Submitted:
30 November 2023
Posted:
01 December 2023
You are already at the latest version
Abstract
Keywords:
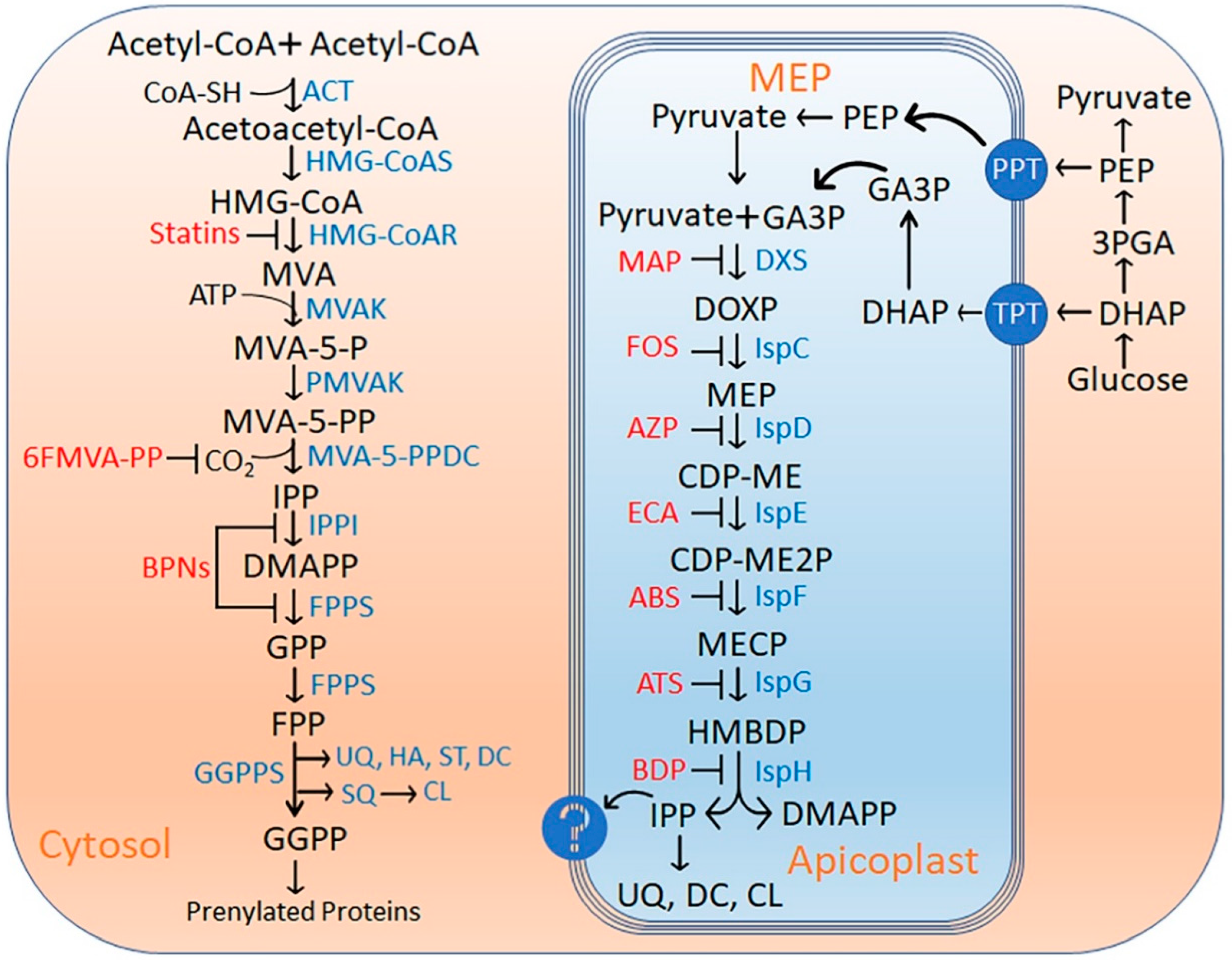
1. Introduction
2. Results and Discussion
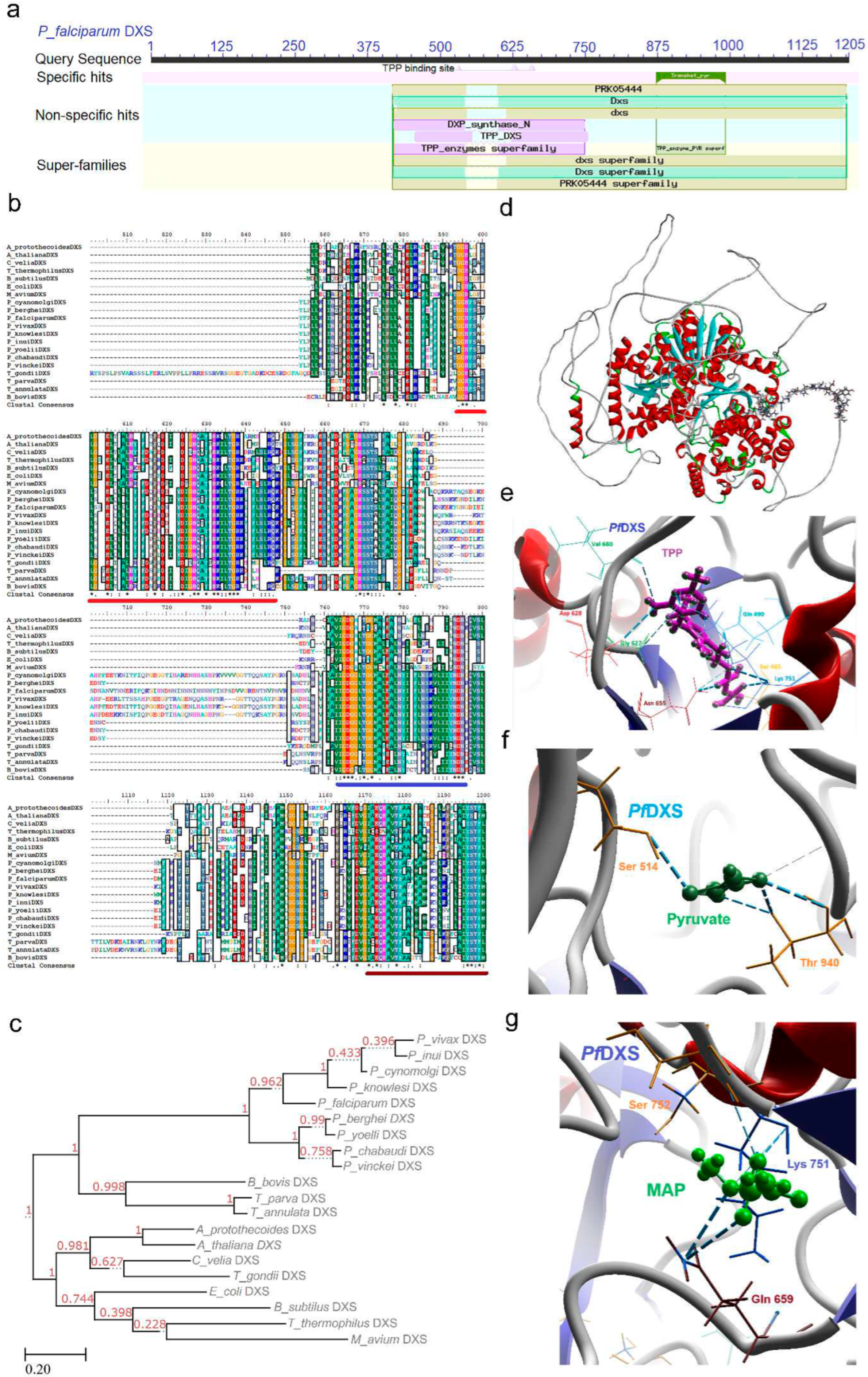
2.1. DXS (DXP Synthase) Enzyme
2.1.1. PfDXS Enzyme’s Structural Insight
2.1.2. PfDXS Enzyme’s Inhibitors Studies
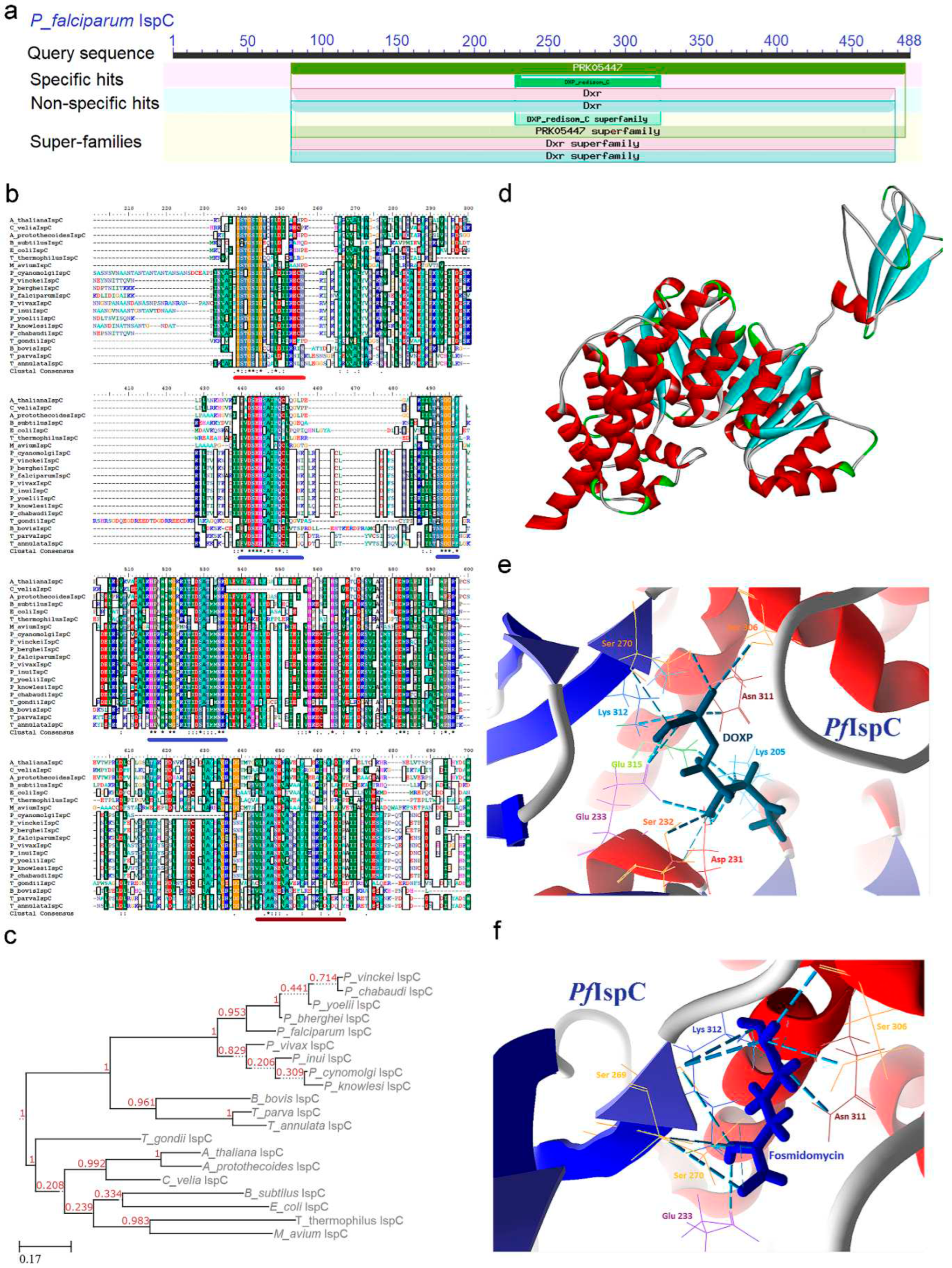
2.2. IspC (1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase) Enzyme
2.2.1. PfIspC Enzyme’s Structural Insight
2.2.2. PfIspC Enzyme’s Inhibitors Studies
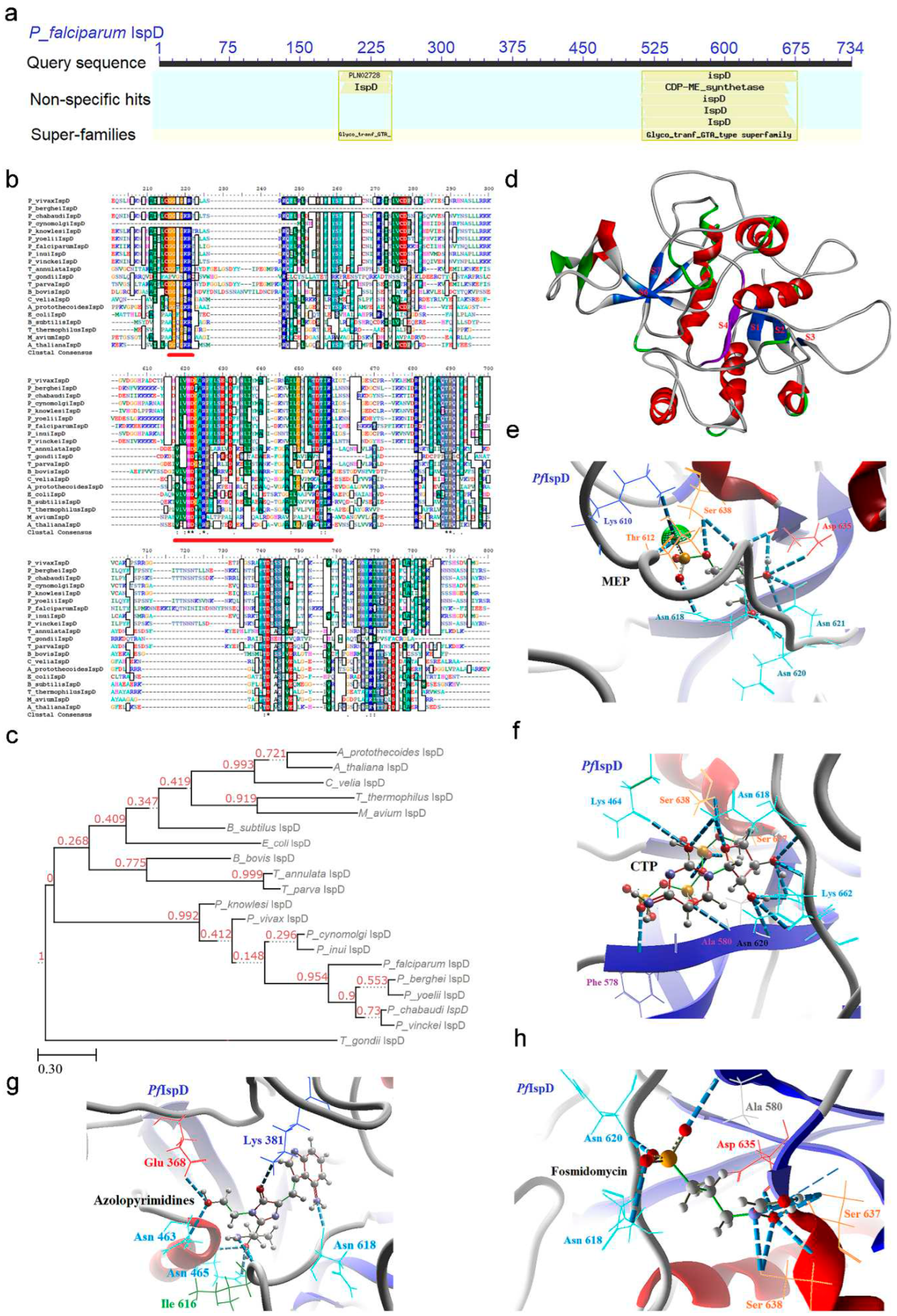
2.3. IspD [2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (YgbP)] Enzyme
2.3.1. PfIspD Enzyme’s Structural Insight
2.3.2. PfIspD Enzyme’s Inhibitors Studies
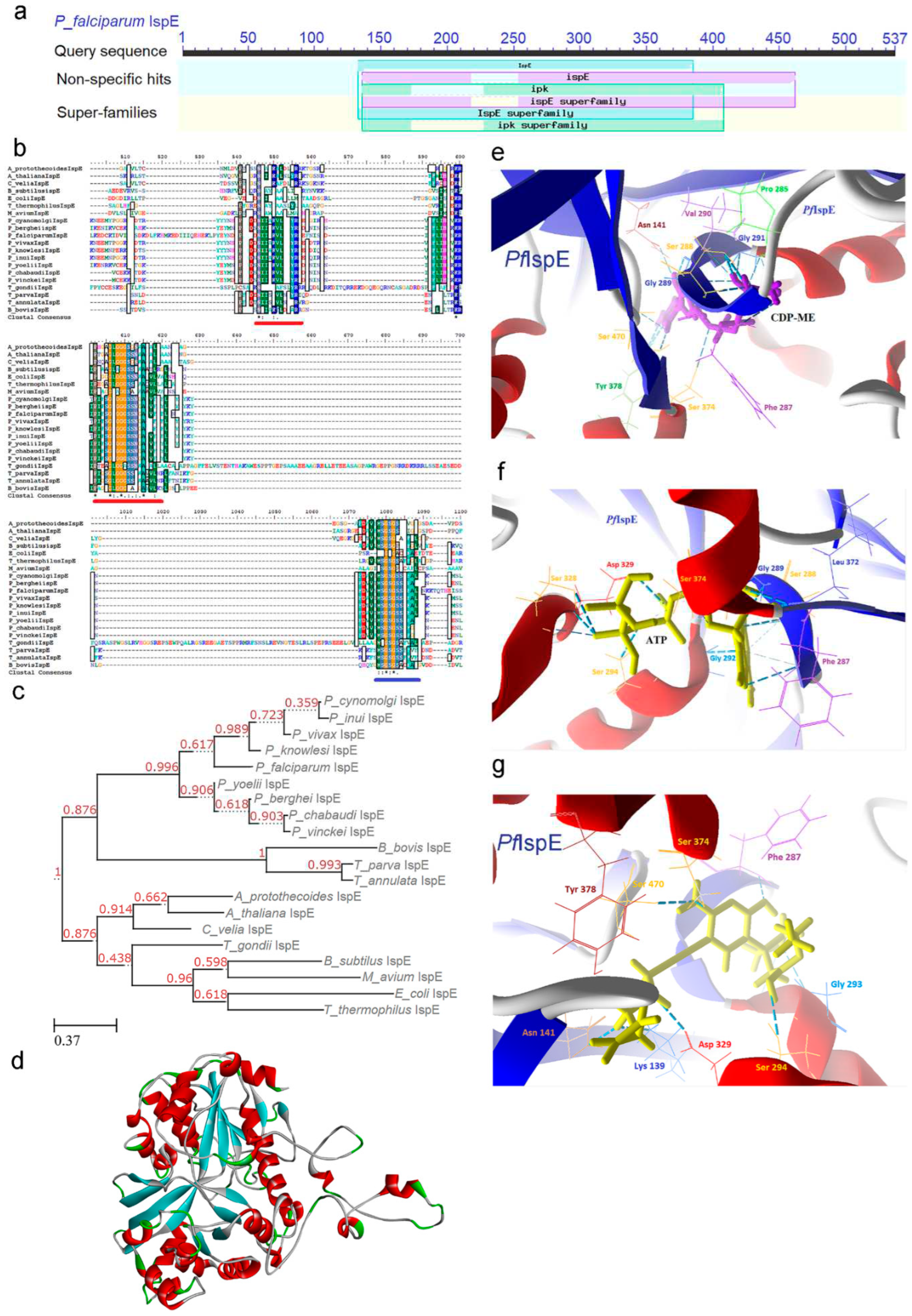
2.4. IspE [4-(cytidine-5-diphospho)-2-C-methyl-D-erythritol kinase (CMK)] Enzyme
2.4.1. PfIspE Enzyme’s Structural Insight
2.4.2. PfIspE Enzyme’s Inhibitors Studies
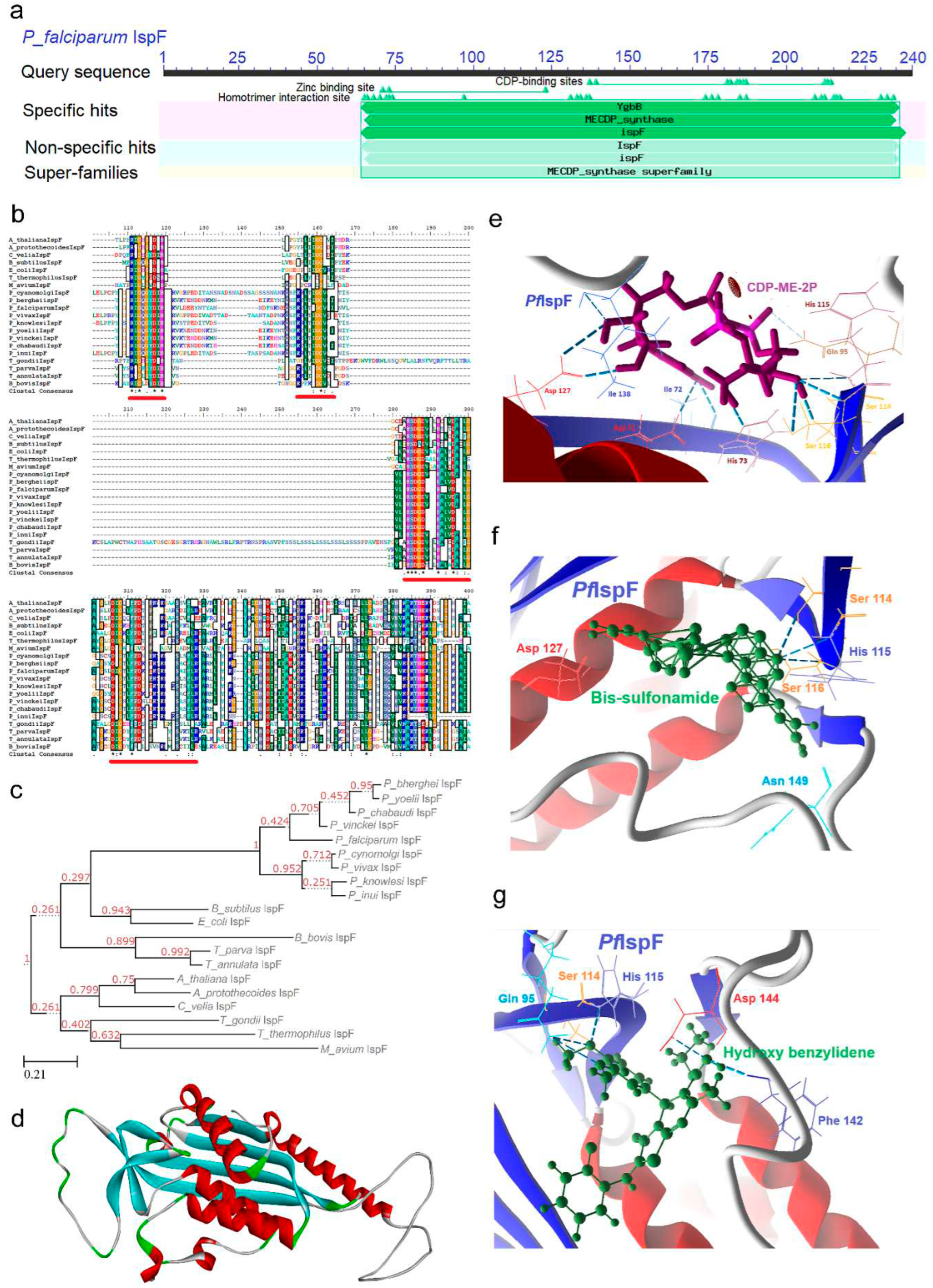
2.5. IspF [2C-Methyl-D-erythritol-2, 4-cyclodiphosphate synthase (ygbB)] Enzyme
2.5.1. PfIspF Enzyme’s Structural Insight
2.5.2. PfIspF Enzyme’s Inhibitors Studies
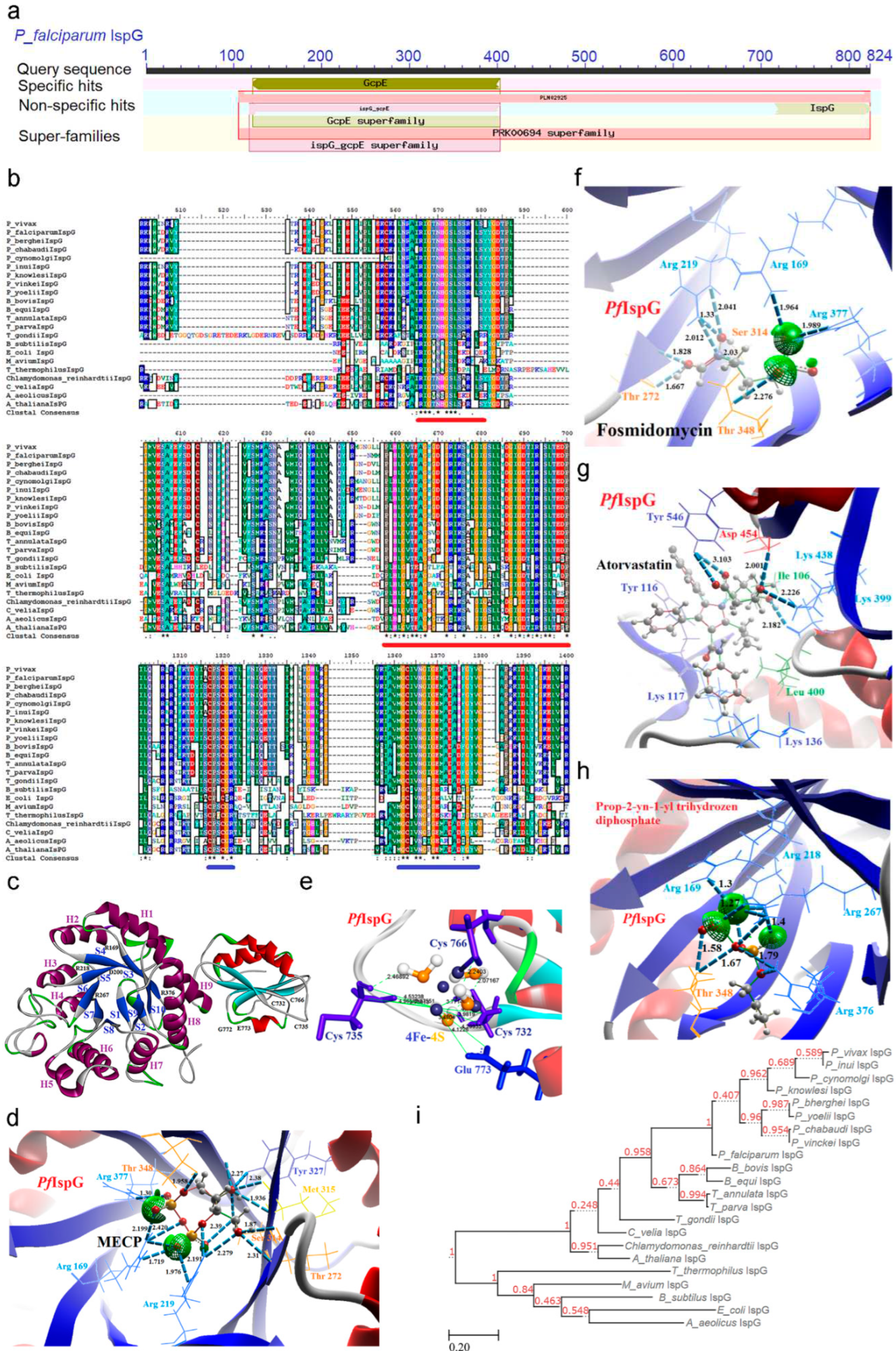
2.6. IspG [4-Hydroxy-3-methyl-2-(E)-butenyl-4-diphosphate synthase (gcpE)] Enzyme
2.6.1. PfIspG Enzyme’s Structural Insight
2.6.2. PfIspG Enzyme’s Inhibitors Studies
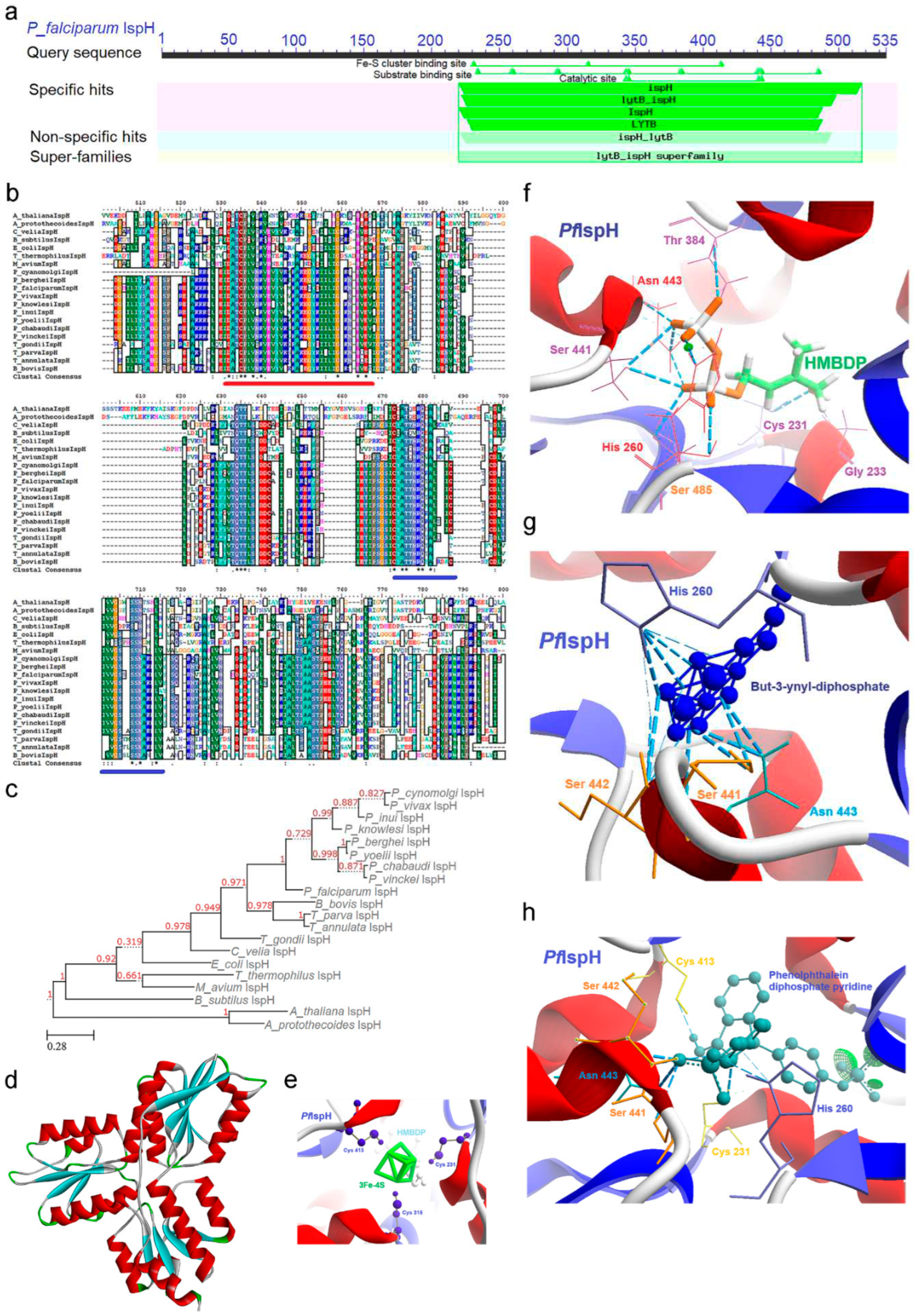
2.7. IspH [4-Hydroxy-3-methyl-2-(E)-butenyl-4-diphosphate reductase (lytB)] Enzyme
2.7.1. PfIspH Enzyme’s Structural Insight
2.7.2. PfIspH Enzyme’s Inhibitors Studies
3. Materials and Methods
3.1. Exploring Orthologous Sequences of MEP Pathway Enzymes
3.2. Conserved Domain Architecture and Signature Motifs
3.3. Phylogenetic Tree Construction and Analysis
3.4. Protein Structure Prediction
3.5. Investigating Substrate Docking and In-Silico Drug Inhibition Strategies
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. World Malaria Report 2022.
- Janouškovec, J., Horák, A., Oborník, M., Lukeš, J., Keeling, P. J. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci U S A. 2010, 107, 10949-10954. https://doi.org/10.1073/pnas.1003335107. [CrossRef]
- Goodman, C.D., Su, V., McFadden, G.I. The effects of anti-bacterials on the malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 2007, 152(2), 181-191. https://doi.org/10.1016/j.molbiopara.2007.01.005. [CrossRef]
- Ralph, S.A., van Dooren, G.G., Waller, R.F., Crawford, M.J., Fraunholz, M.J., Foth, B.J., Tonkin, C.J., Roos, D.S., McFadden, G.I. Metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004, 2, 203-216. https://doi.org/10.1038/nrmicro843. [CrossRef]
- Saggu, G.S., Garg, S., Pala, Z.R., Yadav, S.K., Kochar, S.K., Kochar, D.K., Saxena, V. Characterization of 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase (IspG) from Plasmodium vivax and it’s potential as an antimalarial drug target. Int J Biol Macromol. 2017, 96, 466-473. https://doi.org/10.1016/j.ijbiomac.2016.12.033. Epub 2016 Dec 20. [CrossRef]
- Couto, A.S., Kimura, E.A., Peres, V.J., Uhrig, M.L., Katzin, A.M. Active isoprenoid pathway in the intra-erythrocytic stages of Plasmodium falciparum presence of dolichols of 11 and 12 isoprene units. Biochem J. 1999, 341, 629-637.
- Zhang, B., Watts, K.M., Hodge, D., Kemp, L.M., Hunstad, D.A., Hicks, L.M., Odom, A.R. A second target of the antimalarial and antibacterial agent fosmidomycin revealed by cellular metabolic profiling. Biochemistry. 2011, 50, 3570-3577. https://doi.org/10.1021/bi200113y. Epub 2011 Apr 11. [CrossRef]
- Saggu, G.S., Garg, S., Pala, Z.R., Kochar, S.K., Saxena, V. Deciphering the role of IspD (2 C methyl D erythritol 4 phosphate cytidyltransferase) enzyme as a potential therapeutic drug target against Plasmodium vivax. Gene. 2018, 675, 240-253. https://doi.org/10.1016/j.gene.2018.06.084. Epub 2018 Jun 26. [CrossRef]
- Yeh, E., and DeRisi, J.L. Chemical Rescue of Malaria Parasites Lacking an Apicoplast Defines Organelle Function in Blood-Stage Plasmodium falciparum. PLoS Biol. 2011, 9, e1001138. https://doi.org/10.1371/journal.pbio.1001138. [CrossRef]
- Uddin, T., McFadden, G.I., Goodman, C.D. Validation of putative apicoplast-targeting drugs using a chemical supplementation assay in cultured human malaria parasites. Antimicrob Agents Chemother. 2018, 62(1), 10-1128. https://doi.org/10.1128/AAC.01161-17. [CrossRef]
- Zhang, M., Wang, C., Oberstaller, J., Thomas, P., Otto, T.D., Casandra, D., Adams, J.H. The apicoplast link to fever-survival and artemisinin-resistance in the malaria parasite. Nat Commun. 2021, 12(1), 4563. https://doi.org/10.1038/s41467-021-24814-1. [CrossRef]
- Swift, R.P., Rajaram, K., Elahi, R., Liu, H.B., Prigge, S.T. Roles of Ferredoxin-Dependent Proteins in the Apicoplast of Plasmodium falciparum Parasites. mBio. 2021, 13(1), e0302321-e0302321. https://doi.org/10.1128/mbio.03023-21. Epub 2022 Feb 15. [CrossRef]
- Saggu, G.S. Apicoplast Journey and Its Essentiality as a Compartment for Malaria Parasite Survival. Front Cell Infect Microbiol. 2022, 12, 881825. https://doi.org/10.3389/fcimb.2022.881825. [CrossRef]
- Painter, H.J., Morrisey, J.M., Mather, M.W., Vaidya, A.B. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature. 2007, 446(7131), 88-91. https://doi.org/10.1038/nature05572. [CrossRef]
- Guggisberg, A.M., Park, J., Edwards, R.L., Kelly, M.L., Hodge, D.M., Tolia, N.H., Odom, A.R. A sugar phosphatase regulates the methylerythritol phosphate (MEP) pathway in malaria parasites. Nat Commun. 2014, 5(1), 4467. doi: 10.1038/ncomms5467. [CrossRef]
- Jomaa, H., Wiesner, J., Sanderbrand, S., Altincicek, B., Weidemeyer, C., Hintz, M., Turbachova, I., Eberl, M., Zeidler, J., Lichtenthaler, H.K. Soldati, D. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science. 1999, 285(5433), 1573-1576. https://doi.org/10.1126/science.285.5433.1573. [CrossRef]
- Van Dooren, G.G., Stimmler, L.M., McFadden, G.I. Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol Rev. 2006, 30(4), 596-630. https://doi.org/10.1111/j.1574-6976.2006.00027.x. [CrossRef]
- Simão-Gurge, R.M., Wunderlich, G., Cricco, J.A., Cubillos, E.F.G., Doménech-Carbó, A., Cebrián-Torrejón, G., Katzin, A.M. Biosynthesis of heme O in intraerythrocytic stages of Plasmodium falciparum and potential inhibitors of this pathway. Sci Rep. 2019, 9(1), 19261. https://doi.org/10.1038/s41598-019-55506-y. [CrossRef]
- Bofill Verdaguer, I., Sussmann, R.A., Santiago, V.F., Palmisano, G., Moura, G.C., Mesquita, J.T., Crispim, M. Isoprenoid alcohols utilization by malaria parasites. Front Chem. 2022, 10, 1035548. https://doi.org/10.3389/fchem.2022.1035548. [CrossRef]
- Okada, M., Rajaram, K., Swift, R.P., Mixon, A., Maschek, J.A., Prigge, S.T., Sigala, P.A. Critical role for isoprenoids in apicoplast biogenesis by malaria parasites. Elife. 2022, 11, e73208. doi: 10.7554/eLife.73208. [CrossRef]
- Labaied, M., Jayabalasingham, B., Bano, N., Cha, S.J., Sandoval, J., Guan, G., Coppens, I. Plasmodium salvages cholesterol internalized by LDL and synthesized de novo in the liver. Cell Microbiol. 2011, 13, 569–586. DOI: 10.1111/j.1462-5822.2010.01555.x. [CrossRef]
- Banerjee, T., Jaijyan, D.K., Surolia, N., Singh, A.P., Surolia, A. Apicoplast triose phosphate transporter (TPT) gene knockout is lethal for Plasmodium. Mol Biochem Parasitol. 2012, 186(1), 44-50. https://doi.org/10.1016/j.molbiopara.2012.09.008. [CrossRef]
- Kuzuyama, T., Takagi, M., Kaneda, K. Studies on the nonmevalonate pathway: conversion of 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol to its 2-phospho derivative by 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase. Tetrahedron Lett. 2000, 41, 2925-2928. DOI: 10.1016/S0040-4039(00)00295-1. [CrossRef]
- Estévez, J.M., Cantero, A., Reindl, A., Reichler, S., León, P. 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J Biol Chem. 2001, 276(25), 22901-22909. DOI: 10.1074/jbc.M100854200. [CrossRef]
- Bahl, A., Brunk, B., Crabtree, J., Fraunholz, M.J., Gajria, B., Grant, G.R., Ginsburg, H., Gupta, D., Kissinger, J.C., and Labo, P. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003, 31, 212-215. https://doi.org/10.1093/nar/gkg081. [CrossRef]
- Bringer-Meyer, S., Sahm, H. Junctions of catabolic and anabolic pathways in Zymomonas mobilis: phosphoenolpyruvate carboxylase and malic enzyme. Appl Microbiol Biotechnol. 1989, 31, 529-536. https://doi.org/10.1007/bf00270789. [CrossRef]
- Sprenger, G.A., Schorken, U., Wiegert, T., Grolle, S., Graff, A.A.D., Taylor, S.V., Begley, T.P., Bringer-Meyer, S., Sahm, H. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc Natl Acad Sci U S A. 1997, 94, 12857-12862. https://doi.org/10.1073/pnas.94.24.12857. [CrossRef]
- Brammer, L.A., Smith, J.M., Wade, H., Meyers, C.F. 1-Deoxy-D-xylulose-5-phosphate synthase catalyzes a novel random sequential mechanism. J Biol Chem. 2011, 286(42), 36522-36531. https://doi.org/10.1074/jbc.M111.259747. doi: 10.1074/jbc.M111.259747. [CrossRef]
- Xiang, S., Usunow, G., Lange, G., Busch, M., Tong, L. Crystal structure of 1-deoxy-D-xylulose 5-phosphate synthase, a crucial enzyme for isoprenoids biosynthesis. J Biol Chem. 2007, 282(4), 2676-2682. https://doi.org/10.1074/jbc.M610235200. [CrossRef]
- Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., Tunyasuvunakool, K., Bates, R., Žídek, A., Potapenko, A. Bridgland, A., Highly accurate protein structure prediction with AlphaFold. Nature. 2021, 596(7873), 583-589. https://doi.org/10.1038/s41586-021-03819-2. [CrossRef]
- Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., Yuan, D., Stroe, O., Wood, G., Laydon, A. Žídek, A., AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, 439-444. https://doi.org/10.1093/nar/gkab1061. [CrossRef]
- Frank, R.A.W., Leeper, F.J., Luisi, B.F. Structure, mechanism and catalytic duality of thiamine-dependent enzymes. Cell Mol Life Sci. 2007, 64, 892-905. https://doi.org/10.1007/s00018-007-6423-5. [CrossRef]
- Eubanks, L.M., Poulter, C.D. Rhodobacter capsulatus 1-deoxy-D-xylulose 5-phosphate synthase: steady-state kinetics and substrate binding. Biochemistry. 2003, 42(4), 1140-1149. https://doi.org/10.1021/bi0205303. [CrossRef]
- Gierse, R.M., Reddem, E.R., Alhayek, A., Baitinger, D., Hamid, Z., Jakobi, H., Groves, M.R. Identification of a 1-deoxy-D-xylulose-5-phosphate synthase (DXS) mutant with improved crystallographic properties. Biochem Biophysical Res Commun. 2021, 539, 42-47. https://doi.org/10.1016/j.bbrc.2020.12.069. [CrossRef]
- Chen, P.Y.T., DeColli, A.A., Meyers, C.L.F., Drennan, C.L. X-ray crystallography–based structural elucidation of enzyme-bound intermediates along the 1-deoxy-d-xylulose 5-phosphate synthase reaction coordinate. J Biol Chem. 2019, 294(33), 12405-12414. https://doi.org/10.1074/jbc.RA119.009321. [CrossRef]
- Mac Sweeney, A., Lange, R., Fernandes, R.P., Schulz, H., Dale, G.E., Douangamath, A., Proteau, P.J. Oefner, C., The crystal structure of E. coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation. J Mol Biol. 2005, 345(1), 115-127. https://doi.org/10.1016/j.jmb.2004.10.030. [CrossRef]
- Nair, S.C., Brooks, C.F., Goodman, C.D., Sturm, A., McFadden, G.I., Sundriyal, S., Anglin, J.L., Song, Y., Moreno, S.N.J. Apicoplast isoprenoid precursor synthesis and the molecular basis of fosmidomycin resistance in Toxoplasma gondii. J Exp Med. 2011, 208, 1547-1559. https://doi.org/10.1084/jem.20110039. [CrossRef]
- Wang, X., Edwards, R.L., Ball, H., Johnson, C., Haymond, A., Girma, M., Dowd, C.S. MEPicides: α, β-unsaturated fosmidomycin analogues as DXR inhibitors against malaria. J Med Chem. 2018, 61(19), 8847-8858. https://doi.org/10.1021/acs.jmedchem.8b01026. [CrossRef]
- Phillips, A.M.F., Nogueira, F., Murtinheira, F., Barros, M.T. Synthesis and antimalarial evaluation of prodrugs of novel fosmidomycin analogues. Bioorg Med Chem Lett. 2015, 25(10), 2112-2116. https://doi.org/10.1016/j.bmcl.2015.03.077. [CrossRef]
- Girma, M.B., Ball, H.S., Wang, X., Brothers, R.C., Jackson, E.R., Meyers, M.J., Couch, R.D. Mechanism of Action of N-Acyl and N-Alkoxy Fosmidomycin Analogs: Mono-and Bisubstrate Inhibition of IspC from Plasmodium falciparum, a Causative Agent of Malaria. ACS omega. 2021, 6(42), 27630-27639. https://doi.org/10.1021/acsomega.1c01711. [CrossRef]
- Roca, C., Avalos-Padilla, Y., Prieto-Simón, B., Iglesias, V., Ramírez, M., Imperial, S., Fernàndez-Busquets, X. Selection of an Aptamer against the Enzyme 1-deoxy-D-xylulose-5-phosphate Reductoisomerase from Plasmodium falciparum. Pharmaceutics. 2022, 14(11), 2515. https://doi.org/10.3390/pharmaceutics14112515. [CrossRef]
- Gabrielsen, M., Rohdich, F., Eisenreich, W., Gräwert, T., Hecht, S., Bacher, A., Hunter, W. N. Biosynthesis of isoprenoids: a bifunctional IspDF enzyme from Campylobacter jejuni. Eur J Biochem. 2004, 271(14), 3028-3035. https://doi.org/10.1111/j.1432-1033.2004.04234.x. [CrossRef]
- Gabrielsen, M., Bond, C.S., Hallyburton, I., Hecht, S., Bacher, A., Eisenreich, W., Hunter, W.N. Hexameric assembly of the bifunctional methylerythritol 2, 4-cyclodiphosphate synthase and protein-protein associations in the deoxy-xylulose-dependent pathway of isoprenoid precursor biosynthesis. J Biol Chem. 2004, 279(50), 52753-52761. https://doi.org/10.1074/jbc.M408895200. [CrossRef]
- Liu, Y.L., Guerra, F., Wang, K., Wang, W., Li, J., Huang, C., Zhu, W., Houlihan, K., Li, Z., Zhang, Y. Nair, S.K., Structure, function and inhibition of the two-and three-domain 4Fe-4S IspG proteins. Proc Natl Acad Sci U S A. 2012, 109(22), 8558-8563. https://doi.org/10.1073/pnas.1121107109. [CrossRef]
- Lillo, A.M., Tetzlaff, C.N., Sangari, F.J., Cane, D.E. Functional expression and characterization of EryA, the erythritol kinase of Brucella abortus, and enzymatic synthesis of L-erythritol-4-phosphate. Bioorg Med Chem Lett. 2003, 13(4), 737-739. https://doi.org/10.1016/s0960-894x(02)01032-6. [CrossRef]
- Witschel, M.C., Höffken, H.W., Seet, M., Parra, L., Mietzner, T., Thater, F., Niggeweg, R., Röhl, F., Illarionov, B., Rohdich, F. Kaiser, J. Inhibitors of the herbicidal target IspD: allosteric site binding. Angew Chem Int Ed Engl. 2011, 50(34), 7931-7935. https://doi.org/10.1002/anie.201102281. [CrossRef]
- Wu, W., Herrera, Z., Ebert, D., Baska, K., Cho, S.H., DeRisi, J.L., Yeh, E.A. Chemical rescue screen identifies a Plasmodium falciparum apicoplast inhibitor targeting MEP isoprenoid precursor biosynthesis. Antimicrob Agents Chemother. 2015, 59(1), 356-364. https://doi.org/10.1128/AAC.03342-14. [CrossRef]
- Yao, Z.K., Krai, P.M., Merino, E.F., Simpson, M.E., Slebodnick, C., Cassera, M.B., Carlier, P.R. Determination of the active stereoisomer of the MEP pathway-targeting antimalarial agent MMV008138, and initial structure–activity studies. Bioorg Med Chem Lett. 2015, 25(7), 1515-1519. https://doi.org/10.1016/j.bmcl.2015.02.020. [CrossRef]
- Imlay, L.S., Armstrong, C.M., Masters, M.C., Li, T., Price, K.E., Edwards, R.L., Mann, K.M., Li, L.X., Stallings, C.L., Berry, N.G. Plasmodium IspD (2-C-Methyl-D-erythritol 4-Phosphate Cytidyltransferase), an Essential and Drug able Antimalarial Target. ACS Infect Dis. 2015, 1(4), 157-167. https://doi.org/10.1021/id500047s. [CrossRef]
- Bowman, J.D., Merino, E.F., Brooks, C.F., Striepen, B., Carlier, P.R., Cassera, M.B. Antiapicoplast and gametocytocidal screening to identify the mechanisms of action of compounds within the malaria box. Antimicrob Agents Chemother. 2014, 58(2), 811-819. https://doi.org/10.1128/AAC.01500-13. [CrossRef]
- Ghavami, M., Merino, E.F., Yao, Z.K., Elahi, R., Simpson, M.E., Fernández-Murga, M.L., Cassera, M.B. Biological studies and target engagement of the 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase (IspD)-targeting antimalarial agent (1 R, 3 S)-MMV008138 and analogs. ACS Infect Dis. 2018, 4(4), 549-559. https://doi.org/10.1021/acsinfecdis.7b00159. [CrossRef]
- Hirsch, A.K.H., Lauw, S., Gersbach, P., Schweizer, W.B., Rohdich, F., Eisenreich, W., Bacher, A., Diederich, F. Nonphosphate inhibitors of IspE protein, a kinase in the non-mevalonate pathway for isoprenoid biosynthesis and a potential target for antimalarial therapy. Chem Med Chem. 2007, 2, 806-810. https://doi.org/10.1002/cmdc.200700014. [CrossRef]
- Ahn C.S., Pai H.S. Physiological function of IspE, a plastid MEP pathway gene for isoprenoid biosynthesis, in organelle biogenesis and cell morphogenesis in Nicotiana benthamiana. Plant Mol Biol. 2008, 66(5):503-517. https://doi.org/10.1007/s11103-007-9286-0. [CrossRef]
- Bitok, J.K., Meyers, C.F. 2 C-Methyl-D-erythritol 4-phosphate enhances and sustains cyclodiphosphate synthase IspF activity. ACS Chem Biol. 2012, 7(10), 1702-1710. https://doi.org/10.1021/cb300243w. [CrossRef]
- O’Rourke, P.E., Kalinowska-Tłuścik, J., Fyfe, P.K., Dawson, A., Hunter, W.N. Crystal structures of IspF from Plasmodium falciparum and Burkholderia cenocepacia: comparisons inform antimicrobial drug target assessment. BMC Struct Biol. 2014, 14(1), 1. https://doi.org/10.1186/1472-6807-14-1. [CrossRef]
- Crane, C.M., Kaiser, J., Ramsden, N.L., Lauw, S., Rohdich, F., Eisenreich, W., Hunter, W.N, Bacher, A., Diederich, F. Fluorescent Inhibitors for IspF, an Enzyme in the Non-Mevalonate Pathway for Isoprenoid Biosynthesis and a Potential Target for Antimalarial Therapy. Angew Chem Int Ed Engl. 2006, 45(7), 1069-1074. https://doi.org/10.1002/anie.200503003. [CrossRef]
- Geist, J.G., Lauw, S., Illarionova, V., Illarionov, B., Fischer, M., Gräwert, T., Rohdich, F., Eisenreich, W., Kaiser, J., Groll, M. Scheurer, C. Thiazolopyrimidine inhibitors of 2-methylerythritol 2, 4-cyclodiphosphate synthase (IspF) from Mycobacterium tuberculosis and Plasmodium falciparum. Chem Med Chem. 2010, 5(7), 1092-1101. https://doi.org/10.1002/cmdc.201000083. [CrossRef]
- Pala, Z.R., Saxena, V., Saggu, G.S., Garg, S. Recent advances in the [Fe–S] cluster biogenesis (SUF) pathway functional in the apicoplast of Plasmodium. Trends Parasitol. 2018, 34(9), 800-809. https://doi.org/10.1016/j.pt.2018.05.010. [CrossRef]
- Quitterer, F., Frank, A., Wang, K., Rao, G., O'Dowd, B., Li, J. Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE). J Mol Biol. 2015, 427, 2220-2228. https://doi.org/10.1016/j.jmb.2015.04.002. [CrossRef]
- Rekittke, I., Olkhova, E., Wiesner, J., Demmer, U., Warkentin, E., Jomaa, H., Ermler, U. Structure of the (E)-4-hydroxy-3-methyl-but-2-enyl-diphosphate reductase from Plasmodium falciparum. FEBS Lett. 2013, 587(24), 3968-3972. https://doi.org/10.1016/j.febslet.2013.10.029. [CrossRef]
- Larkin, M.A., Blackshields, G., Brown, N., Chenna, R., McGettigan, P.A., McWilliam, H., Valentin, F., Wallace, I.M., Wilm, A., Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007, 23, 2947-2948. https://doi.org/10.1093/bioinformatics/btm404. [CrossRef]
- Foth, B.J., Ralph, S.A., Tonkin, C.J., Struck, N.S., Fraunholz, M., Roos, D.S., Cowman, A.F., McFadden, G. I. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science. 2003, 299, 705-708. https://doi.org/10.1126/science.1078599. [CrossRef]
- Petersen, T.N., Brunak, S., von Heijne, G., and Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011, 8, 785-786. https://doi.org/10.1038/nmeth.1701. [CrossRef]
- Marchler-Bauer, A., Derbyshire, M.K., Gonzales, N.R., Lu, S., Chitsaz, F., Geer, L.Y., Geer, R.C., He, J., Gwadz, M., Hurwitz, D.I. Lanczycki, C.J. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015, 43, 222-226. https://doi.org/10.1093/nar/gku1221. [CrossRef]
- Paysan-Lafosse, T., Blum, M., Chuguransky, S., Grego, T., Pinto, B.L., Salazar, G.A., Bateman, A. InterPro in 2022. Nucleic Acids Res. 2023, 51, 418-427. https://doi.org/10.1093/nar/gkac993. [CrossRef]
- Sigrist, C.J., Cerutti, L., Hulo, N., Gattiker, A., Falquet, L., Pagni, M., Bairoch, A., Bucher, P. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 2002, 3, 265-274. https://doi.org/10.1093/bib/3.3.265. [CrossRef]
- Jones, D.T., Taylor, W.R., Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Computer applications in the biosciences: Comput Appl Biosci. 1992, 8(3), 275-282. https://doi.org/10.1093/bioinformatics/8.3.275. [CrossRef]
- Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018 1;35(6):1547-1549. https://doi.org/10.1093/molbev/msy096. [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985, 783-791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x. [CrossRef]
- Söding, J., Biegert, A., Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, 244-248. https://doi.org/10.1093/nar/gki408. [CrossRef]
- Sali, A. Blundell, T.L. Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol. 1993, 234(3), 779-815. https://doi.org/10.1006/jmbi.1993.1626. [CrossRef]
- Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A.E., Berendsen, H.J. GROMACS: fast, flexible, and free. J Comput Chem. 2005, 26, 1701-1718. https://doi.org/10.1002/jcc.20291. [CrossRef]
- Yang, J., Roy, A., Zhang, Y. BioLiP: a semi-manually curated database for biologically relevant ligand–protein interactions. Nucleic Acids Res. 2013, 41, 1096-1103. https://doi.org/10.1093/nar/gks966. [CrossRef]
- Thomsen, R., Christensen, M.H. Mol Dock: a new technique for high-accuracy molecular docking. J Med Chem. 2006, 49, 3315-3321. https://doi.org/10.1021/jm051197e. [CrossRef]
- Antonova-Koch, Y., Meister, S., Abraham, M., Luth, M.R., Ottilie, S., Lukens, A.K., Winzeler, E.A. Open-source discovery of chemical leads for next-generation chemoprotective antimalarials. Science. 2018, 362(6419), eaat9446. https://doi.org/10.1126/science.aat9446. [CrossRef]
| Gene name | Features | P. falciparum | P. cynomolgi | P. berghei | T. gondii | P. vivax | E. coli | B. subtilus | A. thaliana |
|---|---|---|---|---|---|---|---|---|---|
| DXS | Accession No. | PF3D7_1337200 | PCYB_122560 | PBANKA_1351000 | TGRH88_022190 | PVX_082790 | EDV65838 | AGG61826 | AEE83625 |
| Gene size (bp) / Encoded protein size (KDa) |
3618/132 | 2964/108.5 | 2532/92 | 4239/155.3 | 3336/122.2 | 1863/68.2 | 1302/47.6 | 2154/78.8 | |
| CDD region | TPP DXS | ||||||||
| (425-750) | (367-695) | (200-425) | (600-925) | (360-679) | (45-285) | (40-280) | (115-365) | ||
| TPP PYR DXS | |||||||||
| (825-990) | (775-925) | (500-650) | (1020-1175) | (757-913) | (320-480) | (325-475) | (400-560) | ||
| Transketolase C | |||||||||
| (1065-1175) | (765-920) | (700-825) | (1300-1380) | (955-1097) | (495-605) | (493-615) | (578-700) | ||
| Signature motif R-x(3)-[LIVMTA]- [DENQSTHKF]-x(5,6)-[GSN]-G-H-[PLIVMF]-[GSTA]-x(2)-[LIMC]-[GS] |
935-951 (GEDGATHQGIYDLSYLG) |
874-890 (GEDGATHQGIYDLTFLG) |
597-613 (GEDGATHQGIYDLSYLG) |
1111- 1132 (GPDGSTHQ GSFDLAYLG) |
860-876 (GEDGATHQGIYDLTFLG) |
28-47 (RQFLITSL SASGGHIG PNLG) |
36-55 (RRYLLD SVSRSSG HFASGL G) |
102-121 (RSDVIFNV SKTGGHLG SSLG) |
|
| IspC | Accession No. | PF3D7_1467300 | PCYB_124770 | PBANKA_1330600 | TGME49_214850 | PVX_117100 | EDV66507 | AGG61028 | Q9XFS9 |
| Gene size (bp) / Encoded protein size (KDa) |
1467/53.6 | 1557/56.9 | 1245/45.6 | 1899/69.5 | 1587/58 | 1197/43.7 | 1152/42.1 | 1434/52.4 | |
| CDD region | DXP reductoisom | ||||||||
| (80-210) | (135-262) | (30-165) | (190-320) | (118-250) | (4-132) | (5-130) | (85-210) | ||
| DXP redisom C | |||||||||
| (225-322) | (280-375) | (175-270) | (382-470) | (265-361) | (146-238) | (145-225) | (225-307) | ||
| DXPR C | |||||||||
| (360-480) | (410-480) | (305-410) | (500-620) | (396-516) | (270-390) | (260-380) | (340-465) | ||
| Signature motif | Not identified | ||||||||
| IspD | Accession No. | PF3D7_0106900 | PCYB_021710 | PBANKA_0206400 | TGME49_306260 | PVX_081425 | EDV68085 | AGG59433 | P69834 |
| Gene size (bp) / Encoded protein size (KDa) |
2205/80.7 | 1248/45.6 | 867/31.7 | 1137/41.5 | 1857/67.9 | 711/25.9 | 699/25.5 | 909/33.2 | |
| CDD region | CDP-ME Synthetase (GTA type super family) | ||||||||
| (510-675) | (140-350) | (95-235) | (255-365) | (191-558) | (7-222) | (5-220) | (80-292) | ||
| Signature motif [IVT]-[LIVMC]-[IVT]- [HS]-D-[SGAV]-[AV]-R |
512-519 (ILIHDGAR) |
221-228 (ILVHDGAR) |
94-101 (ILVHDAAR) |
265-272 (VMIHDAAR) |
424-431 (ILVHDGAR) |
102-109 (VLVHDAAR) | 100-107 (VLVHDGAR) |
175-182 (VCIHDSAR) |
|
| IspE | Accession No. | PF3D7_0503100 | PCYB_103980 | PBANKA_1102800 | TGME49_306550 | PVX_097660 | YP_489475 | AGG59389 | AAG01340 |
| Gene size (bp) / Encoded protein size (KDa) |
1614/59 | 1581/57.8 | 303/12.1 | 3648/133.6 | 1524/55.7 | 852/31.1 | 870/31.7 | 1152/42.1 | |
| CDD region | IspE (135-380) | PRK00650 (135-495) |
PRK02534 (6-70) |
PHA03247 (135-350) |
PRK00650 (131-497) |
GHMP kinases N | |||
| (92-148) | (85-142) | (160-215) | |||||||
| GHMP kinases C | |||||||||
| (188-262) | (195-275) | ||||||||
| Signature motif | Not identified | ||||||||
| IspF | Accession No. | PF3D7_0209300 | PCYB_042060 | PBANKA_0306400 | TGME49_255690 | PVX_003920 | YP_490955 | AGG59434 | NP_850971 |
| Gene size (bp) / Encoded Protein size (KDa) |
723/26.4 | 840/30.6 | 540/19.6 | 1017/37.1 | 864/31.5 | 480/17.4 | 477/17.3 | 696/25.4 | |
| CDD region | MECDP Synthase | ||||||||
| (65-235) | (90-270) | (5-175) | (175-305) | (106-282) | (2-154) | (3-155) | (75-227) | ||
| Signature motif S-[DN]-[GA]-D-LIVAP]-[LIVAG]-x-H-[STAC]-x(2)-[DNT]-[SAG]-x(2)-[SGA] |
116-131 (SDGDIIYHSIVDSILG) |
152-167 (SDGDVVFHALVDALLG) |
56-71 (SDGDIIYHALVDSILG) |
186-201 (SDGDVLLHAVCDAVFG) |
163-178 (SDGDVIFHALVDALLG) |
35-50 (SDGDVALHALTDALLG) |
36-51 (SDADVLLHTVADACLG) |
109-124 (SDGDVLLHCVVDAILG) |
|
| IspG | Accession No. | PF3D7_1022800 | PCYB_061710 | PBANKA_0507000 | TGME49_262430 | PVX_111575 | EDV66700 | AGG61906 | AAL91150 |
| Gene size (bp) / Encoded protein size (KDa) |
2475/90.6 | 1797/65.7 | 2460/90 | 3852/141.3 | 2463/90.2 | 1119/40.9 | 1134/41.4 | 2226/81.5 | |
| CDD region | GcpE super family | ||||||||
| (125-400 & 720-820) |
(30-180 & 490-590) |
(125-400 & 660-810) |
(385-680 & 1150-1280) | (110-385 & 712-813) | (1-360) | (1-360) | (87-270 & 600-700) |
||
| Signature motif | Not identified | ||||||||
| IspH | Accession No. | PF3D7_0104400 | PCYB_021940 | PBANKA_0208700 | TGME49_227420 | PVX_081535 | EDV65216 | WP_003237046 | Q94B35 |
| Gene size (bp) / Encoded protein size (KDa) |
1608/58.8 | 1311/47.9 | 1050/38.3 | 2157/78.9 | 1281/46.8 | 951/34.7 | 945/34.5 | 1401/51.2 | |
| CDD region | LytB IspH | ||||||||
| (225-495) | (135-390) | (30-310) | (390-660) | (112-388) | (2-282) | (2-282) | (115-450) | ||
| Signature motif [LIVMAC]-[LIVFYWA]- {DYP}-[DN]-P-P-[FYW] |
497-503 (LLTNPPF) |
110-118 (CGSGACGGC) |
304-310 (VLTNPPY) |
601-609 (VVGSEASSN) |
387-393 (VLTGEPF) |
219-227) (VVGSKNSSN) |
220-228 (VVGDPKSNN) |
373-381 (VVGGWNSSN) |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





