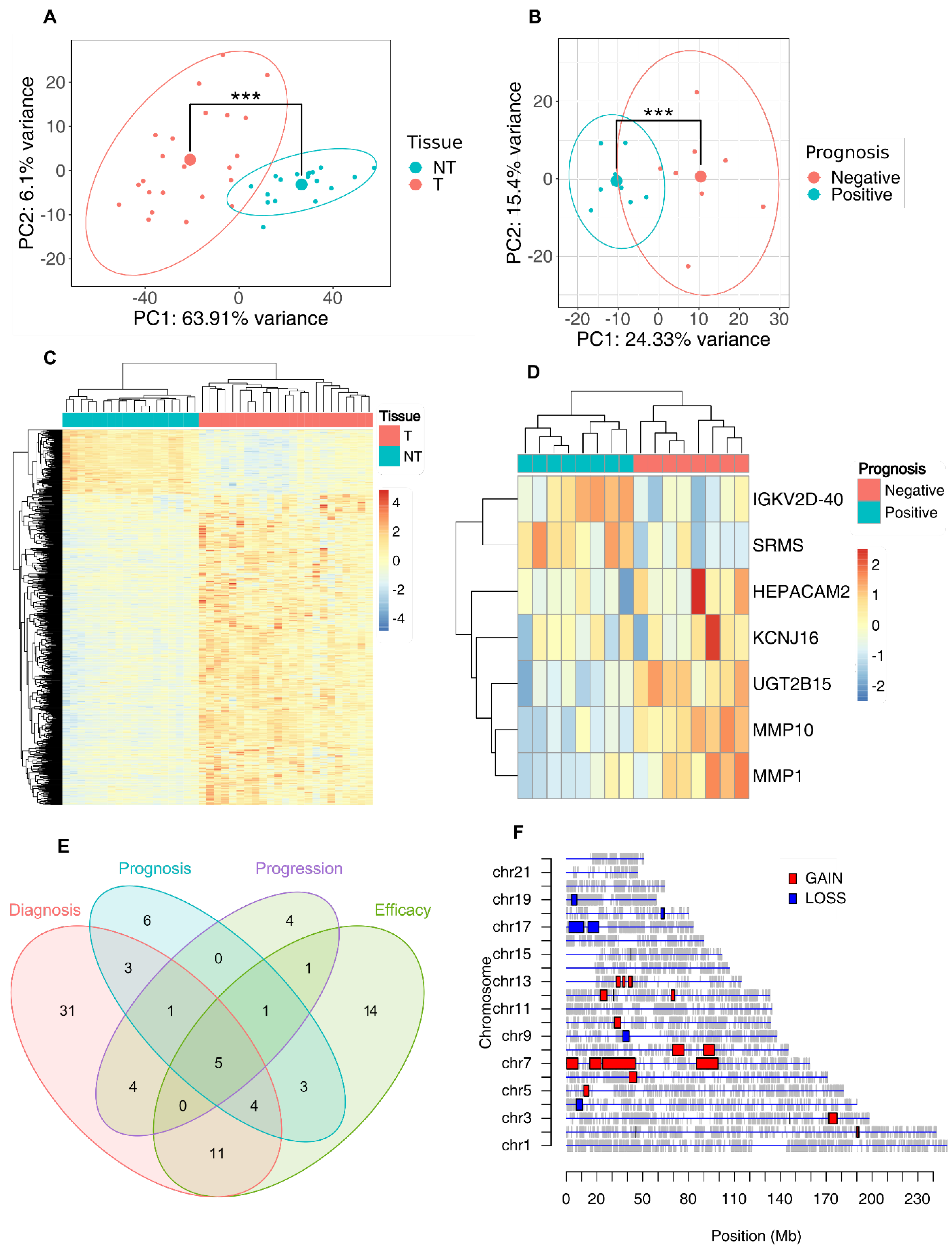
3.3. Whole Transcriptome Profiling of Esophageal Adenocarcinoma Tissues for the Identification of a Prognostic Signature
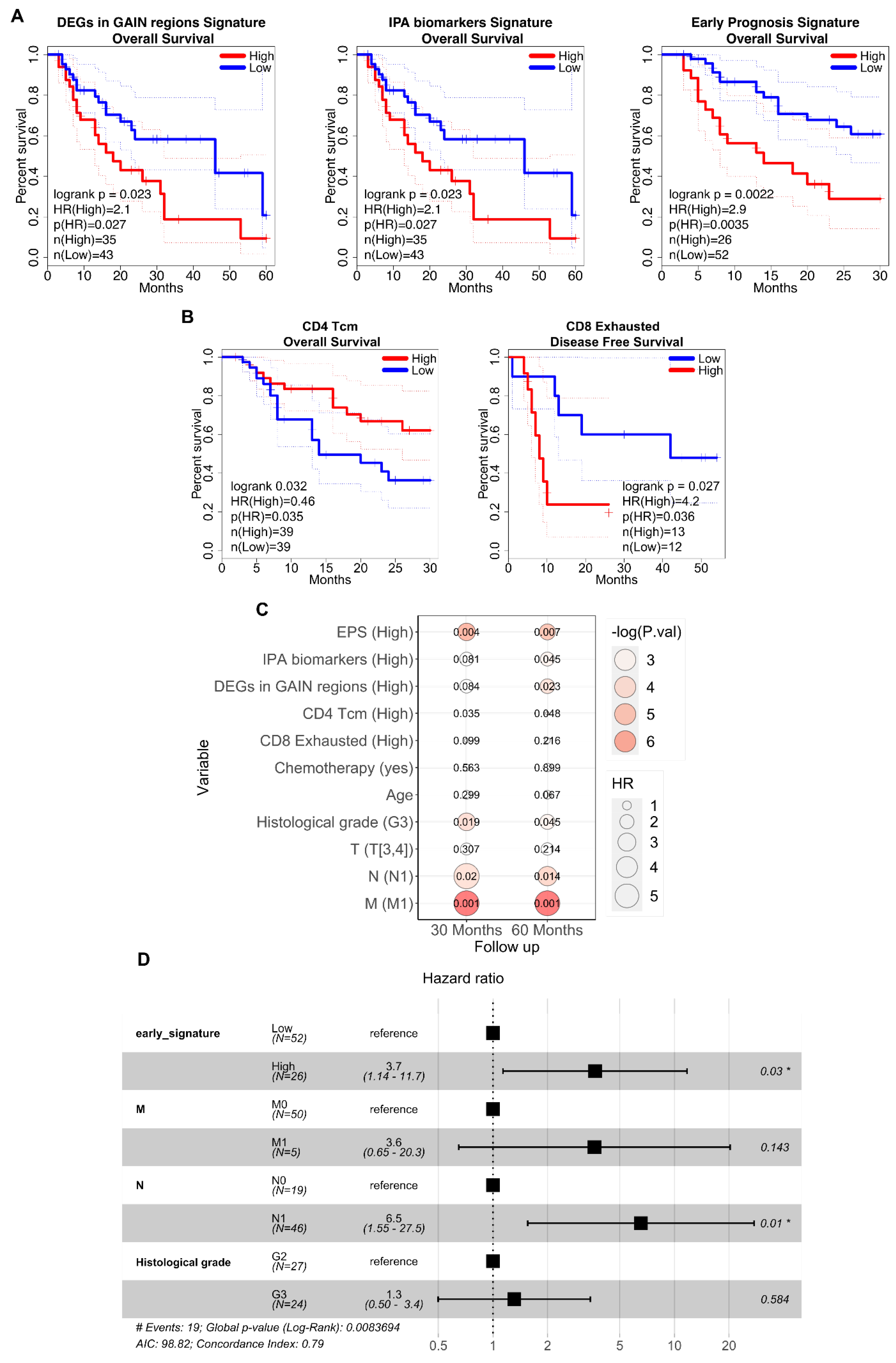
3.4. Association between the Prognostic Signatures and Patients’ Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coleman HG, et al. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology. 2018;154(2):390-405. [CrossRef]
- Fridman WH, et al. The immune contexture in human tumours: Impact on clinical outcome. Nature Reviews Cancer. Published online 2012. [CrossRef]
- Fassan M, et al. PD-L1 expression, CD8+ and CD4+ lymphocyte rate are predictive of pathological complete response after neoadjuvant chemoradiotherapy for squamous cell cancer of the thoracic esophagus. Cancer Medicine. 2019;8(13):6036-6048. [CrossRef]
- Hong, M. , Tao, S., Zhang, L. et al. RNA sequencing: new technologies and applications in cancer research. J Hematol Oncol 13, 166 (2020). [CrossRef]
- Chung W, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nature Communications. 2017;8. [CrossRef]
- Peng J, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Research. 2019;29(9):725-738. [CrossRef]
- Lambrechts D, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nature Medicine. 2018;24(8):1277-1289. [CrossRef]
- Savas P, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nature Medicine. 2018;24(7):986-993. [CrossRef]
- JH L, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162(1):184-197. [CrossRef]
- McCarthy DJ, Campbell KR, Lun ATL, Wills QF. Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33(8):1179-1186. [CrossRef]
- Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, Marioni JC. EmptyDrops: Distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biology. 2019;20(1):63. [CrossRef]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology. Published online 2018. [CrossRef]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature Biotechnology. 2015;33(5):495-502. [CrossRef]
- Croft W, Evans RPT, Pearce H, Elshafie M, Griffiths EA, Moss P. The single cell transcriptional landscape of esophageal adenocarcinoma and its modulation by neoadjuvant chemotherapy. Mol Cancer. 2022 Oct 17;21(1):200. [CrossRef] [PubMed]
- Li X, Wang K, Lyu Y, et al. Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis. Nature Communications. 2020;11(1):1-14. [CrossRef]
- Zheng, Y. , Chen, Z., Han, Y. et al. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat Commun 11, 6268 (2020). [CrossRef]
- van de Sande B, Flerin C, Davie K, et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nature Protocols. 2020;15(7):2247-2276. [CrossRef]
- Lambert SA, Jolma A, Campitelli LF, et al. Erratum: The Human Transcription Factors (Cell (2018) 172(4) (650–665), (S0092867418301065) (10.1016/j.cell.2018.01.029)). Cell. 2018;175(2):598-599. [CrossRef]
- Moerman T, Aibar Santos S, Bravo González-Blas C, et al. GRNBoost2 and Arboreto: Efficient and scalable inference of gene regulatory networks. Bioinformatics. 2019;35(12):2159-2161. [CrossRef]
- Lugli E, Zanon V, Mavilio D, Roberto A. FACS analysis of memory T lymphocytes. In: Methods in Molecular Biology. Vol 1514. Humana Press Inc.; 2017:31-47. [CrossRef]
- Brummelman J, Haftmann C, Núñez NG, et al. Development, application and computational analysis of high-dimensional fluorescent antibody panels for single-cell flow cytometry. Nature Protocols. 2019;14(7). [CrossRef]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183-1186. [CrossRef]
- Dobin A, Davis CA, Schlesinger F, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21. [CrossRef]
- Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923-930. [CrossRef]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12). [CrossRef]
- Chiesa M, Colombo GI, Piacentini L. DaMiRseq -An R/Bioconductor package for data mining of RNA-Seq data: Normalization, feature selection and classification. Bioinformatics. 2018;34(8):1416-1418. [CrossRef]
- Ferrari F, Solari A, Battaglia C, Bicciato S. PREDA: An R-package to identify regional variations in genomic data. Bioinformatics. 2011;27(17):2446-2447. [CrossRef]
- Therneau, TM. A Package for Survival Analysis in R. 2020. Available from: https://CRAN.R-project.
- Salem ME, et al. Comparative Molecular Analyses of Esophageal Squamous Cell Carcinoma, Esophageal Adenocarcinoma, and Gastric Adenocarcinoma. The Oncologist. 2018;23(11):1319-1327. [CrossRef]
- Killcoyne S, et al. Genomic copy number predicts esophageal cancer years before transformation. Nature Medicine. 2020;26(11):1726-1732. [CrossRef]
- Karagoz K, Lehman HL, Stairs DB, Sinha R, Arga KY. Proteomic and Metabolic Signatures of Esophageal Squamous Cell Carcinoma. Current cancer drug targets. Published online , 2016. 2 February.
- Chen X, Li D, Wang N, et al. Bioinformatic analysis suggests that UGT2B15 activates the Hippo YAP signaling pathway leading to the pathogenesis of gastric cancer. Oncology Reports. Published online , 2018. 26 July. [CrossRef]
- Li D, Yin Y, He M, Wang J. Identification of Potential Biomarkers Associated with Prognosis in Gastric Cancer via Bioinformatics Analysis. Medical Science Monitor. 2021;27. [CrossRef]
- Quilty F, Byrne AM, Aird J, et al. Impact of Deoxycholic Acid on Oesophageal Adenocarcinoma Invasion: Effect on Matrix Metalloproteinases. International Journal of Molecular Sciences. 2020;21(21). [CrossRef]
- Peng H hua, Zhang X, Cao P guo. MMP-1/PAR-1 signal transduction axis and its prognostic impact in esophageal squamous cell carcinoma. Brazilian Journal of Medical and Biological Research. 2012;45(1):86. [CrossRef]
- Wang Z, Hao Y, Lowe AW. The Adenocarcinoma-Associated Antigen, AGR2, Promotes Tumor Growth, Cell Migration, and Cellular Transformation. Cancer Research. 2008;68(2). [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




