Submitted:
27 November 2023
Posted:
30 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and method
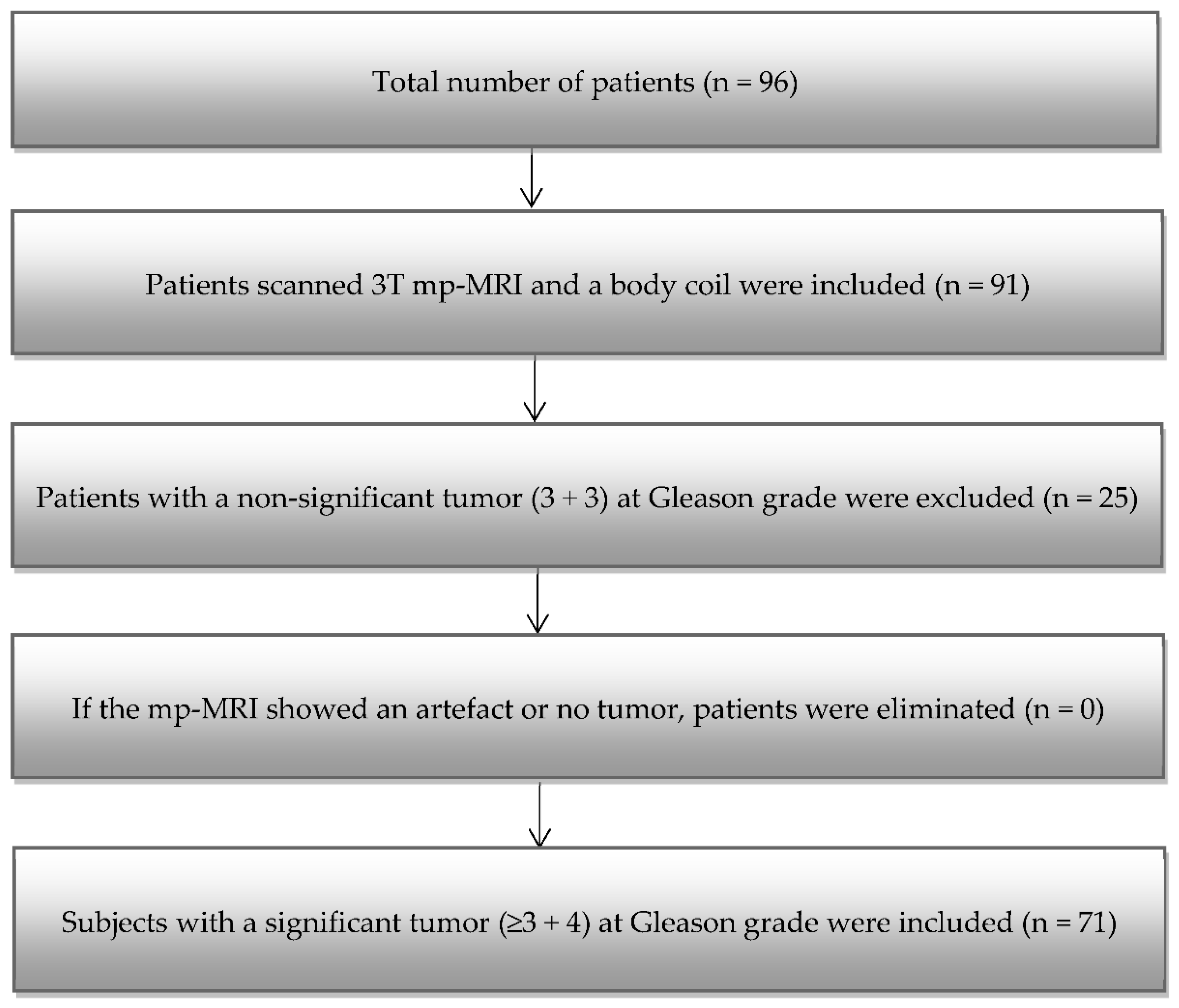
2.1. Patient Group
2.2. Multiparametric Magnetic Resonance Imaging
2.3. Segmentation
2.4. Feature extraction
2.5. Feature selection
2.5.1. Classification and Prediction
2.6. Statistical analysis
3. Result
3.1. Patients
3.2. Association between radiomic attributes and significant versus non-tumor regions
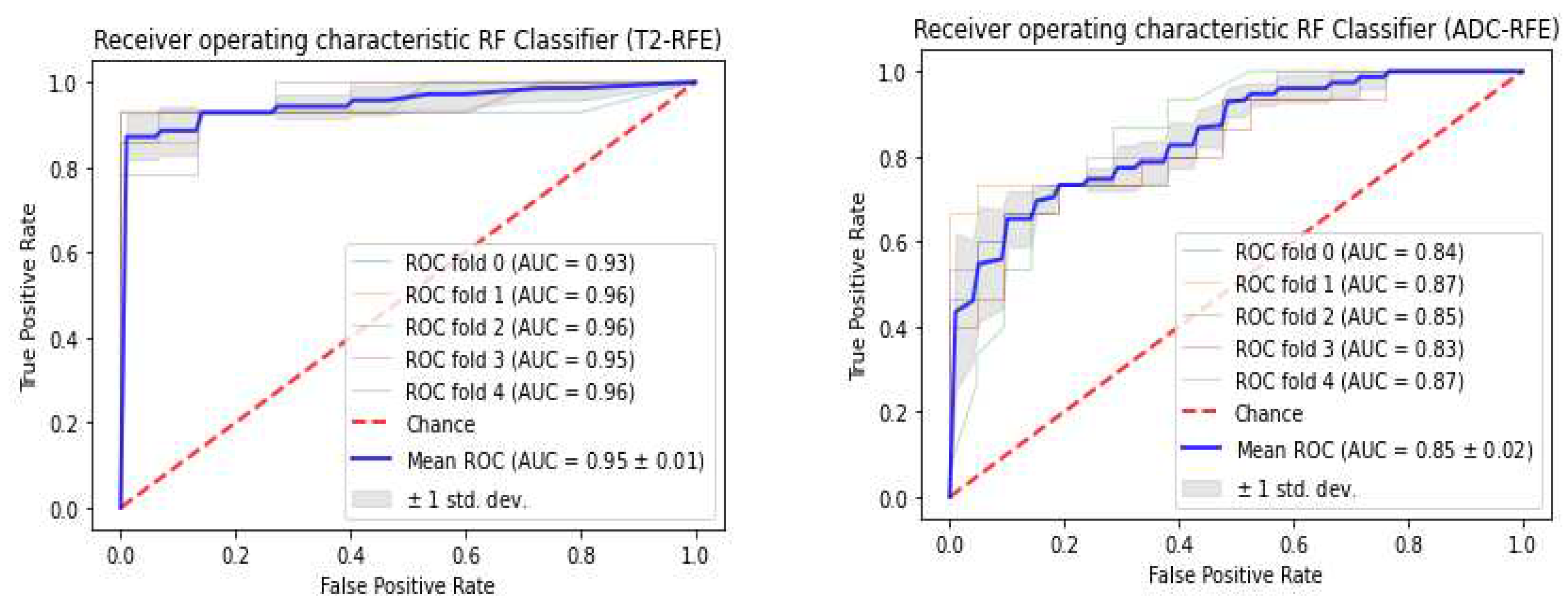
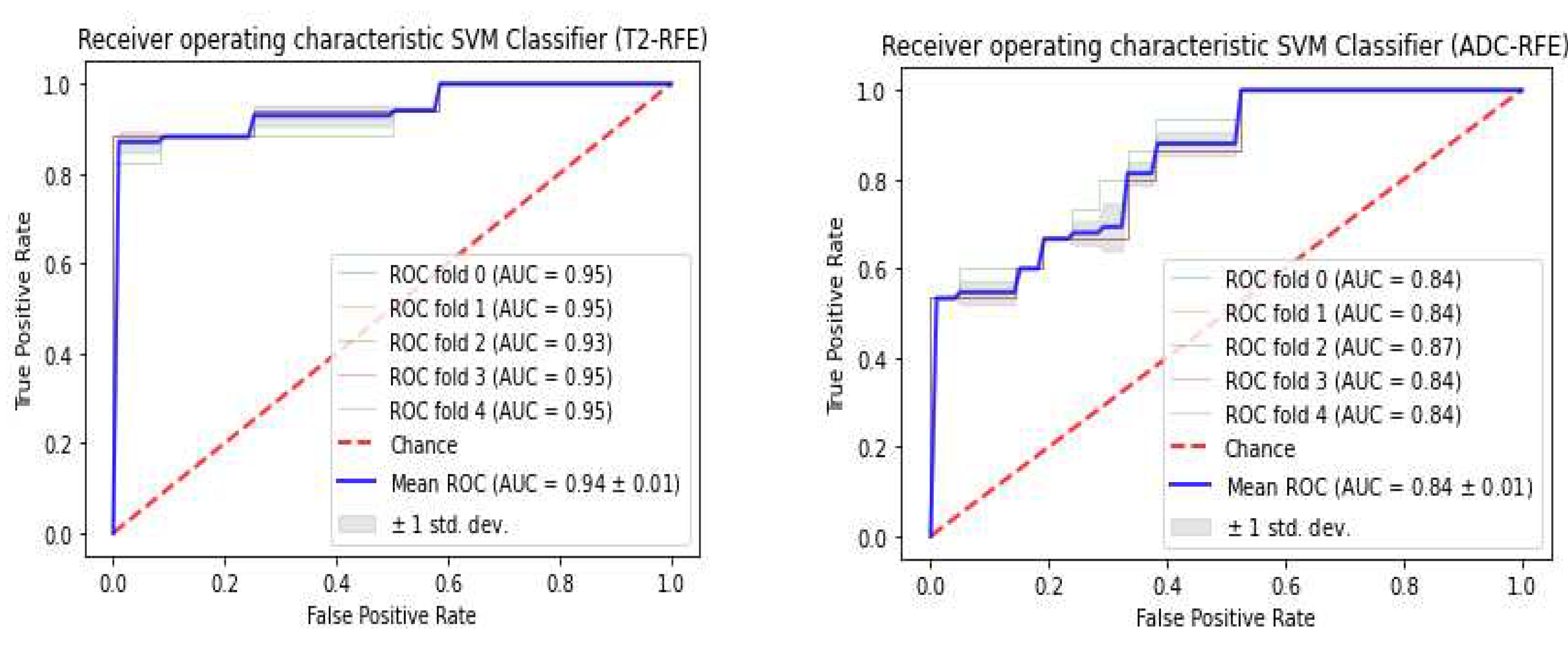
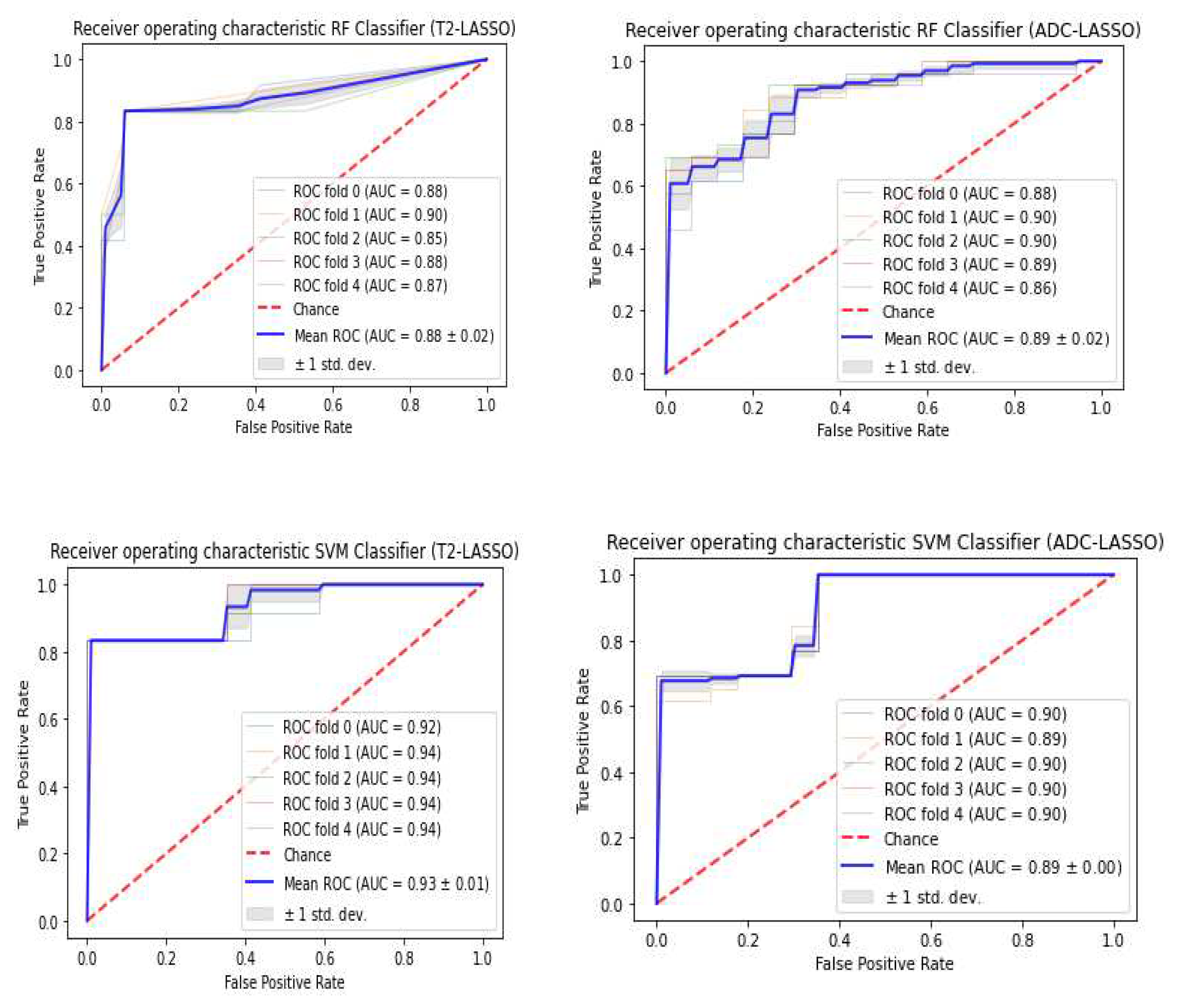
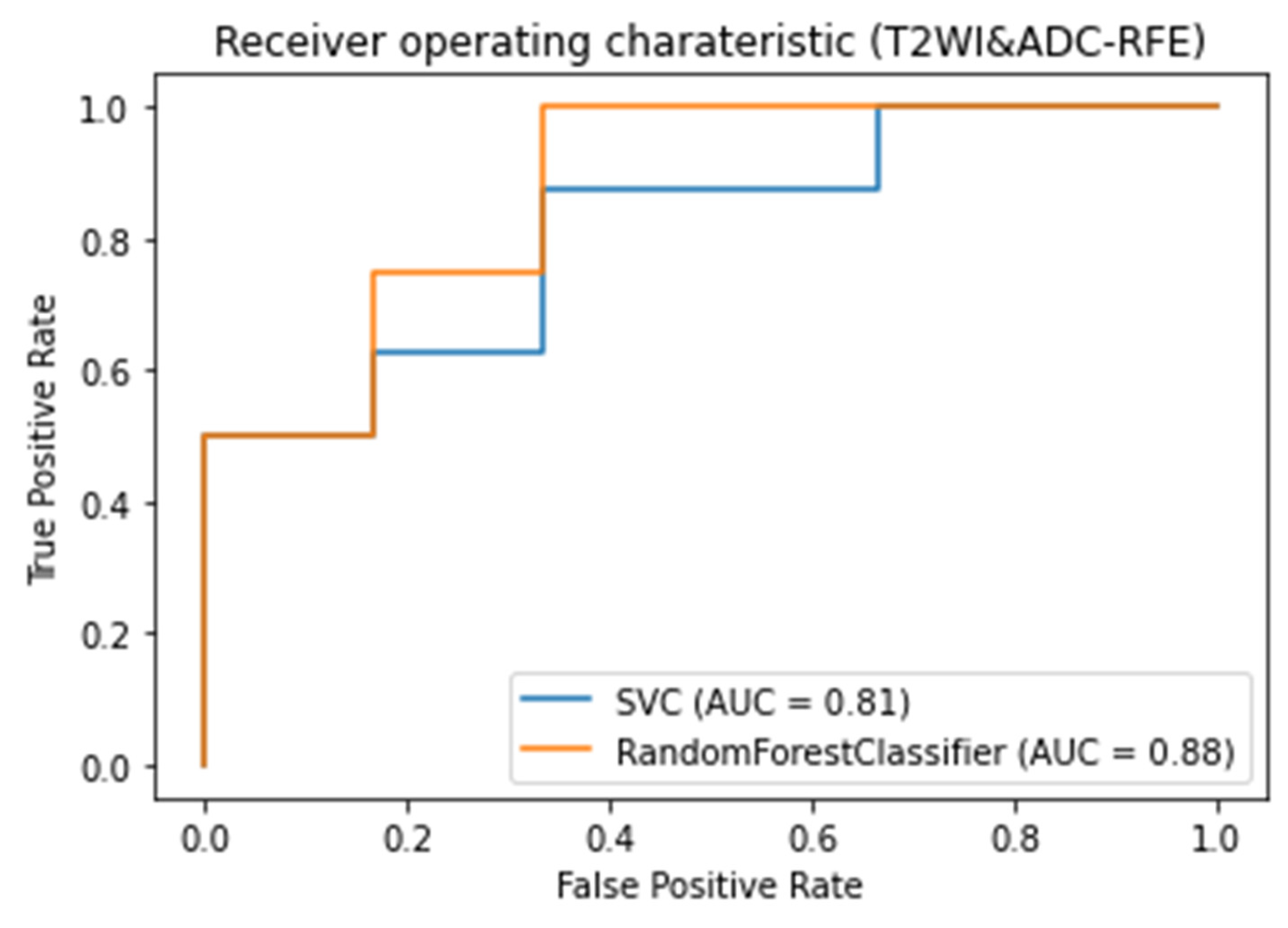
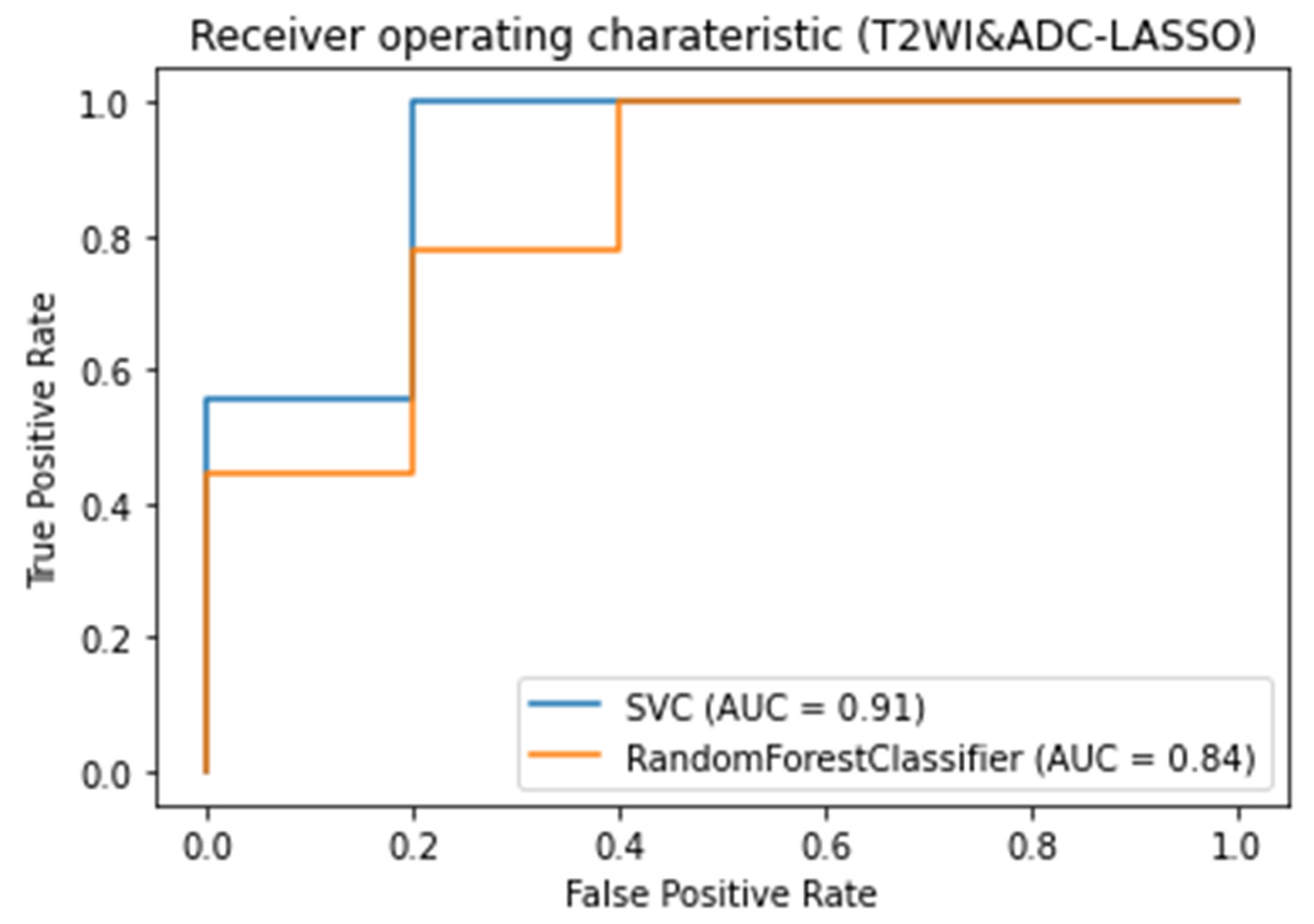
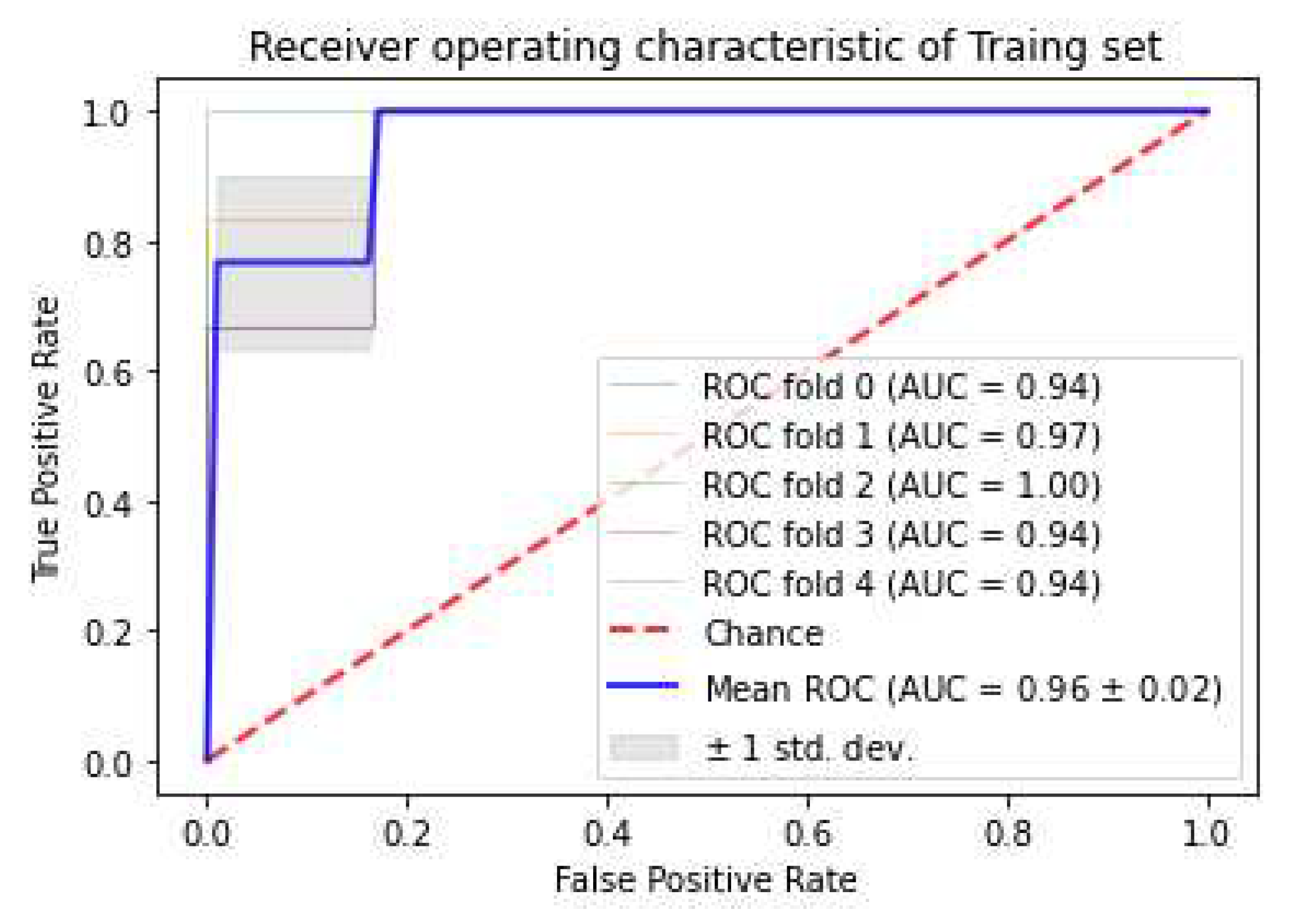
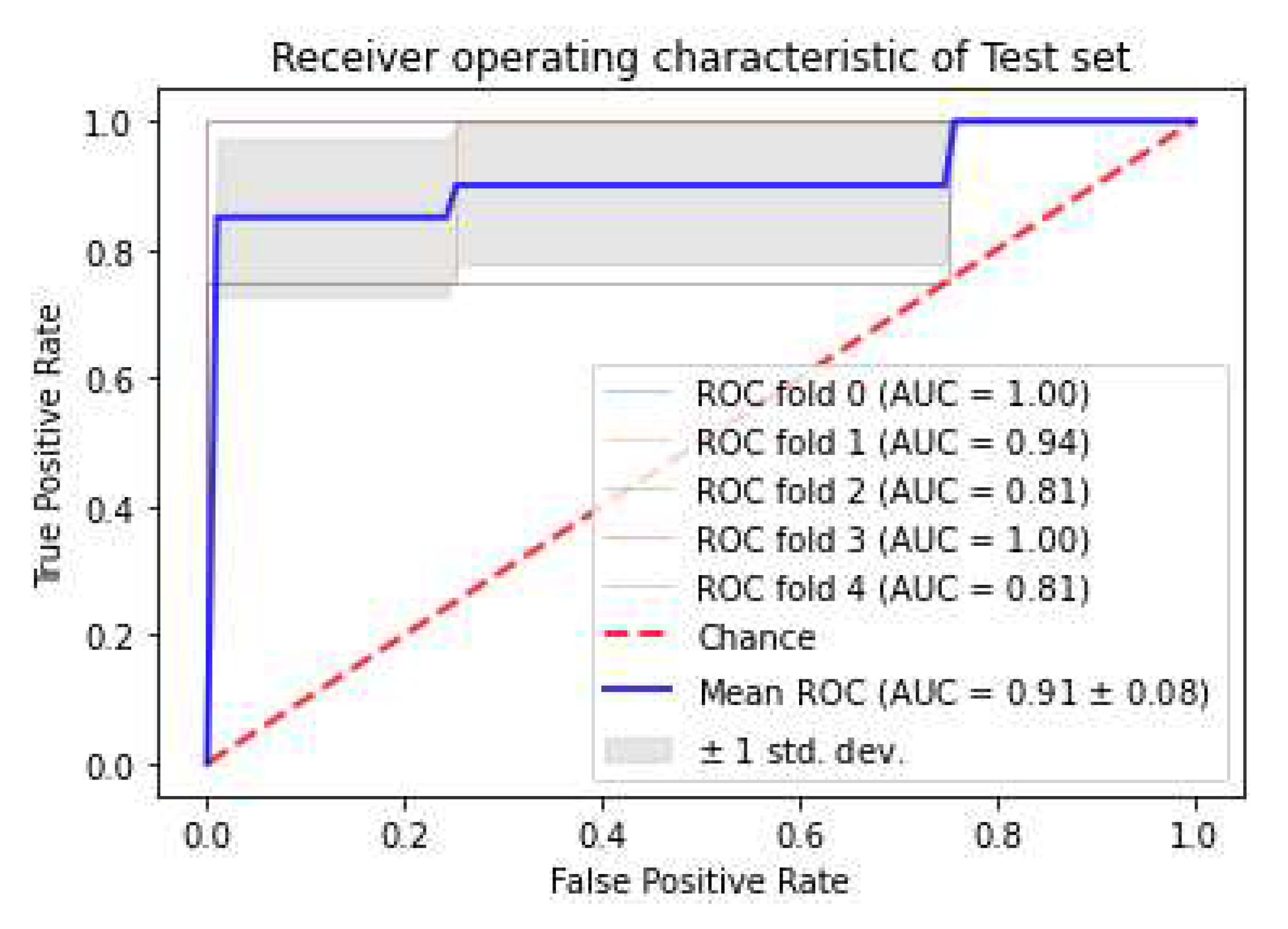
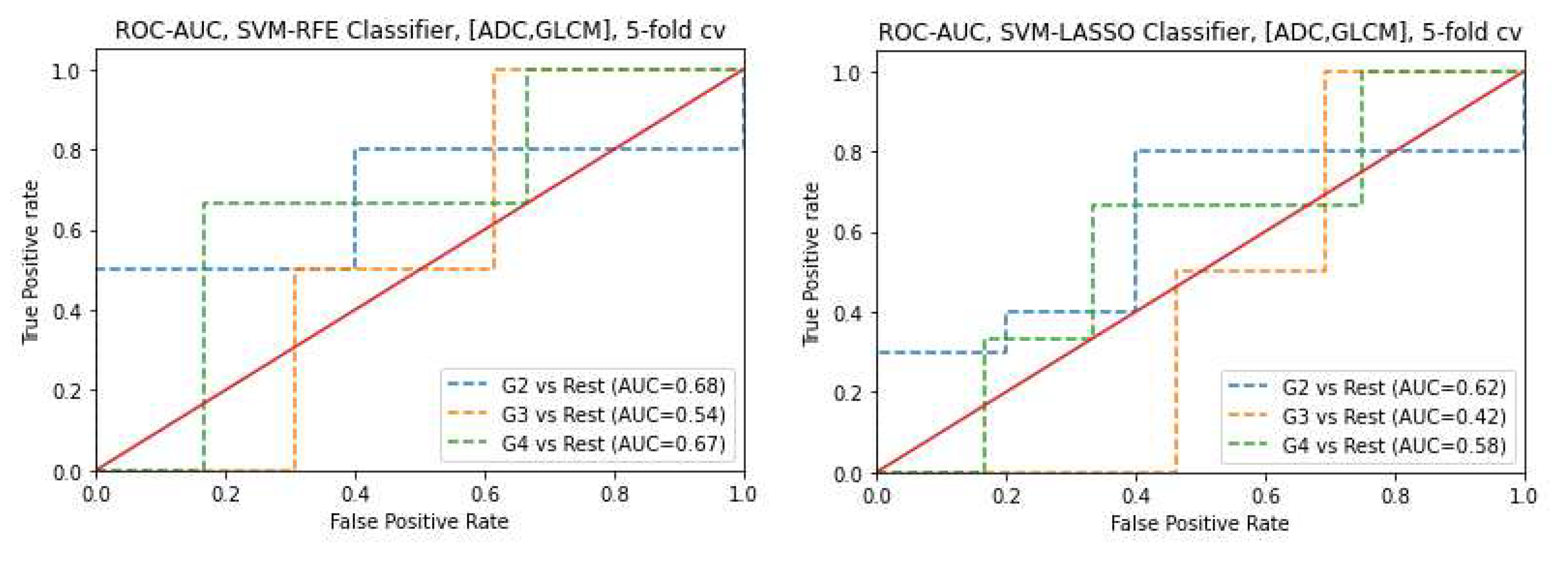
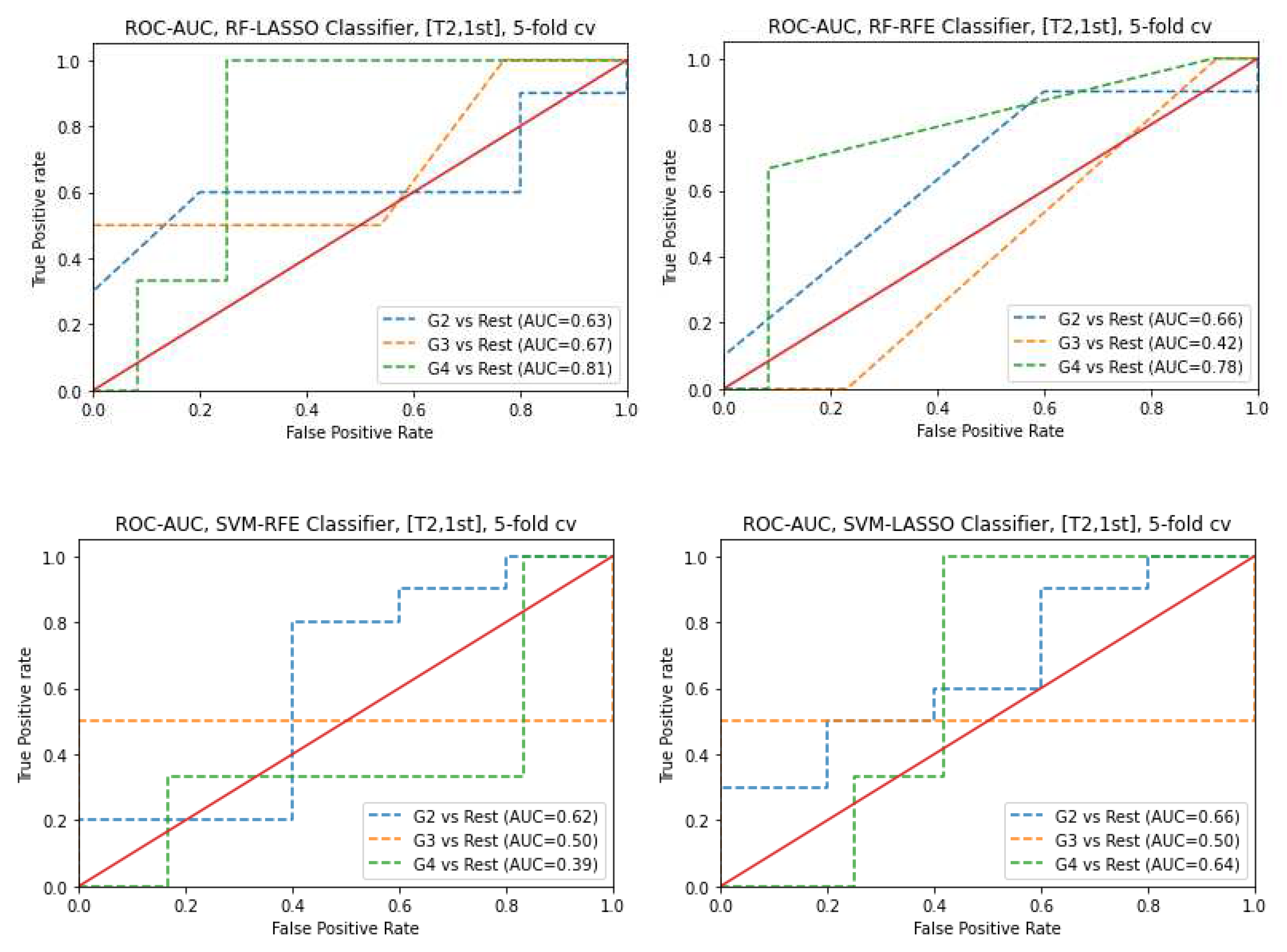
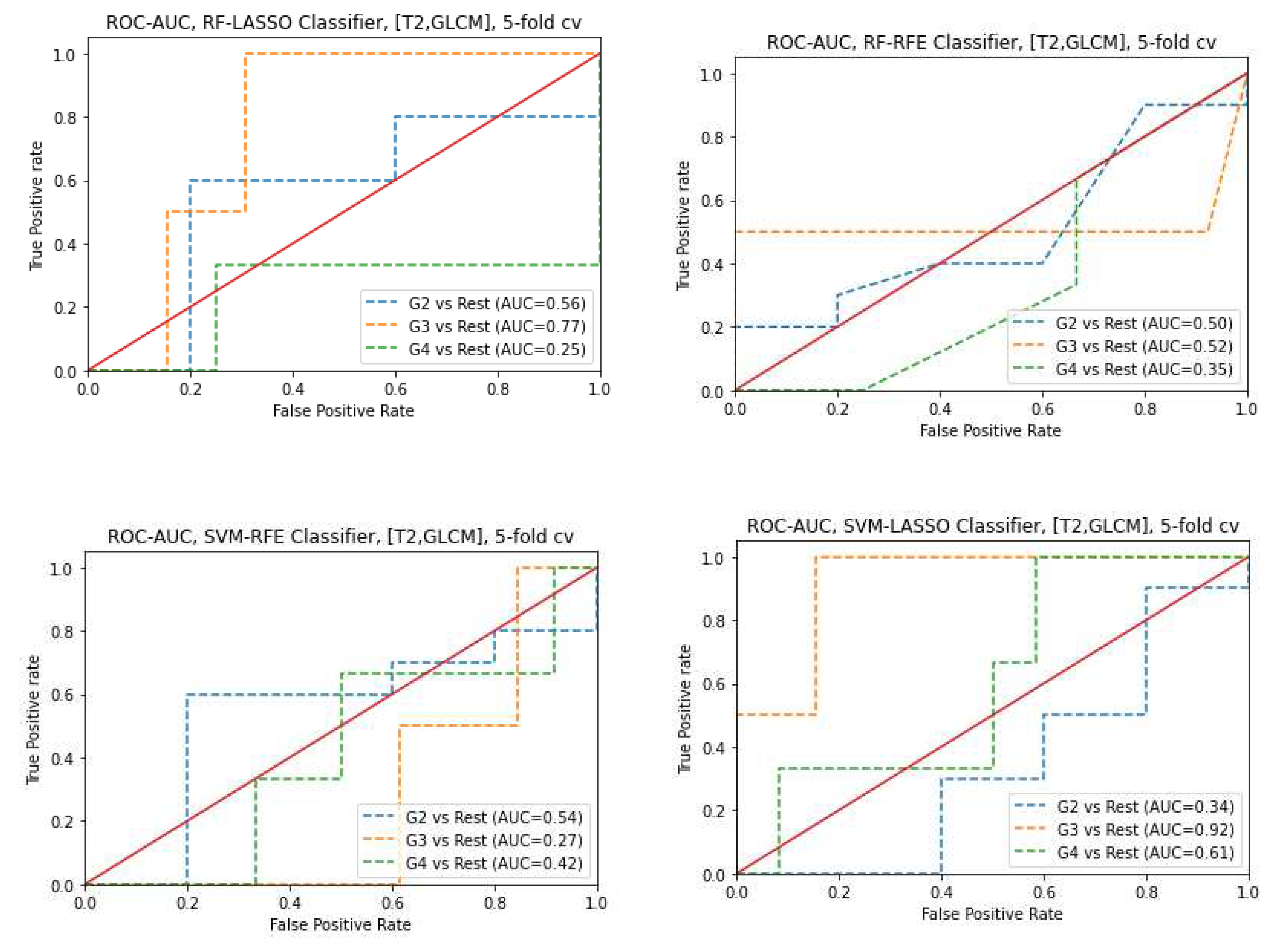
3.3. Classifiers and Feature Selection Performance
3.4. Model strength and performance variations
3.5. Association between GS and Radiomics Attributes
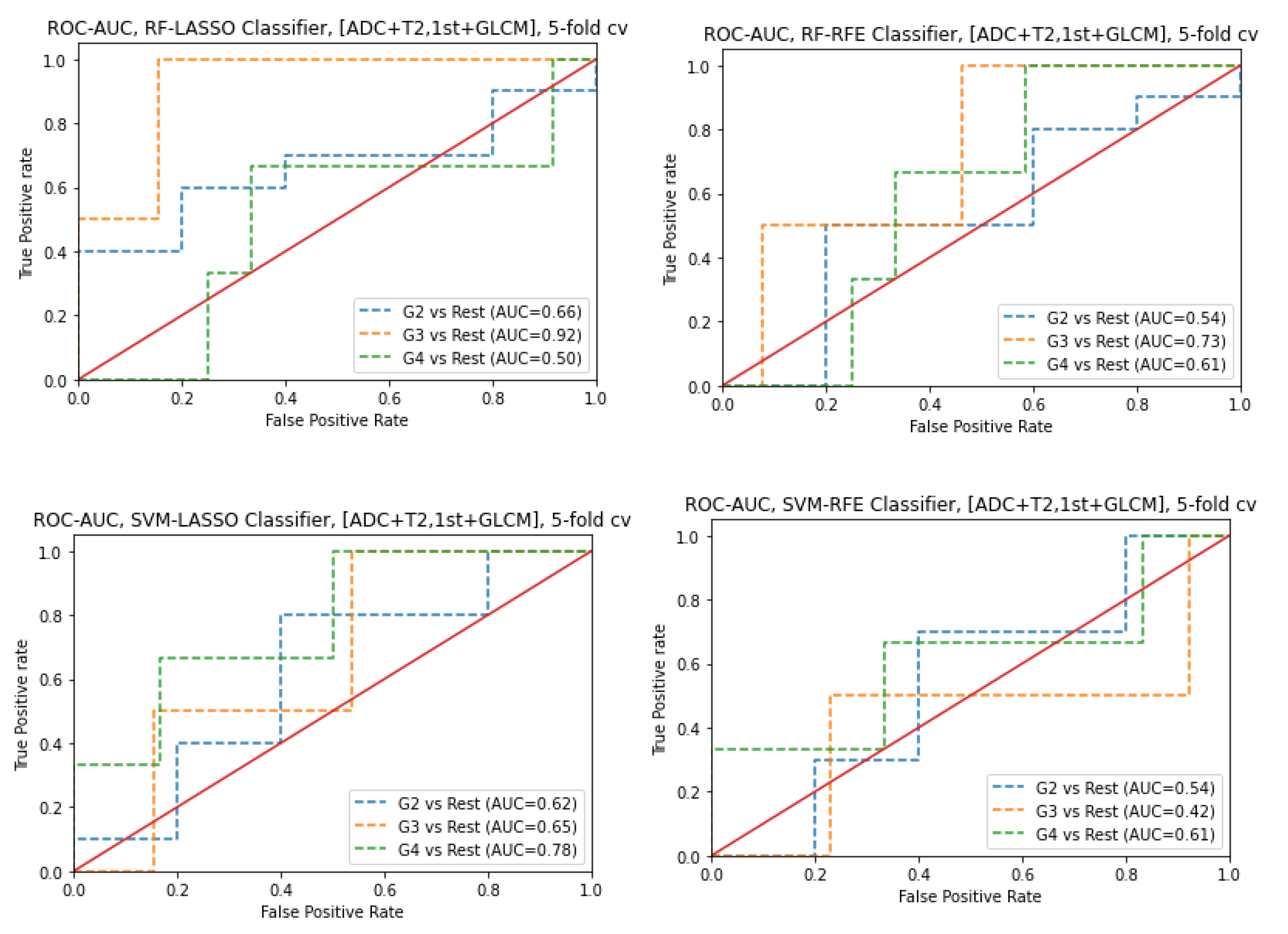
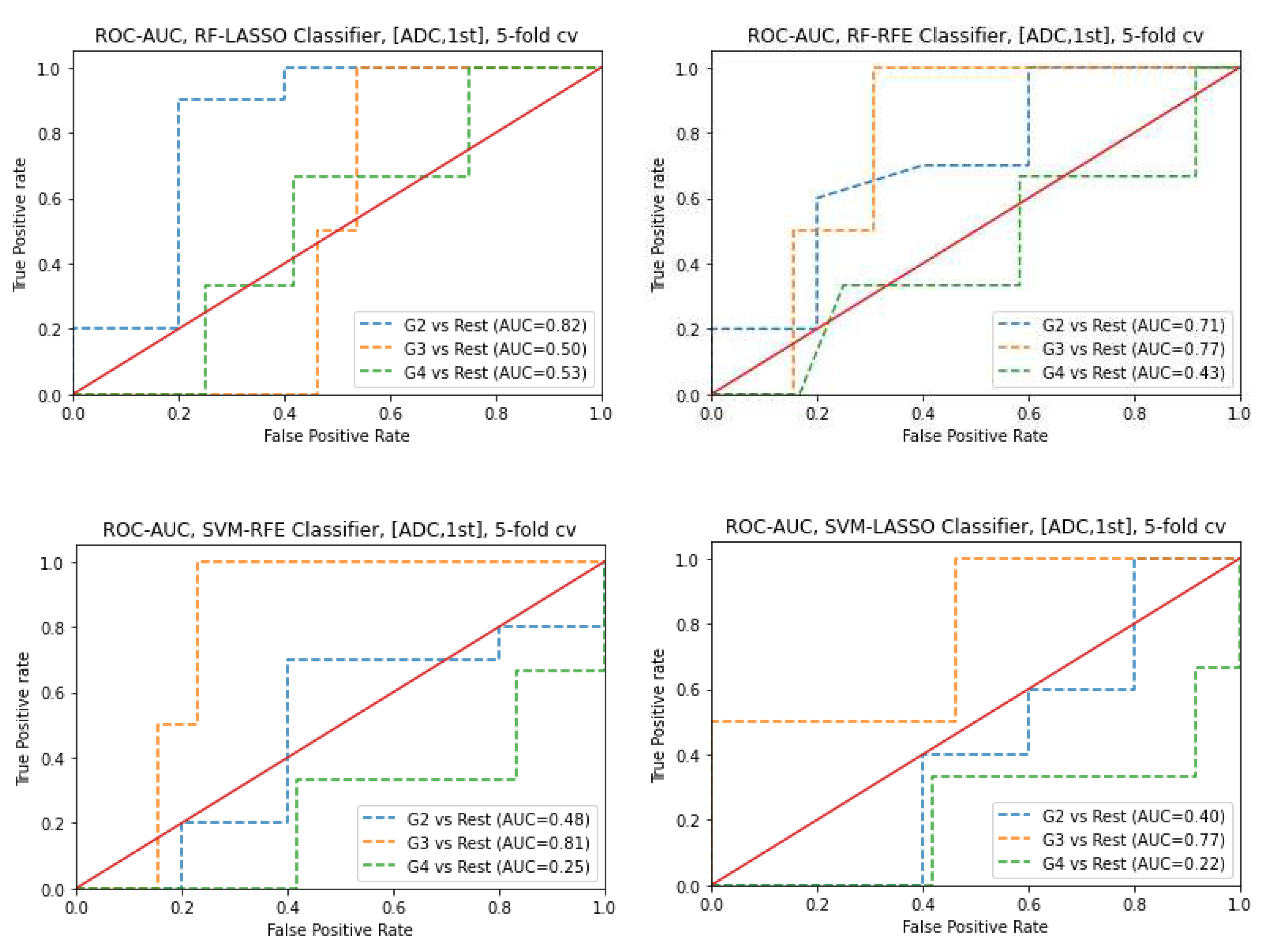
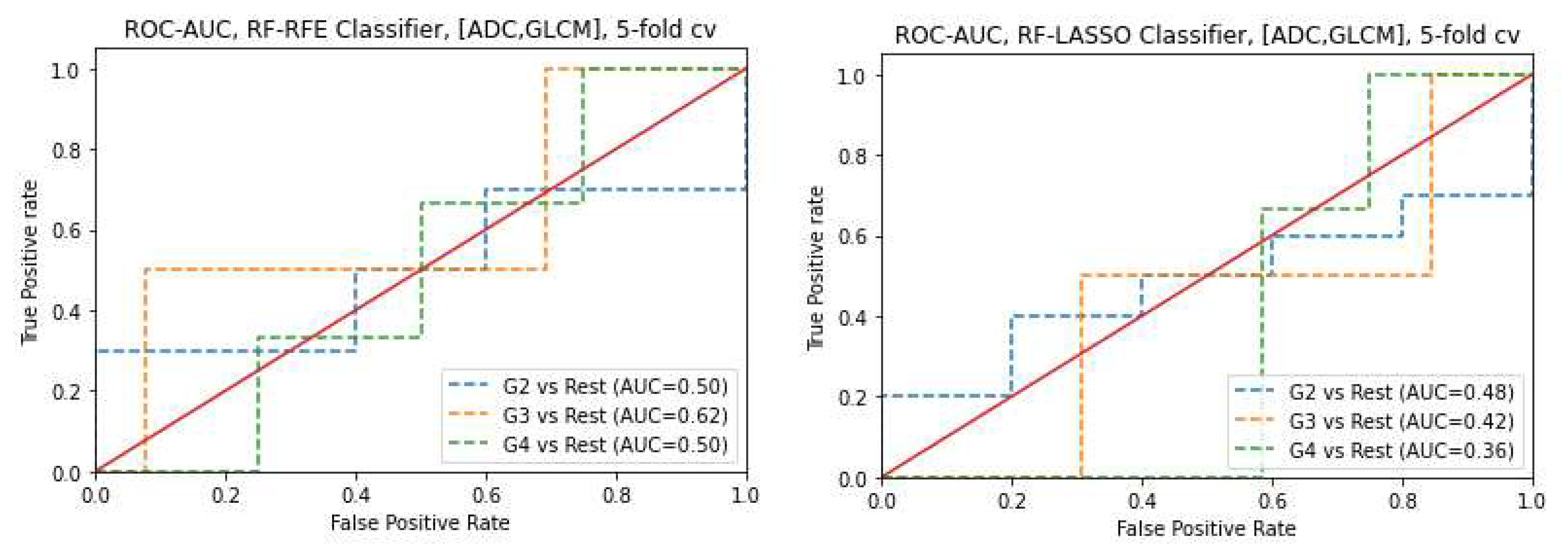
3.6. Prediction of Gleason score
4. Discussion
- Significant cancer versus non-tumor regions
- GS prediction
5. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgements
Conflicts of Interest
References
- Johnson, L.M., et al., Multiparametric MRI in prostate cancer management. Nature reviews Clinical oncology, 2014. 11(6): p. 346. [CrossRef]
- Zhang, Y., Chen, W., Yue, X., Shen, J., Gao, C., Pang, P., Cui, F. and Xu, M., 2020. Development of a novel, multi-parametric, MRI-based radiomic nomogram for differentiating between clinically significant and insignificant prostate cancer. Frontiers in oncology, 10, p.888. [CrossRef]
- Bray, F., et al., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians, 2018. 68(6): p. 394-424. [CrossRef]
- Rawla, P., Epidemiology of prostate cancer. World journal of oncology, 2019. 10(2): p. 63.
- Sekhoacha, M., Riet, K., Motloung, P., Gumenku, L., Adegoke, A. and Mashele, S., 2022. Prostate cancer review: Genetics, diagnosis, treatment options, and alternative approaches. Molecules, 27(17), p.5730. [CrossRef]
- Wang, J., et al., Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. European radiology, 2017. 27(10): p. 4082-4090. [CrossRef]
- Zhu, X., Shao, L., Liu, Z., Liu, Z., He, J., Liu, J., Ping, H. and Lu, J., 2023. MRI-derived radiomics models for diagnosis, aggressiveness, and prognosis evaluation in prostate cancer. Journal of Zhejiang University-SCIENCE B, 24(8), pp.663-681.
- Sun, Y., et al., Multiparametric MRI and radiomics in prostate cancer: a review. Australasian physical & engineering sciences in medicine, 2019. 42(1): p. 3-25. [CrossRef]
- Turkbey, B. and P.L. Choyke, Multiparametric MRI and prostate cancer diagnosis and risk stratification. Current opinion in urology, 2012. 22(4): p. 310. [CrossRef]
- Gibbs, P., M.D. Pickles, and L.W. Turnbull, Diffusion imaging of the prostate at 3.0 tesla. Investigative radiology, 2006. 41(2): p. 185-188. [CrossRef]
- Cabarrus, M.C. and A.C. Westphalen, Multiparametric magnetic resonance imaging of the prostate—a basic tutorial. Translational andrology and urology, 2017. 6(3): p. 376. [CrossRef]
- Radulescu, E., et al., Abnormalities in fronto-striatal connectivity within language networks relate to differences in grey-matter heterogeneity in Asperger syndrome. Neuroimage Clin, 2013. 2: p. 716-26. [CrossRef]
- Katahira, K., et al., Ultra-high-b-value diffusion-weighted MR imaging for the detection of prostate cancer: evaluation in 201 cases with histopathological correlation. European radiology, 2011. 21(1): p. 188-196. [CrossRef]
- Rosenkrantz, A.B., et al., Computed diffusion-weighted imaging of the prostate at 3 T: impact on image quality and tumour detection. European radiology, 2013. 23(11): p. 3170-3177. [CrossRef]
- Nagarajan, R., et al., Correlation of Gleason scores with diffusion-weighted imaging findings of prostate cancer. Advances in urology, 2012. 2012. [CrossRef]
- Hambrock, T., et al., Prostate cancer: computer-aided diagnosis with multiparametric 3-T MR imaging—effect on observer performance. Radiology, 2013. 266(2): p. 521-530. [CrossRef]
- Lambin, P., et al., Radiomics: extracting more information from medical images using advanced feature analysis. European journal of cancer, 2012. 48(4): p. 441-446. [CrossRef]
- Alic, L., W.J. Niessen, and J.F. Veenland, Quantification of heterogeneity as a biomarker in tumor imaging: a systematic review. PloS one, 2014. 9(10): p. e110300. [CrossRef]
- Ghezzo, S., Bezzi, C., Presotto, L., Mapelli, P., Bettinardi, V., Savi, A., Neri, I., Preza, E., Gajate, A.M.S., De Cobelli, F. and Scifo, P., 2022. State of the art of radiomic analysis in the clinical management of prostate cancer: A systematic review. Critical Reviews in Oncology/Hematology, 169, p.103544. [CrossRef]
- Nioche, C., et al., LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer research, 2018. 78(16): p. 4786-4789. [CrossRef]
- Fisher, R., L. Pusztai, and C. Swanton, Cancer heterogeneity: implications for targeted therapeutics. British journal of cancer, 2013. 108(3): p. 479-485. [CrossRef]
- Gillies, R.J., P.E. Kinahan, and H. Hricak, Radiomics: images are more than pictures, they are data. Radiology, 2016. 278(2): p. 563-577. [CrossRef]
- Bi, W.L., et al., Artificial intelligence in cancer imaging: clinical challenges and applications. CA: a cancer journal for clinicians, 2019. 69(2): p. 127-157. [CrossRef]
- Van Griethuysen, J.J., et al., Computational radiomics system to decode the radiographic phenotype. Cancer research, 2017. 77(21): p. e104-e107.
- De Santi, B., Salvi, M., Giannini, V., Meiburger, K.M., Marzola, F., Russo, F., Bosco, M. and Molinari, F., 2020, July. Comparison of Histogram-based Textural Features between Cancerous and Normal Prostatic Tissue in Multiparametric Magnetic Resonance Images. In 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) (pp. 1671-1674). IEEE.
- Woźnicki, P., Westhoff, N., Huber, T., Riffel, P., Froelich, M.F., Gresser, E., von Hardenberg, J., Mühlberg, A., Michel, M.S., Schoenberg, S.O. and Nörenberg, D., 2020. Multiparametric MRI for prostate cancer characterization: Combined use of radiomics model with PI-RADS and clinical parameters. Cancers, 12(7), p.1767. [CrossRef]
- Kniep, H.C., et al., Radiomics of brain MRI: utility in prediction of metastatic tumor type. Radiology, 2019. 290(2): p. 479-487. [CrossRef]
- Lin, M., et al., Prostate lesion delineation from multiparametric magnetic resonance imaging based on locality alignment discriminant analysis. Medical physics, 2018. 45(10): p. 4607-4618. [CrossRef]
- Yin, P., et al., Comparison of radiomics machine-learning classifiers and feature selection for differentiation of sacral chordoma and sacral giant cell tumour based on 3D computed tomography features. European radiology, 2019. 29(4): p. 1841-1847. [CrossRef]
- Maniruzzaman, M., et al., Statistical characterization and classification of colon microarray gene expression data using multiple machine learning paradigms. Computer methods and programs in biomedicine, 2019. 176: p. 173-193. [CrossRef]
- Zhang, Y., et al., Radiomics analysis for the differentiation of autoimmune pancreatitis and pancreatic ductal adenocarcinoma in 18F-FDG PET/CT. Medical physics, 2019. 46(10): p. 4520-4530. [CrossRef]
- Zhang, X., et al., Optimizing a machine learning based glioma grading system using multi-parametric MRI histogram and texture features. Oncotarget, 2017. 8(29): p. 47816. [CrossRef]
- Hameed, M., et al., The clinical utility of prostate cancer heterogeneity using texture analysis of multiparametric MRI. International urology and nephrology, 2019. 51(5): p. 817-824. [CrossRef]
- Lavesson, N. and P. Davidsson. Quantifying the impact of learning algorithm parameter tuning. in AAAI. 2006.
- Mantovani, R.G., et al. To tune or not to tune: recommending when to adjust SVM hyper-parameters via meta-learning. in 2015 International joint conference on neural networks (IJCNN). 2015. Ieee.
- Probst, P., B. Bischl, and A.-L. Boulesteix, Tunability: Importance of hyperparameters of machine learning algorithms. arXiv preprint. arXiv:1802.09596, 2018.
- Weerts, H.J., A.C. Mueller, and J. Vanschoren, Importance of tuning hyperparameters of machine learning algorithms. arXiv preprint. arXiv:2007.07588, 2020.
- Valarmathi, R. and T. Sheela, Heart disease prediction using hyper parameter optimization (HPO) tuning. Biomedical Signal Processing and Control, 2021. 70: p. 103033. [CrossRef]
- Montgomery, D.C., Design and analysis of experiments. 2017: John wiley & sons.
- Litjens, G., et al., Computer-aided detection of prostate cancer in MRI. IEEE transactions on medical imaging, 2014. 33(5): p. 1083-1092. [CrossRef]
- Barentsz, J.O., et al., ESUR prostate MR guidelines 2012. European radiology, 2012. 22(4): p. 746-757. [CrossRef]
- Kitajima, K., et al., Prostate cancer detection with 3 T MRI: comparison of diffusion-weighted imaging and dynamic contrast-enhanced MRI in combination with T2-weighted imaging. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine, 2010. 31(3): p. 625-631. [CrossRef]
- Clark, K., et al., The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. Journal of digital imaging, 2013. 26(6): p. 1045-1057. [CrossRef]
- Erickson, B.J., et al., Machine learning for medical imaging. Radiographics, 2017. 37(2): p. 505-515.
- Chatterjee, S., D. Dey, and S. Munshi, Integration of morphological preprocessing and fractal based feature extraction with recursive feature elimination for skin lesion types classification. Computer methods and programs in biomedicine, 2019. 178: p. 201-218. [CrossRef]
- Fan, M., et al., Integration of dynamic contrast-enhanced magnetic resonance imaging and T2-weighted imaging radiomic features by a canonical correlation analysis-based feature fusion method to predict histological grade in ductal breast carcinoma. Physics in Medicine & Biology, 2019. 64(21): p. 215001. [CrossRef]
- Liu, Y., et al., Early prediction of acute xerostomia during radiation therapy for nasopharyngeal cancer based on delta radiomics from CT images. Quantitative imaging in medicine and surgery, 2019. 9(7): p. 1288. [CrossRef]
- Wang, X., et al., Classification of pulmonary lesion based on multiparametric MRI: Utility of radiomics and comparison of machine learning methods. European radiology, 2020. 30(8): p. 4595-4605. [CrossRef]
- Chen, X., et al., Applying a new quantitative image analysis scheme based on global mammographic features to assist diagnosis of breast cancer. Computer methods and programs in biomedicine, 2019. 179: p. 104995. [CrossRef]
- Chen, L., et al., Primary tumor site specificity is preserved in patient-derived tumor xenograft models. Frontiers in genetics, 2019: p. 738. [CrossRef]
- Geetha, R., et al., Cervical cancer identification with synthetic minority oversampling technique and PCA analysis using random forest classifier. Journal of medical systems, 2019. 43(9): p. 1-19. [CrossRef]
- Peng, Y., et al., Quantitative analysis of multiparametric prostate MR images: differentiation between prostate cancer and normal tissue and correlation with Gleason score—a computer-aided diagnosis development study. Radiology, 2013. 267(3): p. 787-796. [CrossRef]
- Chawla, N.V., N. Japkowicz, and A. Kotcz, Special issue on learning from imbalanced data sets. ACM SIGKDD explorations newsletter, 2004. 6(1): p. 1-6.
- Weiss, G.M., Mining with rarity: a unifying framework. ACM Sigkdd Explorations Newsletter, 2004. 6(1): p. 7-19.
- Fehr, D., et al., Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proceedings of the National Academy of Sciences, 2015. 112(46): p. E6265-E6273. [CrossRef]
- Holm, S., A simple sequentially rejective multiple test procedure. Scandinavian journal of statistics, 1979: p. 65-70.
- Epstein, J.I., et al., The 2014 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma. The American journal of surgical pathology, 2016. 40(2): p. 244-252. [CrossRef]
- Fan, X., et al., Multiparametric MRI and Machine Learning Based Radiomic Models for Preoperative Prediction of Multiple Biological Characteristics in Prostate Cancer. Frontiers in oncology, 2022. 12. [CrossRef]
- Hamerla, G., et al., Comparison of machine learning classifiers for differentiation of grade 1 from higher gradings in meningioma: a multicenter radiomics study. Magnetic resonance imaging, 2019. 63: p. 244-249. [CrossRef]
- Larroza, A., et al., Support vector machine classification of brain metastasis and radiation necrosis based on texture analysis in MRI. Journal of magnetic resonance imaging, 2015. 42(5): p. 1362-1368. [CrossRef]
- Rustam, Z. and N. Angie. Prostate Cancer Classification Using Random Forest and Support Vector Machines. in Journal of Physics: Conference Series. 2021. IOP Publishing. [CrossRef]
- Sun, Y., et al., Radiomic features of pretreatment MRI could identify T stage in patients with rectal cancer: preliminary findings. Journal of Magnetic Resonance Imaging, 2018. 48(3): p. 615-621.
- Wang, L., et al., Assessment of biologic aggressiveness of prostate cancer: correlation of MR signal intensity with Gleason grade after radical prostatectomy. Radiology, 2008. 246(1): p. 168-176. [CrossRef]
- Wibmer, A., et al., Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. European radiology, 2015. 25(10): p. 2840-2850. [CrossRef]
- Qiao, X., Gu, X., Liu, Y., Shu, X., Ai, G., Qian, S., Liu, L., He, X. and Zhang, J., 2023. MRI Radiomics-Based Machine Learning Models for Ki67 Expression and Gleason Grade Group Prediction in Prostate Cancer. Cancers, 15(18), p.4536. [CrossRef]
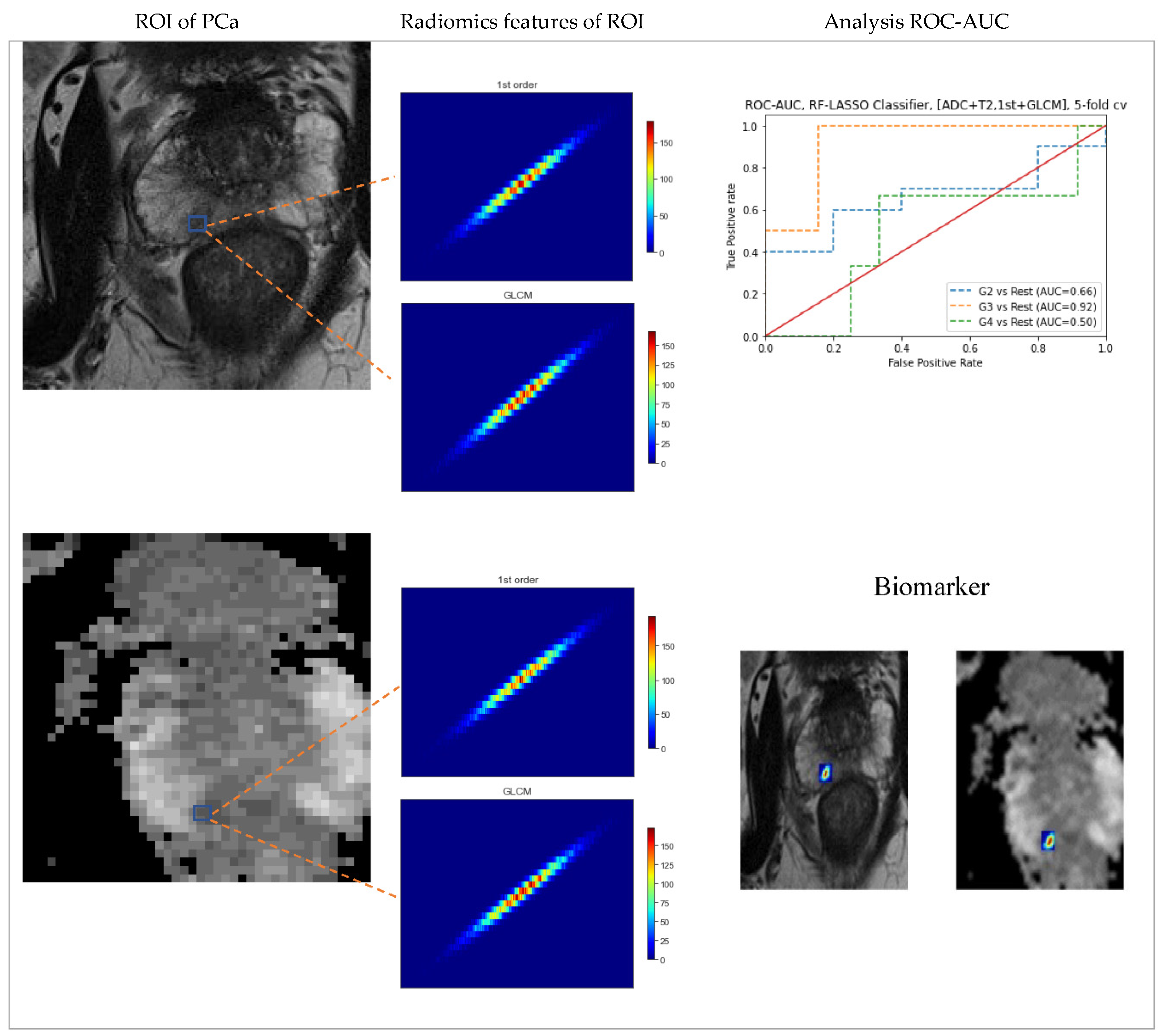
| Sequence parameter | T2WI | ADC |
|---|---|---|
| Repetition time (ms) | 5560 | 2700 |
| Echo time (ms) | 104 | 63 |
| Flip angle (degrees) | 160 | 90 |
| Bandwidth (Hz/px) | 200 | 1500 |
| Phase FoV % | 100 | 65.625 |
| Slice thickness (mm) | 3 | 3 |
| Slice gap (mm) | 3 | 3 |
| Average | 4 | 8 |
| Phase encoding direction | Row | Row |
| Number of acquisitions | 1 | 1 |
| Feature | Median (interquartile 25th,50th, and 75th percentiles) | P | |
|---|---|---|---|
| Significant cancer | nontumor regions | ||
| ADC | |||
| 1st order | |||
| Skewness | 0.37 (0.02, 0.37, 0.69) | 0.03 (-0.29, 0.03, 0.47) | 0.001 |
| Kurtosis | -0.49 (-0.85, -0.49, 0.29) | -0.54 (-0.86, -0.54, -0.12) | 0.52 |
| Entropylog1o | 1.14 (1.09, 1.14, 1.19) | 1.09 (1.05, 1.09, 1.14) | ˂ 0.001 |
| Entropylog2 | 3.80 (3.62, 3.80, 3.97) | 3.62 (3.50, 3.62, 3.80) | ˂ 0.001 |
| Uniformity | 0.07 (0.06, 0.07, 0.08) | 0.08 (0.07, 0.08, 0.10) | ˂ 0.001 |
| GLCM | |||
| JointEntropyLog2 | 6.18 (5.85, 6.18, 6.45) | 6.05(5.83,6.05,6.21) | 0.03 |
| JointEntropyLog10 | 1.86 (1.79, 1.86, 1.94) | 1.82(1.75,1.82,1.87) | 0.006 |
| Angular Second Moment | 0.01 (0.01, 0.1, 0.01) | 0.016 (0.014, 0.016, 0.019) | 0.006 |
| Contrast | 145.64 (107.88, 145.64, 201.96) | 84.02 (59.85, 84.02, 122.08) | ˂ 0.001 |
| Dissimilarity | 9.30 (8.33, 930, 11.36) | 7.51 (6.19, 7.51, 8.61) | ˂ 0.001 |
| Correlation | 0.18 (0.06, 0.18, 0.35) | 0.23 (0.09, 0.23, 0.39) | 0.37 |
| T2WI | |||
| 1st order | |||
| Skewness | 0.07 (-0.20, 0.07, 0.32) | 0.15 (-0.20, 0.15, 0.43) | 0.50 |
| Kurtosis | -0.18 (-0.55, -0.018, 0.43) | -0.34 (-0.59, -0.34, 0.11) | 0.18 |
| Entropylog1o | 1.30 (1.23, 1.30, 1.41) | 1.06 (0.97, 1.06, 1.16) | ˂ 0.001 |
| Entropylog2 | 4.34 (4.11, 4.34, 4.69) | 3.52 (3.24, 3.52, 3.85) | ˂ 0.001 |
| Uniformity | 0.05 (0.04, 0.05, 0.06) | 0.09 (0.07, 0.09, 0.12) | ˂ 0.001 |
| GLCM | |||
| JointEntropyLog2 | 7.50 (6.89, 7.50, 8.16) | 6.45 (5.95,6.45,7.12) | ˂0.001 |
| JointEntropyLog10 | 2.31 (2.12, 2.31, 2.50) | 1.96 (1.79,1.96,2.14) | ˂ 0.001 |
| Angular Second Moment | 0.006 (0.004, 0.006, 0.01) | 0.01 (0.01, 0.01, 0.02) | ˂ 0.001 |
| Contrast | 92.42 (64.22, 92.42, 132.48) | 13.36 (10.08, 13.36, 20.47) | ˂ 0.001 |
| Dissimilarity | 7.62 (6.36, 7.62, 9.07) | 2.88 (2.46, 2.88, 3.60) | ˂ 0.001 |
| Correlation | 0.25 (0.13, 0.25, 0.35) | 0.38 (0.24, 0.38, 0.50) | ˂ 0.001 |
| Feature | r | P |
|---|---|---|
| ADC | ||
| 1st order | ||
| Skewness | 0.315 | ˂0.001 |
| Entropylog1o | 0.305 | ˂0.001 |
| Entropylog2 | 0.305 | ˂0.001 |
| Uniformity | -0.331 | ˂0.001 |
| GLCM | ||
| Angular Second Moment | -0.236 | 0.005 |
| Contrast | 0.376 | ˂0.001 |
| Dissimilarity | 0.468 | ˂0.001 |
| T2WI | ||
| 1st order | ||
| Entropylog1o | 0.561 | ˂0.001 |
| Entropylog2 | 0.561 | ˂0.001 |
| Uniformity | -0.254 | 0.002 |
| GLCM | ||
| JointEntropyLog2 | 0.270 | 0.001 |
| JointEntropyLog10 | 0.269 | 0.001 |
| Contrast | 0.765 | ˂0.001 |
| Dissimilarity | 0.809 | ˂0.001 |
| Correlation | 0.370 | ˂0.001 |
| Feature | Gleason Score Median (interquartile 25th,50th, and 75th percentiles) | P | ||
|---|---|---|---|---|
| G2 | G3 | G4 | ||
| ADC | ||||
| 1st order | ||||
| Skewness | 0.30 (-0.01, 0.30, 0.58) | 0.60 (-0.12, 0.60, 1.24) | 0.39 (0.10, 0.39, 0.75) | 0.92 |
| Kurtosis | -0.49 (-0.87, -0.49, 0.26) | -0.38 (-0.78, -0.38, 1.24) | -0.34 (-0.90, -0.34, 0.49) | 0.81 |
| Entropylog1o | 1.12 (1.08, 1.12, 1.16) | 1.15 (1.09, 1.15, 1.21) | 1.16 (1.09, 1.16, 1.22) | 0.03 |
| Entropylog2 | 3.75(3.61, 3.75, 3.87) | 3.83 (3.62, 3.83, 4.03) | 3.88 (3.64, 3.88, 3.06) | 0.03 |
| Uniformity | 0.07(0.07, 0.07, 0.08) | 0.07 (0.06, 0.07, 0.08) | 0.07 (0.06, 0.07, 0.08) | 0.01 |
| GLCM | ||||
| JointEntropyLog2 | 6.12 (5.87, 6.12, 6.47) | 7.84 (7.42, 7.84, 8.22) | 6.28 (6.11, 6.28, 6.60) | 0.03 |
| JointEntropyLog10 | 1.84 (1.76, 1.84, 1.94) | 2.36 (2.24, 2.36, 2.54) | 1.89 (1.83, 1.89, 1.98) | 0.18 |
| Angular Second Moment | 0.02 (0.01, 0.02, 0.02) | 0.005(0.0037, 0.005, 0.006) | 0.01 (0.01, 0.01, 0.01) | 0.05 |
| Contrast | 132.43 (101.12, 132.43, 182.76) | 83.79 (64.25, 83.79, 128.98) | 149.88 (107.82, 149.88, 220.89) | 0.15 |
| Dissimilarity | 9.01 (8.05, 9.01, 10.79) | 7.09 (6.25, 7.09, 9.03) | 9.84 (8.25, 9.84, 11.96) | 0.14 |
| Correlation | 0.18 (0.02, 0.18, 0.33) | 0.26 (0.14, 0.26, 0.47) | 0.20 (0.05, 0.20, 0.41) | 0.54 |
| T2WI | ||||
| 1st order | ||||
| Skewness | 0.03 (-0.22, 0.03, 0.29) | 0.23 (-0.12, 0.23, 0.47) | -0.03 (-0.26, -0.03, 0.23) | 0.85 |
| Kurtosis | -0.14 (-0.47, -0.14, 0.64) | 0.07 (-0.39, 0.07, 0.27) | -0.62 (-0.76, -0.62, -0.31) | 0.78 |
| Entropylog1o | 1.29 (1.23, 1.29, 1.38) | 1.33 (1.26, 1.33, 1.44) | 1.28 (1.18, 1.28, 1.38) | 0.76 |
| Entropylog2 | 4.31 (4.09, 4.31, 4.61) | 4.42 (4.20, 4.42, 4.47) | 4.28 (3.92, 4.28, 4.61) | 0.76 |
| Uniformity | 0.05 (0.04, 0.05, 0.06) | 0.05 (0.04, 0.05, 0.06) | 0.05 (0.04, 0.05, 0.07) | 0.80 |
| GLCM | ||||
| JointEntropyLog2 | 7.48 (6.87, 7.48, 8.16) | 6.17 (5.75, 6.17, 6.41) | 7.12 (6.79, 7.12, 8.23) | 0.40 |
| JointEntropyLog10 | 2.28 (2.11, 2.28, 2.25) | 1.88 (1.76, 1.88, 2.01) | 2.19 (2.06, 2.19, 2.48) | 0.72 |
| Angular Second Moment | 0.008 (0.004, 0.01, 0.01) | 0.01(0.01, 0.01, 0.02) | 0.01(0.01, 001, 0.01) | 0.69 |
| Contrast | 94.93 (69.61, 94.93, 138.60) | 180.96 (126.01, 180.96, 280.64) | 101.25 (54.24, 101.25, 125.81) | 0.62 |
| Dissimilarity | 7.68 (6.58, 7.68, 9.03) | 9.71 (8.62, 9.71, 13.23) | 7.97 (6.06, 7.97, 9.17) | 0.63 |
| Correlation | 0.24 (0.10, 0.24, 0.34) | 0.24 (0.07, 0.24, 0.33) | 0.22 (0.14, 0.22, 0.31) | 0.78 |
| Feature | r | P |
|---|---|---|
| ADC | ||
| 1st order | ||
| Uniformity | -0.30 | 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





