Submitted:
08 December 2023
Posted:
08 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
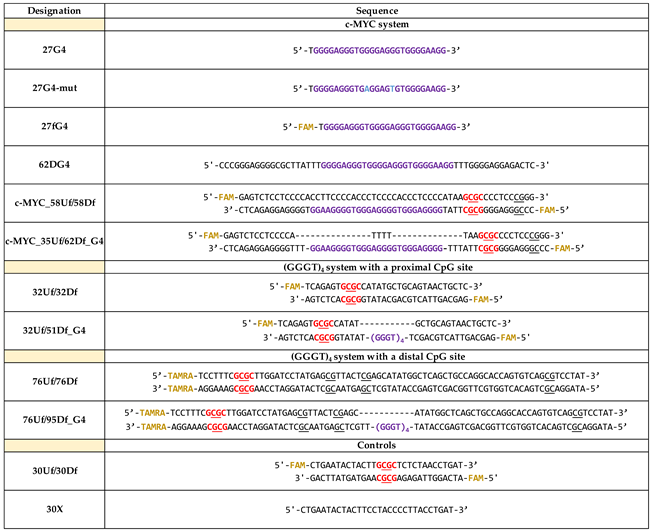
2.1. Synthetic G4-Containing DNA and Control Single- and Double-Stranded Oligonucleotides
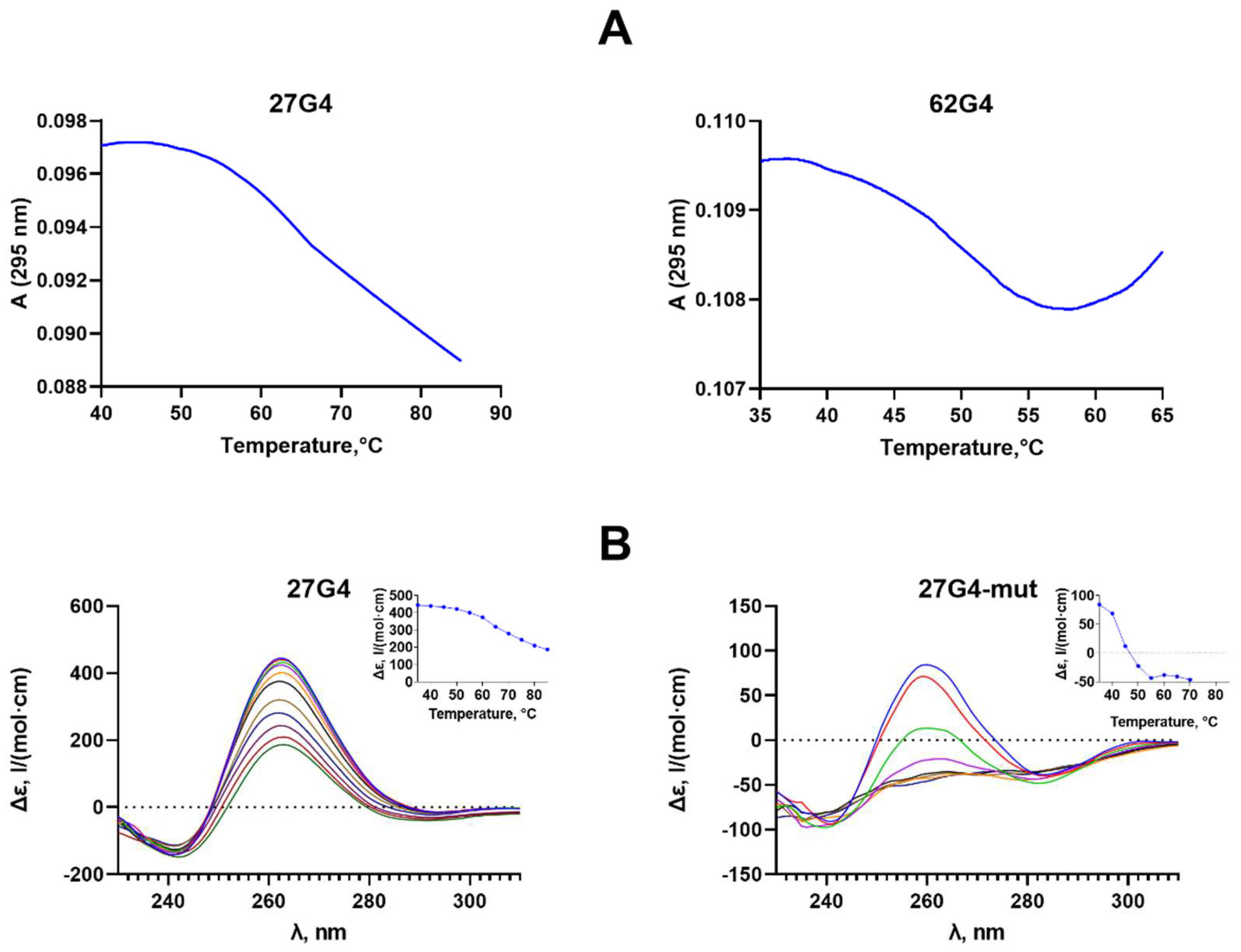
2.2. Formation, Topology and Thermal Stability of c-MYC G4 in the Designed DNA Models
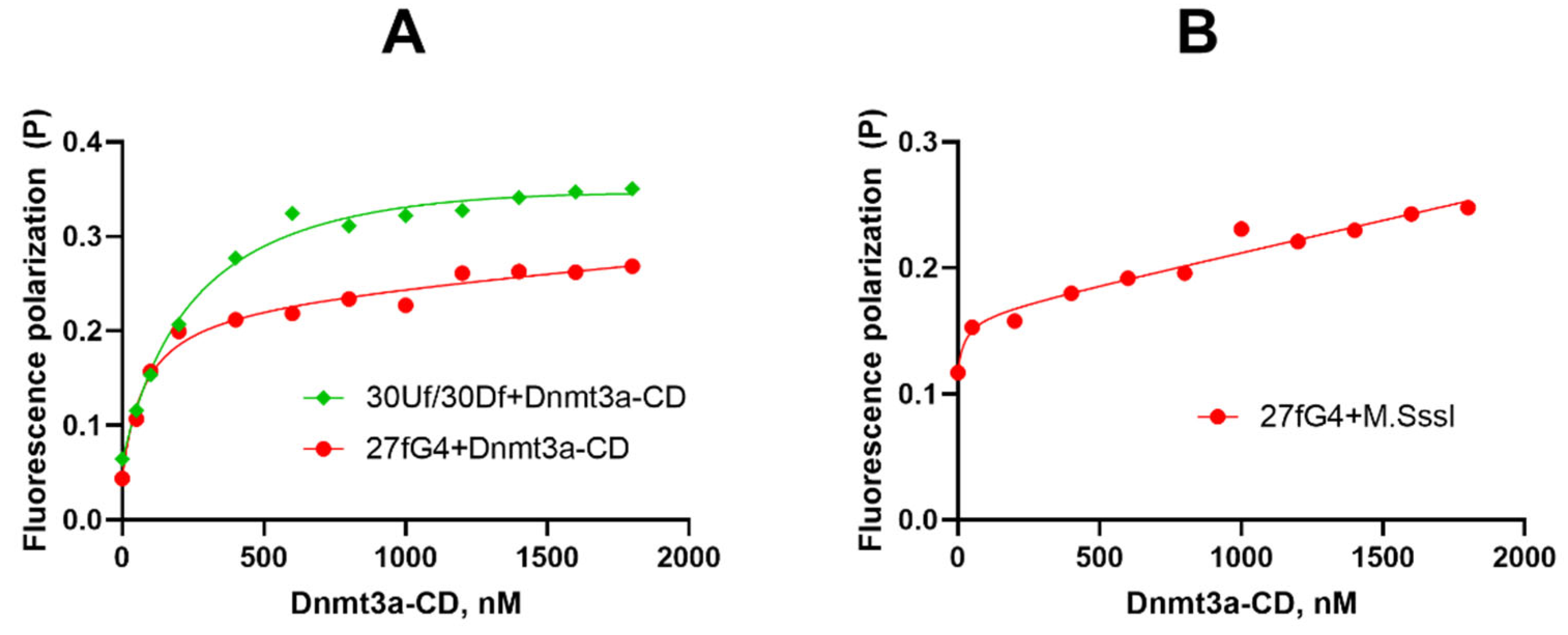
2.3. Dnmt3a-CD Effectively Binds to Oligonucleotide 27fG4
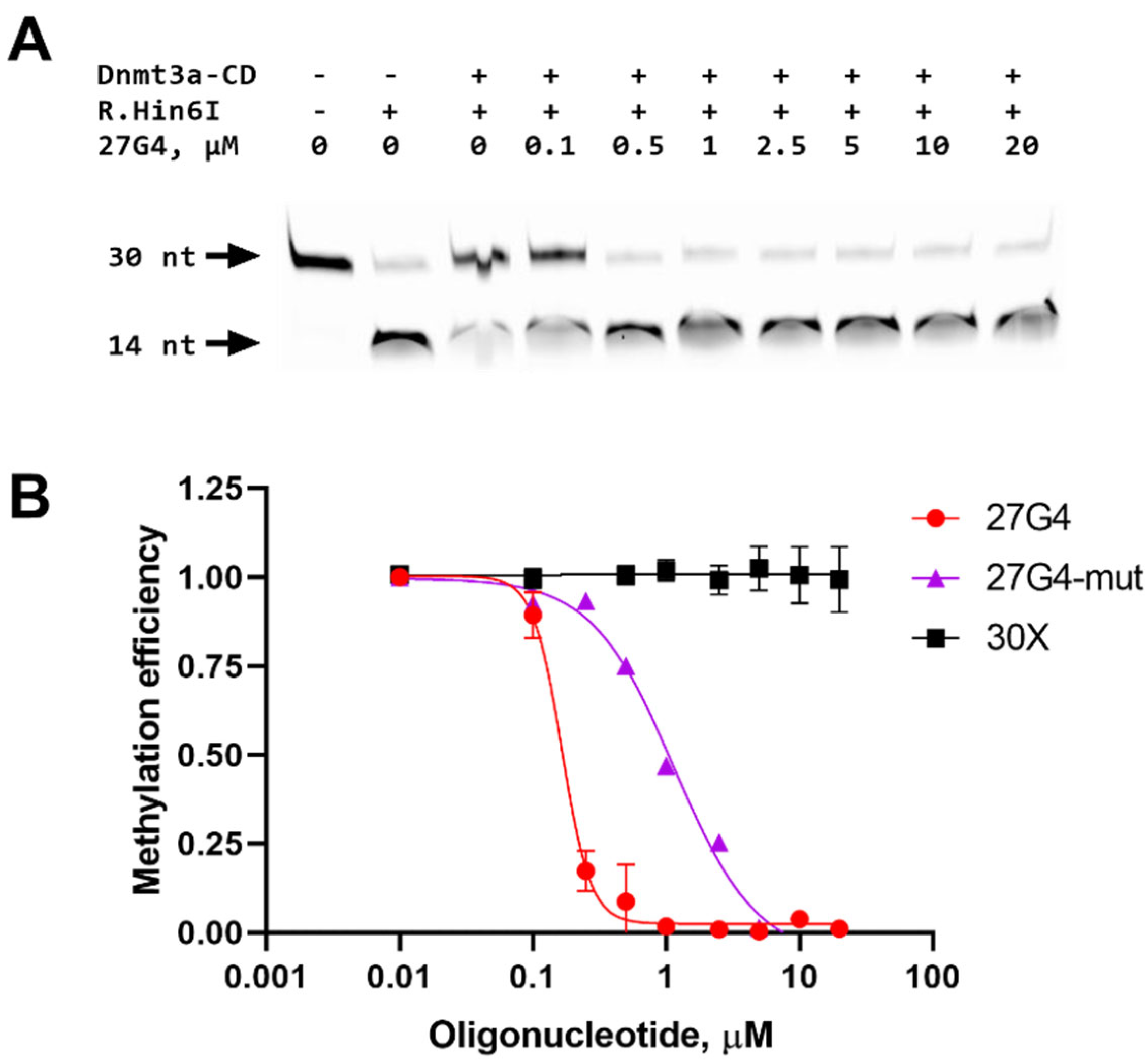
2.4. Inhibition of a 30-bp DNA Duplex Methylation in the Presence of 27G4
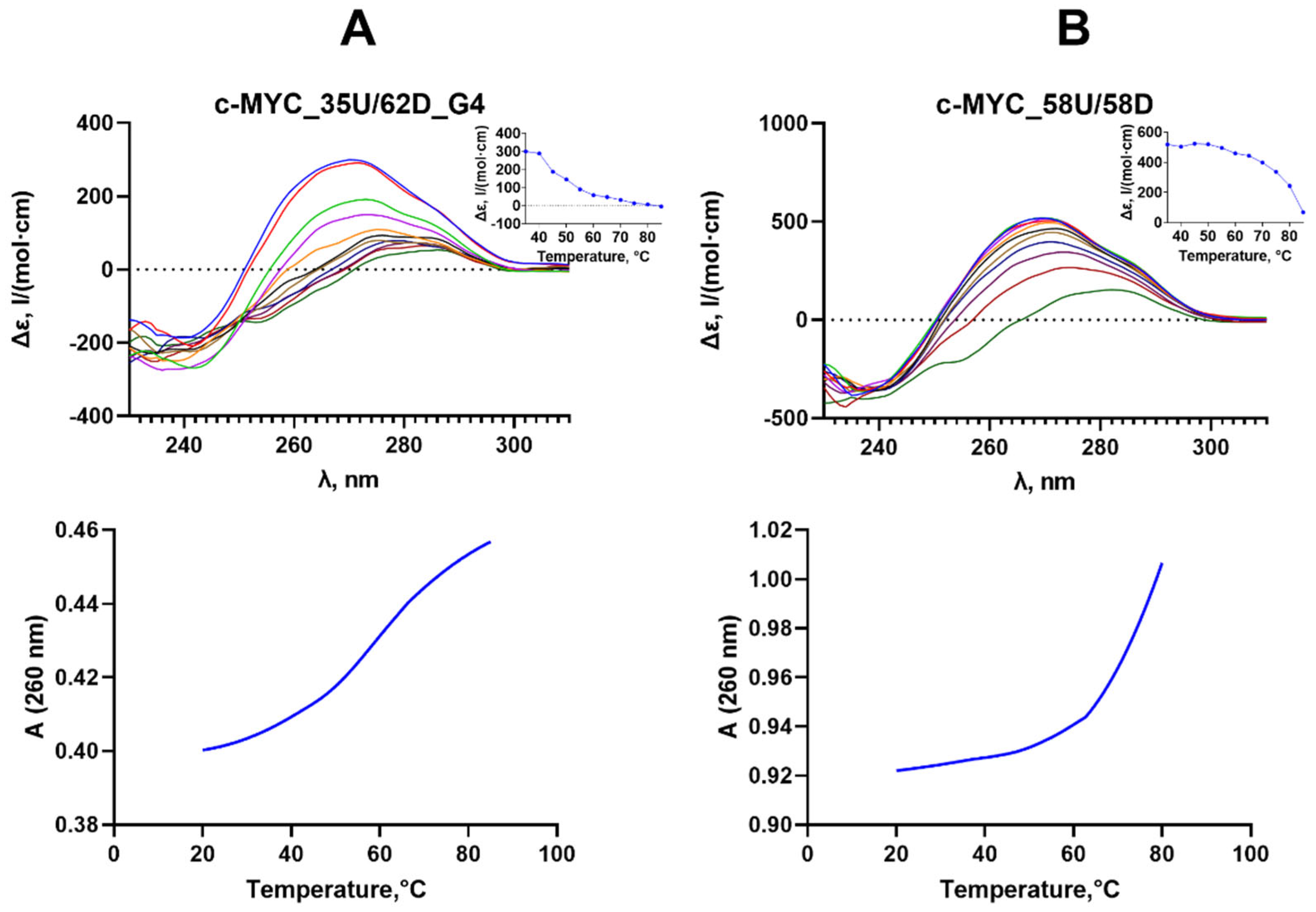
2.5. Effect of c-MYC G4 on Methylation of the c-MYC Promoter Region
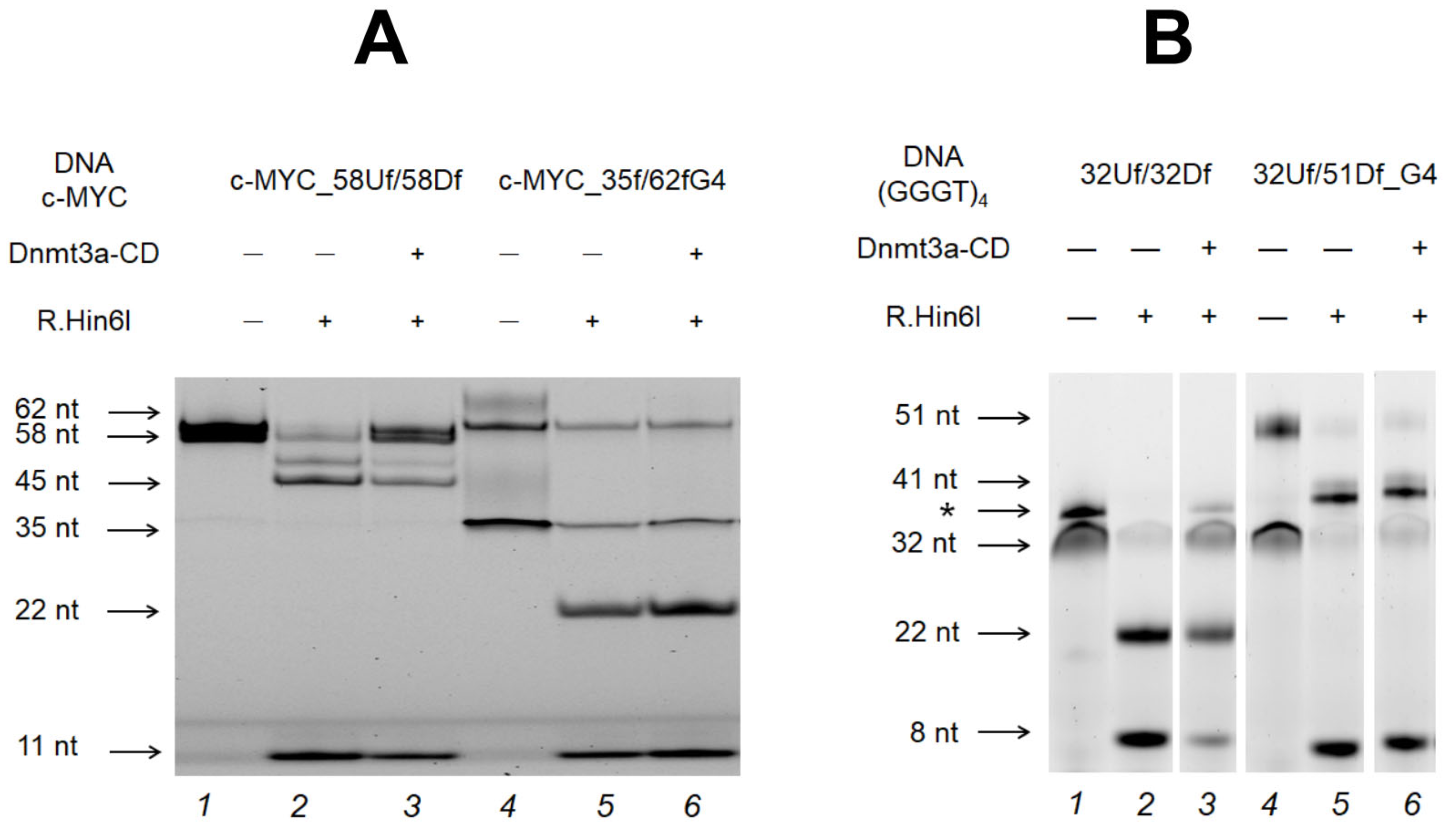
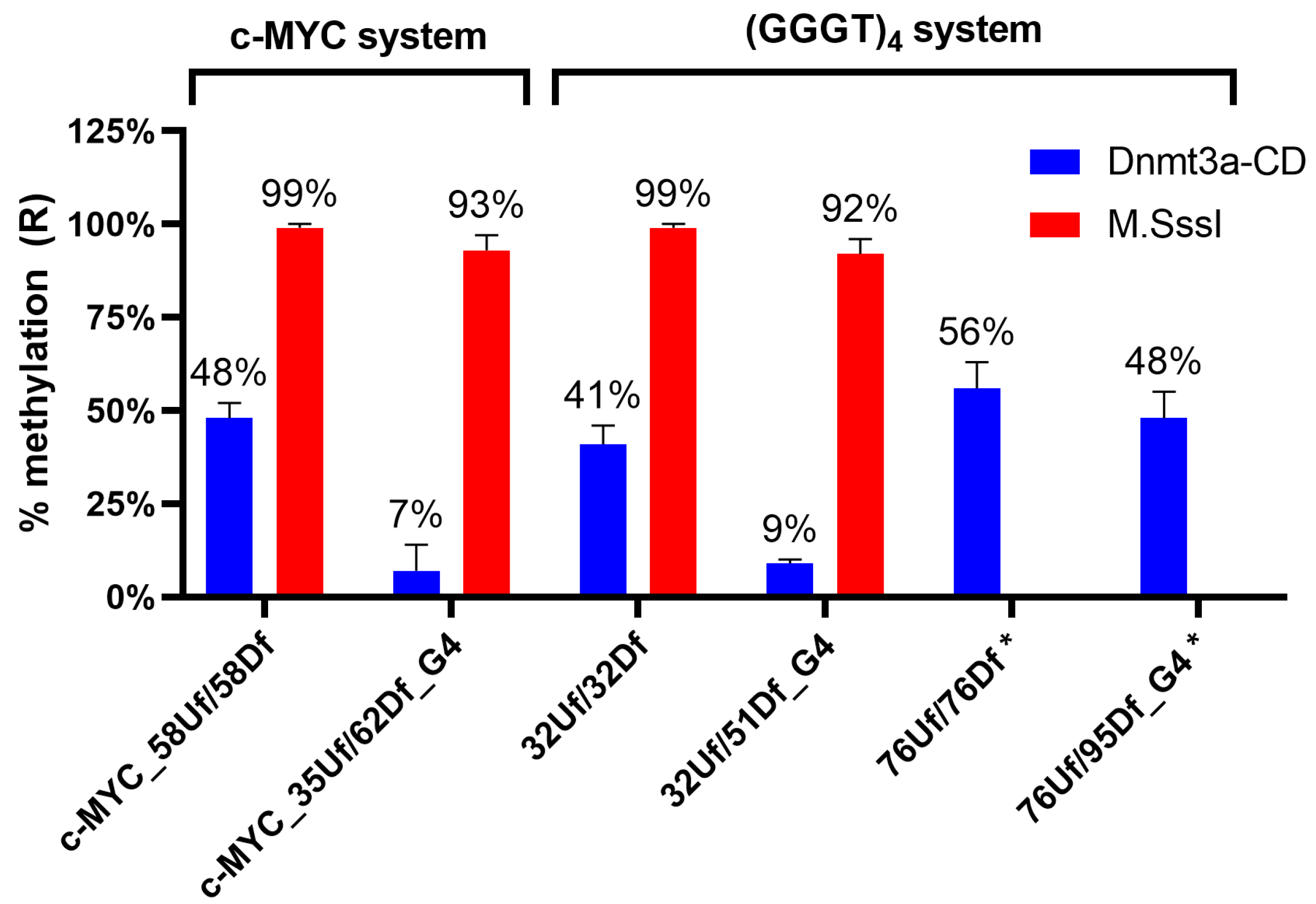
2.6. Influence of the Relative Positions of G4 and CpG Sites within a DNA Duplex on the Methylation Activity of Dnmt3a-CD
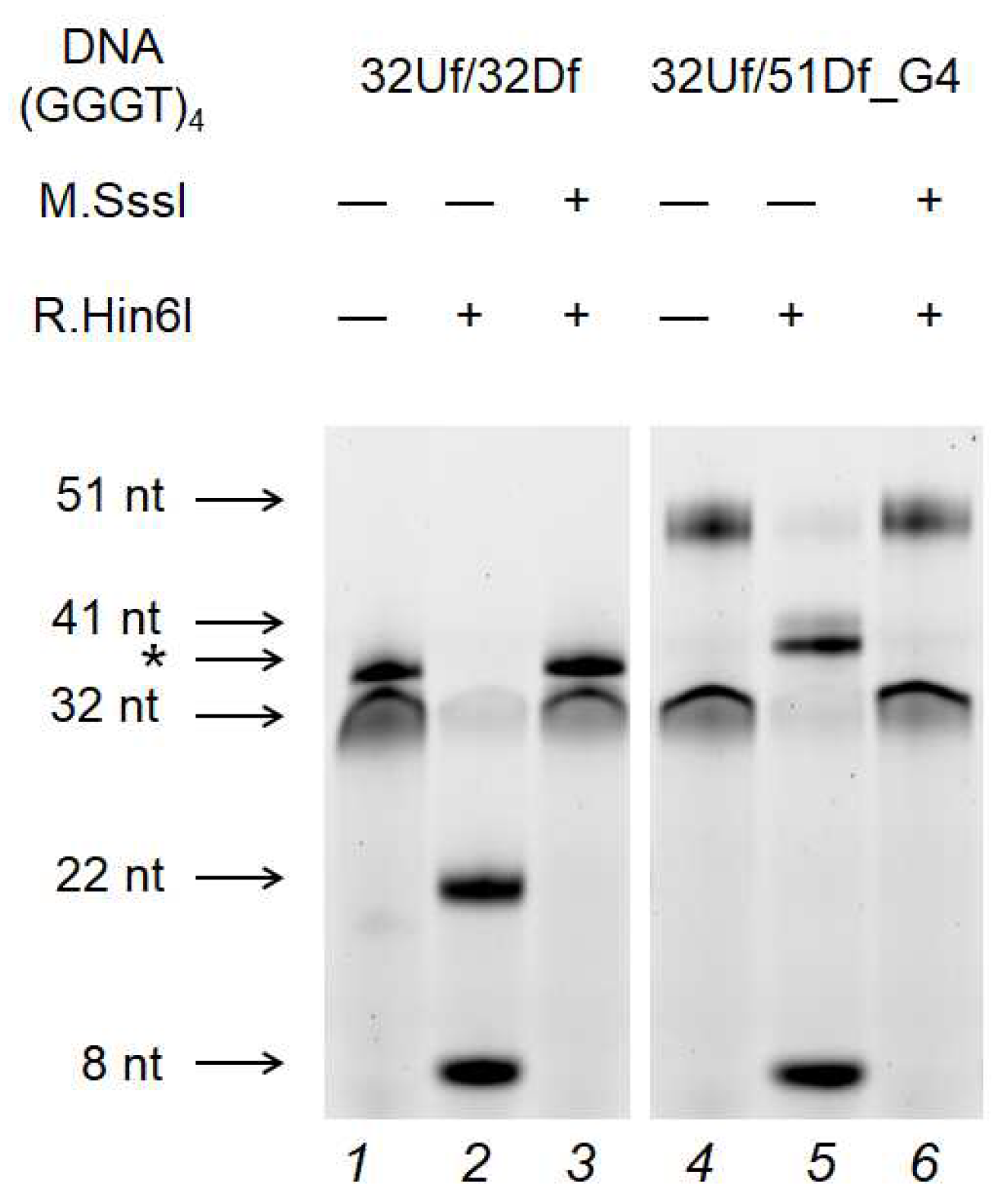
2.7. M.SssI-Mediated Methylation of Model DNA with G4 Structure
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Enzymes
4.3. CD Measurements
4.4. UV Spectroscopy Melting of Oligonucleotides Containing G4 Motifs and DNA Duplexes
4.5. Complex Formation of Dnmt3a-CD and M.SssI with DNA
4.6. Methylation of DNA Duplexes
4.7. Inhibition of Methylation of a Standard DNA Substrate by a G4 Oligonucleotide
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Todd, A.K.; Johnston, M.; Neidle, S. Highly Prevalent Putative Quadruplex Sequence Motifs in Human DNA. Nucleic Acids Res 2005, 33, 2901–2907. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. Prevalence of Quadruplexes in the Human Genome. Nucleic Acids Res 2005, 33, 2908–2916. [Google Scholar] [CrossRef]
- Lane, A.N.; Chaires, J.B.; Gray, R.D.; Trent, J.O. Stability and Kinetics of G-Quadruplex Structures. Nucleic Acids Res 2008, 36, 5482–5515. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, Topology and Structure. Nucleic Acids Res 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed]
- Meier-Stephenson, V. G4-Quadruplex-Binding Proteins: Review and Insights into Selectivity. Biophysical Reviews 2022 14:3 2022, 14, 635–654. [Google Scholar] [CrossRef] [PubMed]
- Ambrus, A.; Chen, D.; Dai, J.; Jones, R.A.; Yang, D. Solution Structure of the Biologically Relevant G-Quadruplex Element in the Human c-MYC Promoter. Implications for G-Quadruplex Stabilization. Biochemistry 2005, 44, 2048–2058. [Google Scholar] [CrossRef]
- Gowher, H.; Jeltsch, A. Mammalian DNA Methyltransferases: New Discoveries and Open Questions. Biochem Soc Trans 2018, 46, 1191–1202. [Google Scholar] [CrossRef]
- Jeltsch, A.; Jurkowska, R.Z. New Concepts in DNA Methylation. Trends Biochem Sci 2014, 39, 310–318. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. G-Quadruplexes in Promoters throughout the Human Genome. Nucleic Acids Res 2007, 35, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Hurley, L.H. Structures, Folding Patterns, and Functions of Intramolecular DNA G-Quadruplexes Found in Eukaryotic Promoter Regions. Biochimie 2008, 90, 1149–1171. [Google Scholar] [CrossRef]
- Chen, L.; Dickerhoff, J.; Sakai, S.; Yang, D. DNA G-Quadruplex in Human Telomeres and Oncogene Promoters: Structures, Functions, and Small Molecule Targeting. Acc Chem Res 2022, 55, 2628–2646. [Google Scholar] [CrossRef]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The Regulation and Functions of DNA and RNA G-Quadruplexes. Nature Reviews Molecular Cell Biology 2020 21:8 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Dolinnaya, N.G.; Ogloblina, A.M.; Yakubovskaya, M.G. Structure, Properties, and Biological Relevance of the DNA and RNA G-Quadruplexes: Overview 50 Years after Their Discovery. Biochemistry (Moscow) 2016, 81, 1602–1649. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Chen, H.; Zhang, Q.; Xu, M.; Yuan, G.; Zhou, J. Exploration of the Structure and Recognition of a G-Quadruplex in the Her2 Proto-Oncogene Promoter and Its Transcriptional Regulation. Sci Rep 2019, 9, 3966. [Google Scholar] [CrossRef] [PubMed]
- Ducani, C.; Bernardinelli, G.; Högberg, B.; Keppler, B.K.; Terenzi, A. Interplay of Three G-Quadruplex Units in the KIT Promoter. J Am Chem Soc 2019, 141, 10205–10213. [Google Scholar] [CrossRef]
- Monsen, R.C.; DeLeeuw, L.W.; Dean, W.L.; Gray, R.D.; Chakravarthy, S.; Hopkins, J.B.; Chaires, J.B.; Trent, J.O. Long Promoter Sequences Form Higher-Order G-Quadruplexes: An Integrative Structural Biology Study of c-Myc, k-Ras and c-Kit Promoter Sequences. Nucleic Acids Res 2022, 50, 4127–4147. [Google Scholar] [CrossRef]
- Jurkowska, R.Z.; Jurkowski, T.P.; Jeltsch, A. Structure and Function of Mammalian DNA Methyltransferases. ChemBioChem 2011, 12, 206–222. [Google Scholar] [CrossRef]
- Wong, K.K.; Lawrie, C.H.; Green, T.M. Oncogenic Roles and Inhibitors of DNMT1, DNMT3A, and DNMT3B in Acute Myeloid Leukaemia. Biomark Insights 2019, 14. [Google Scholar] [CrossRef] [PubMed]
- Varizhuk, A.; Isaakova, E.; Pozmogova, G. DNA G-Quadruplexes (G4s) Modulate Epigenetic (Re)Programming and Chromatin Remodeling. BioEssays 2019, 41, 1900091. [Google Scholar] [CrossRef]
- Halder, R.; Halder, K.; Sharma, P.; Garg, G.; Sengupta, S.; Chowdhury, S. Guanine Quadruplex DNA Structure Restricts Methylation of CpG Dinucleotides Genome-Wide. Mol Biosyst 2010, 6, 2439–2447. [Google Scholar] [CrossRef]
- Rauchhaus, J.; Robinson, J.; Monti, L.; Di Antonio, M. G-Quadruplexes Mark Sites of Methylation Instability Associated with Ageing and Cancer. Genes (Basel) 2022, 13, 1665. [Google Scholar] [CrossRef] [PubMed]
- Cree, S.L.; Fredericks, R.; Miller, A.; Pearce, F.G.; Filichev, V.; Fee, C.; Kennedy, M.A. DNA G-Quadruplexes Show Strong Interaction with DNA Methyltransferases in Vitro. FEBS Lett 2016, 590, 2870–2883. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.-Q.; Ghanbarian, A.T.; Spiegel, J.; Martínez Cuesta, S.; Beraldi, D.; Di Antonio, M.; Marsico, G.; Hänsel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. DNA G-Quadruplex Structures Mold the DNA Methylome. Nat Struct Mol Biol 2018, 25, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Wierstra, I.; Alves, J. The C-myc Promoter: Still MysterY and Challenge. Adv Cancer Res 2008, 99, 113–333. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, R.; Bhattacharya, S.; Dash, J.; Bhattacharya, S. Recent Update on Targeting c-MYC G-Quadruplexes by Small Molecules for Anticancer Therapeutics. J Med Chem 2021, 64, 42–70. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Hurley, L. Structure of the Biologically Relevant G-Quadruplex in The c-MYC Promoter. Nucleosides Nucleotides Nucleic Acids 2006, 25, 951–968. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, A. V.; Monakhova, M. V.; Ogloblina, A.M.; Andreeva, N.A.; Laptev, G.Y.; Polshakov, V.I.; Gromova, E.S.; Zvereva, M.I.; Yakubovskaya, M.G.; Oretskaya, T.S.; et al. Responses of DNA Mismatch Repair Proteins to a Stable G-Quadruplex Embedded into a DNA Duplex Structure. Int J Mol Sci 2020, 21, 8773. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.L.; Lacroix, L. UV Melting of G-Quadruplexes. Curr Protoc Nucleic Acid Chem 2009, 37, 17.1.1–17.1.15. [Google Scholar] [CrossRef] [PubMed]
- Sekibo, D.A.T.; Fox, K.R. The Effects of DNA Supercoiling on G-Quadruplex Formation. Nucleic Acids Res 2017, 45, 12069–12079. [Google Scholar] [CrossRef]
- Zhang, Z.M.; Lu, R.; Wang, P.; Yu, Y.; Chen, D.; Gao, L.; Liu, S.; Ji, D.; Rothbart, S.B.; Wang, Y.; et al. Structural Basis for DNMT3A-Mediated de Novo DNA Methylation. Nature 2018, 554, 387–391. [Google Scholar] [CrossRef]
- Jurkowska, R.Z.; Rajavelu, A.; Anspach, N.; Urbanke, C.; Jankevicius, G.; Ragozin, S.; Nellen, W.; Jeltsch, A. Oligomerization and Binding of the Dnmt3a DNA Methyltransferase to Parallel DNA Molecules: Heterochromatic Localization and Role of Dnmt3L. Journal of Biological Chemistry 2011, 286, 24200–24207. [Google Scholar] [CrossRef] [PubMed]
- Chédin, F. The DNMT3 Family of Mammalian De Novo DNA Methyltransferases. Prog Mol Biol Transl Sci 2011, 101, 255–285. [Google Scholar] [CrossRef] [PubMed]
- Darii, M. V; Cherepanova, N.A.; Subach, O.M.; Kirsanova, O. V; Raskó, T.; Ślaska-Kiss, K.; Kiss, A.; Deville-Bonne, D.; Reboud-Ravaux, M.; Gromova, E.S. Mutational Analysis of the CG Recognizing DNA Methyltransferase SssI: Insight into Enzyme–DNA Interactions. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2009, 1794, 1654–1662. [Google Scholar] [CrossRef] [PubMed]
- Pósfai, J.; Bhagwat, A.S.; Pósfai, G.; Roberts, R.J. Predictive Motifs Derived from Cytosine Methyltransferases. Nucleic Acids Res 1989, 17, 2421–2435. [Google Scholar] [CrossRef] [PubMed]
- Loiko, A.G.; Sergeev, A. V.; Genatullina, A.I.; Monakhova, M. V.; Kubareva, E.A.; Dolinnaya, N.G.; Gromova, E.S. Impact of G-Quadruplex Structures on Methylation of Model Substrates by DNA Methyltransferase Dnmt3a. Int J Mol Sci 2022, 23, 10226. [Google Scholar] [CrossRef] [PubMed]
- Sergeev, A.; Vorobyov, A.; Yakubovskaya, M.; Kirsanova, O.; Gromova, E.S. Novel Anticancer Drug Curaxin CBL0137 Impairs DNA Methylation by Eukaryotic DNA Methyltransferase Dnmt3a. Bioorg Med Chem Lett 2020, 30, 127296. [Google Scholar] [CrossRef]
- Brázda, V.; Hároníková, L.; Liao, J.C.C.; Fojta, M. DNA and RNA Quadruplex-Binding Proteins. Int J Mol Sci 2014, 15, 17493–17517. [Google Scholar] [CrossRef] [PubMed]
- Sergeev, A. V.; Tevyashova, A.N.; Vorobyov, A.P.; Gromova, E.S. The Effect of Antitumor Antibiotic Olivomycin A and Its New Semi-Synthetic Derivative Olivamide on the Activity of Murine DNA Methyltransferase Dnmt3a. Biochemistry (Moscow) 2019, 84, 62–70. [Google Scholar] [CrossRef]
- Zheng, K. wei; Chen, Z.; Hao, Y. hua; Tan, Z. Molecular Crowding Creates an Essential Environment for the Formation of Stable G-Quadruplexes in Long Double-Stranded DNA. Nucleic Acids Res 2010, 38, 327–338. [Google Scholar] [CrossRef]
- Jara-Espejo, M.; Peres Line, S.R. DNA G-Quadruplex Stability, Position and Chromatin Accessibility Are Associated with CpG Island Methylation. FEBS J 2020, 287, 483–495. [Google Scholar] [CrossRef]
- Dai, Y.; Teng, X.; Zhang, Q.; Hou, H.; Li, J. Advances and Challenges in Identifying and Characterizing G-Quadruplex–Protein Interactions. Trends Biochem Sci 2023, 48, 894–909. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowski, M.; Czapinska, H.; Bochtler, M. CpG Underrepresentation and the Bacterial CpG-Specific DNA Methyltransferase M.Mpel. Proceedings of the National Academy of Sciences 2013, 110, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Sergeev, A. V.; Kirsanova, O. V.; Loiko, A.G.; Nomerotskaya, E.I.; Gromova, E.S. Detection of DNA Methylation by Dnmt3a Methyltransferase Using Methyl-Dependent Restriction Endonucleases. Mol Biol 2018, 52, 272–278. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





