Submitted:
11 November 2023
Posted:
13 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
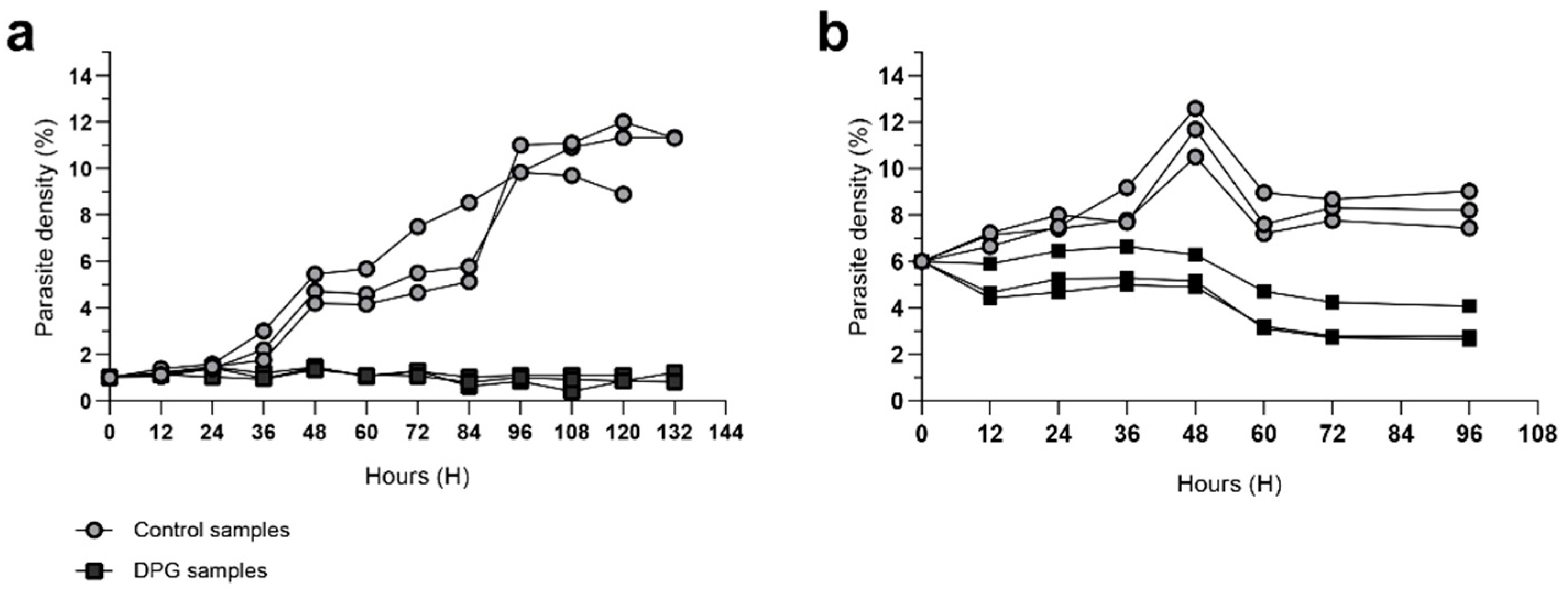
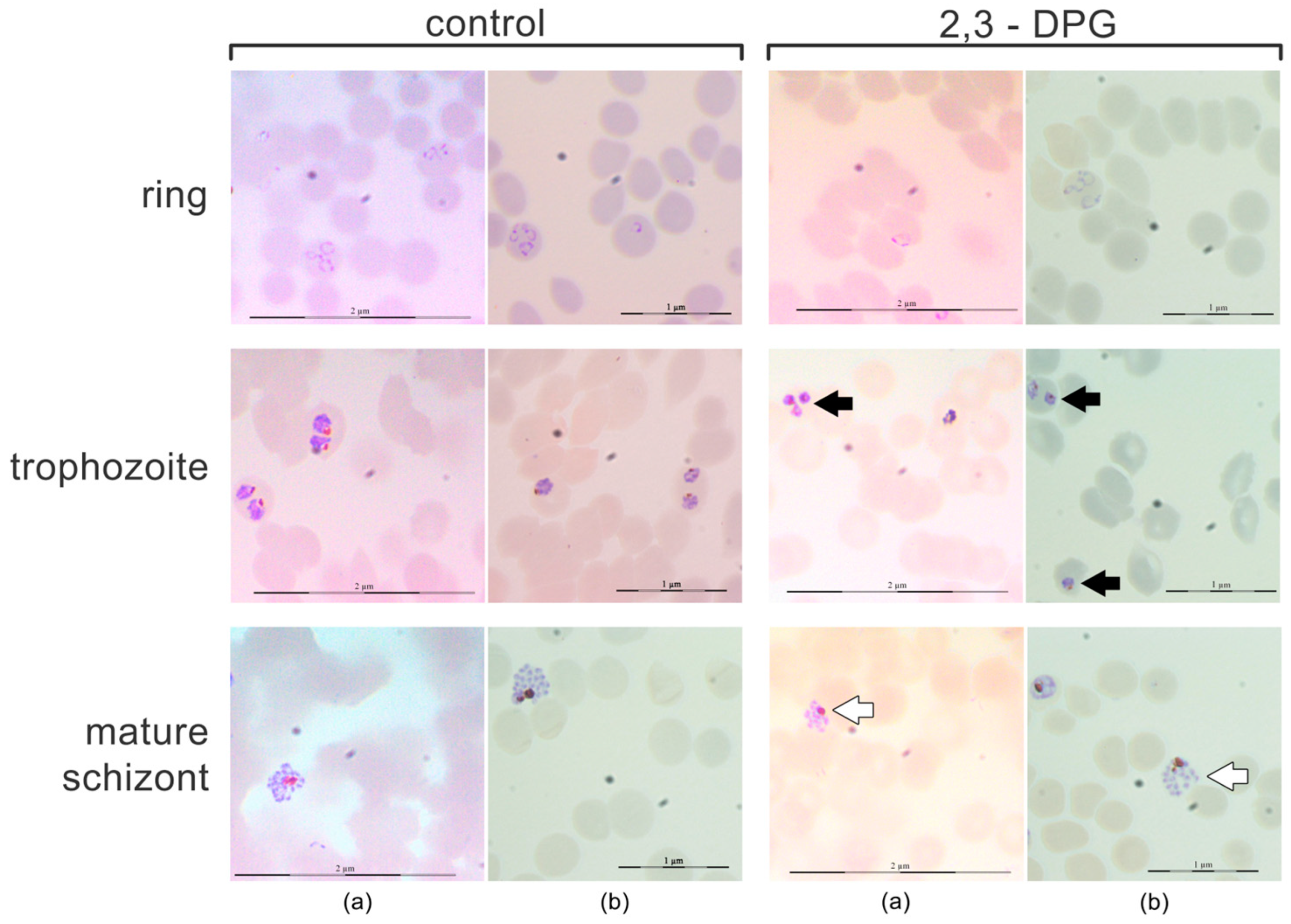
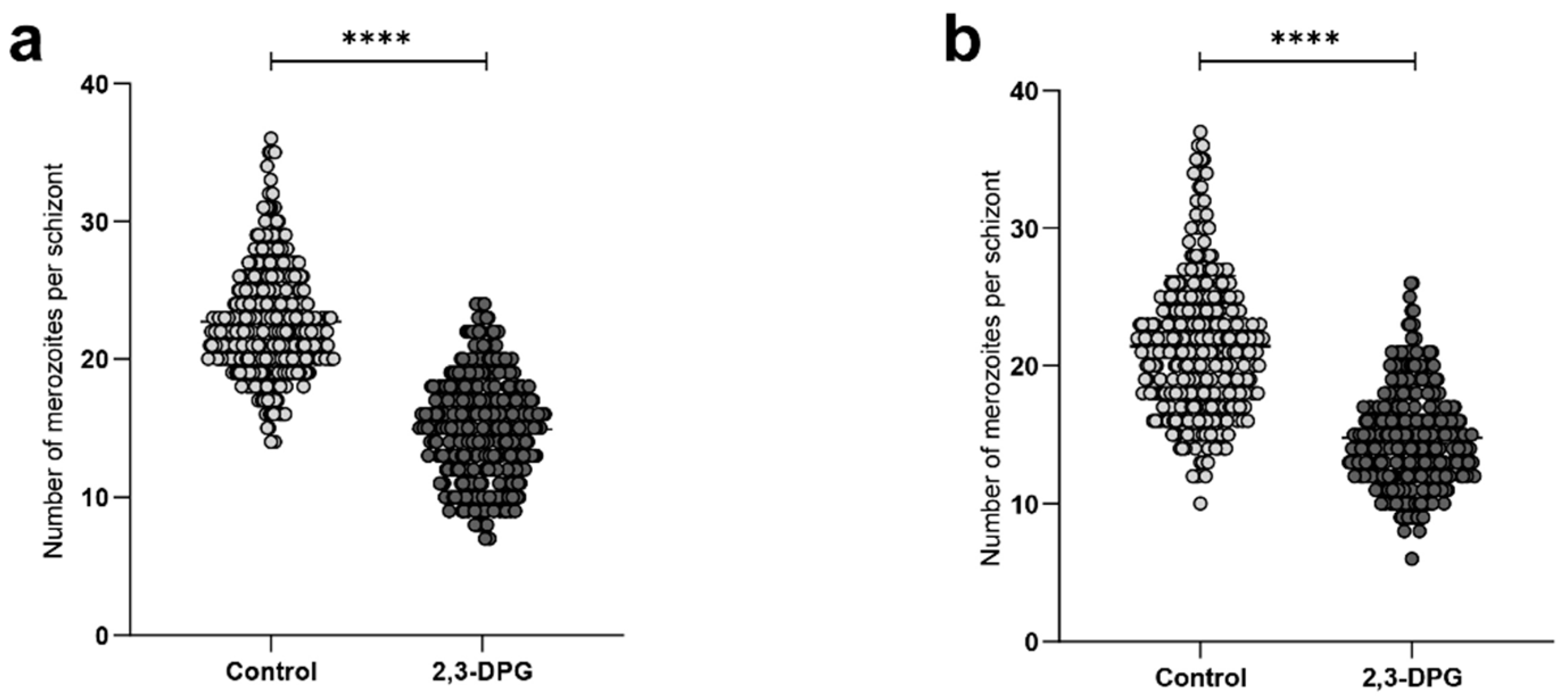
2.1. Effect of 2,3-DPG on the parasite intraerythrocytic development
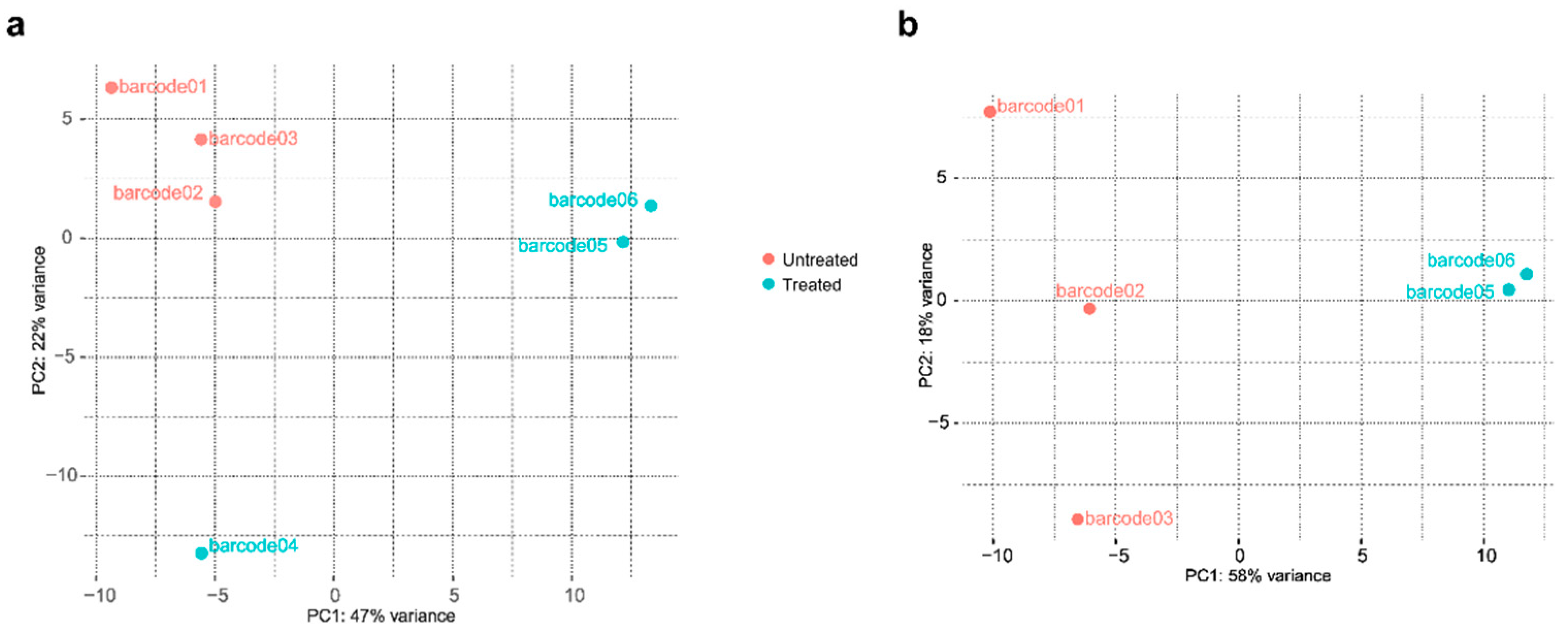
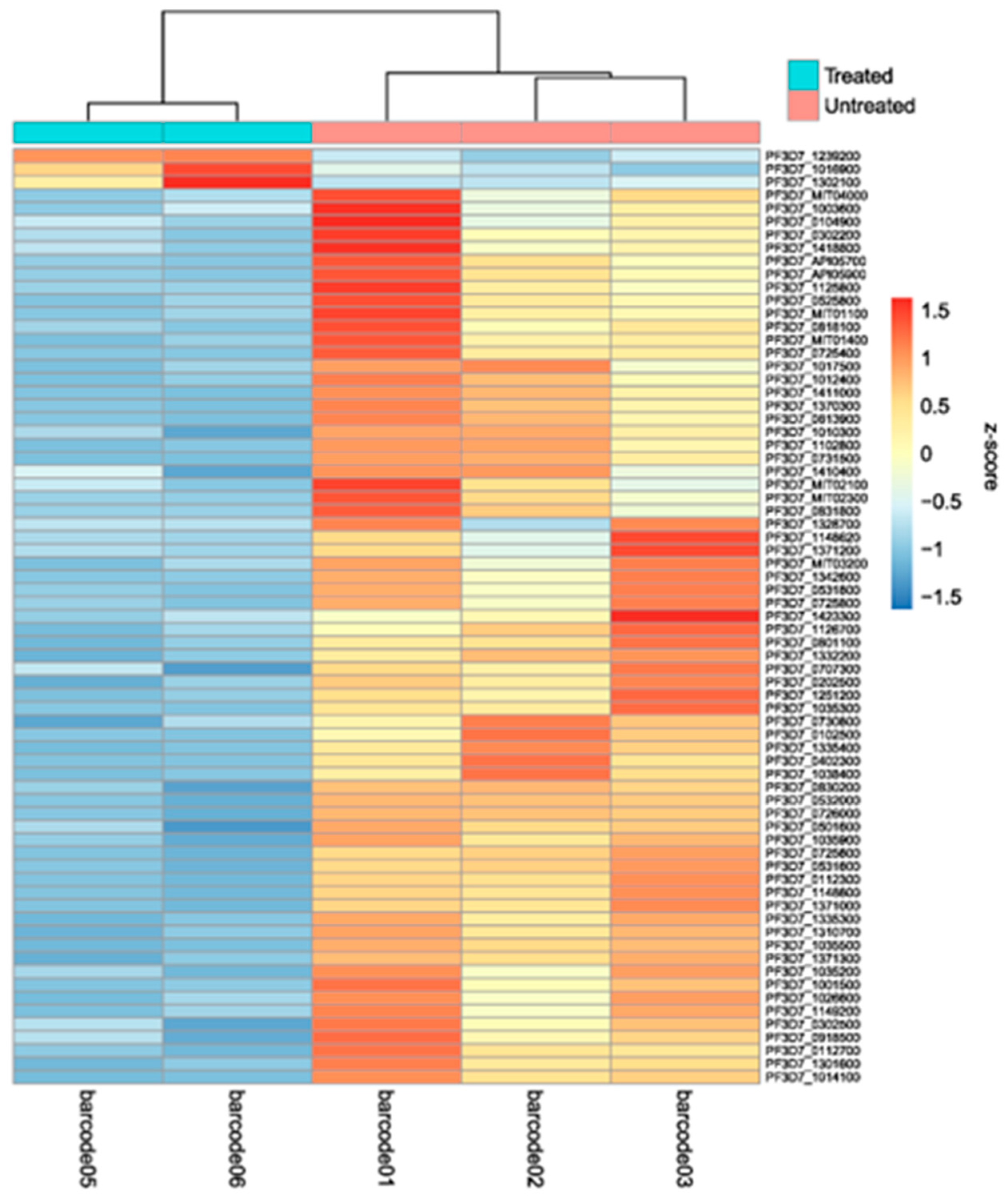
2.2. Differentially expressed genes between treated and untreated cultures
2.2.1. Gene Ontology enrichment analysis
3. Discussion
4. Materials and Methods
4.1. Blood donors
4.2. Plasmodium falciparum in vitro cultures
4.3. Effect of 2,3-DPG on the parasite intraerythrocytic development
4.4. RNA preparation
4.6. Analysis of gene expression
4.7. Statistical analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
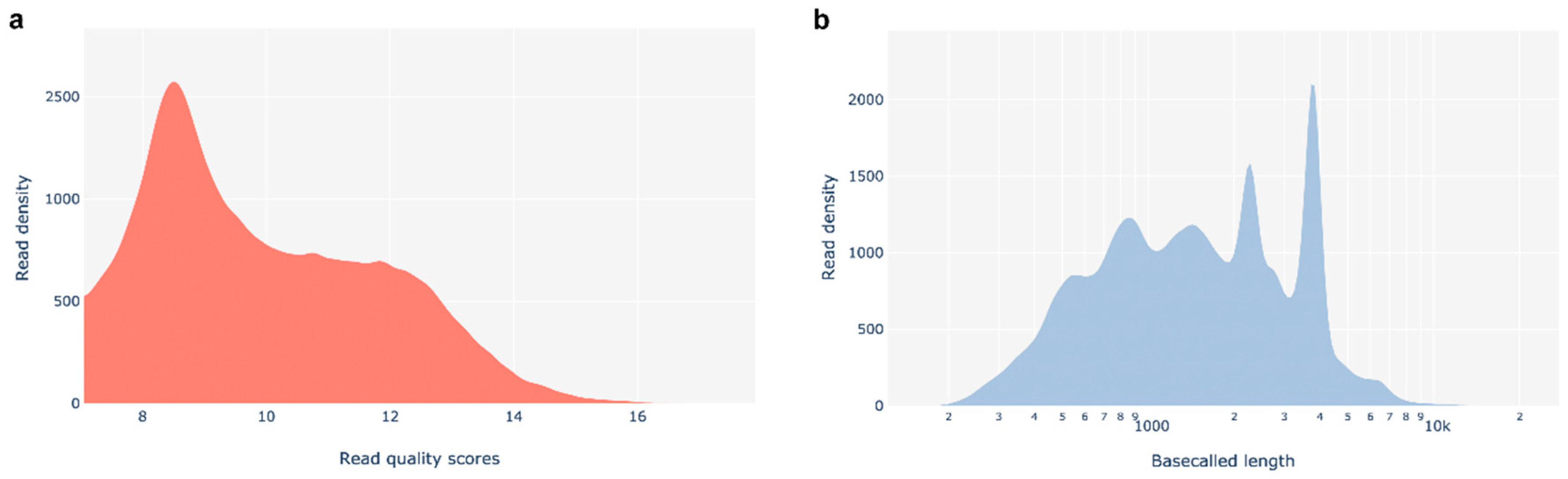
A.1. Transcriptome sequencing analysis – quality control
A.1.1. RNA quality and cDNA libraries sequencing
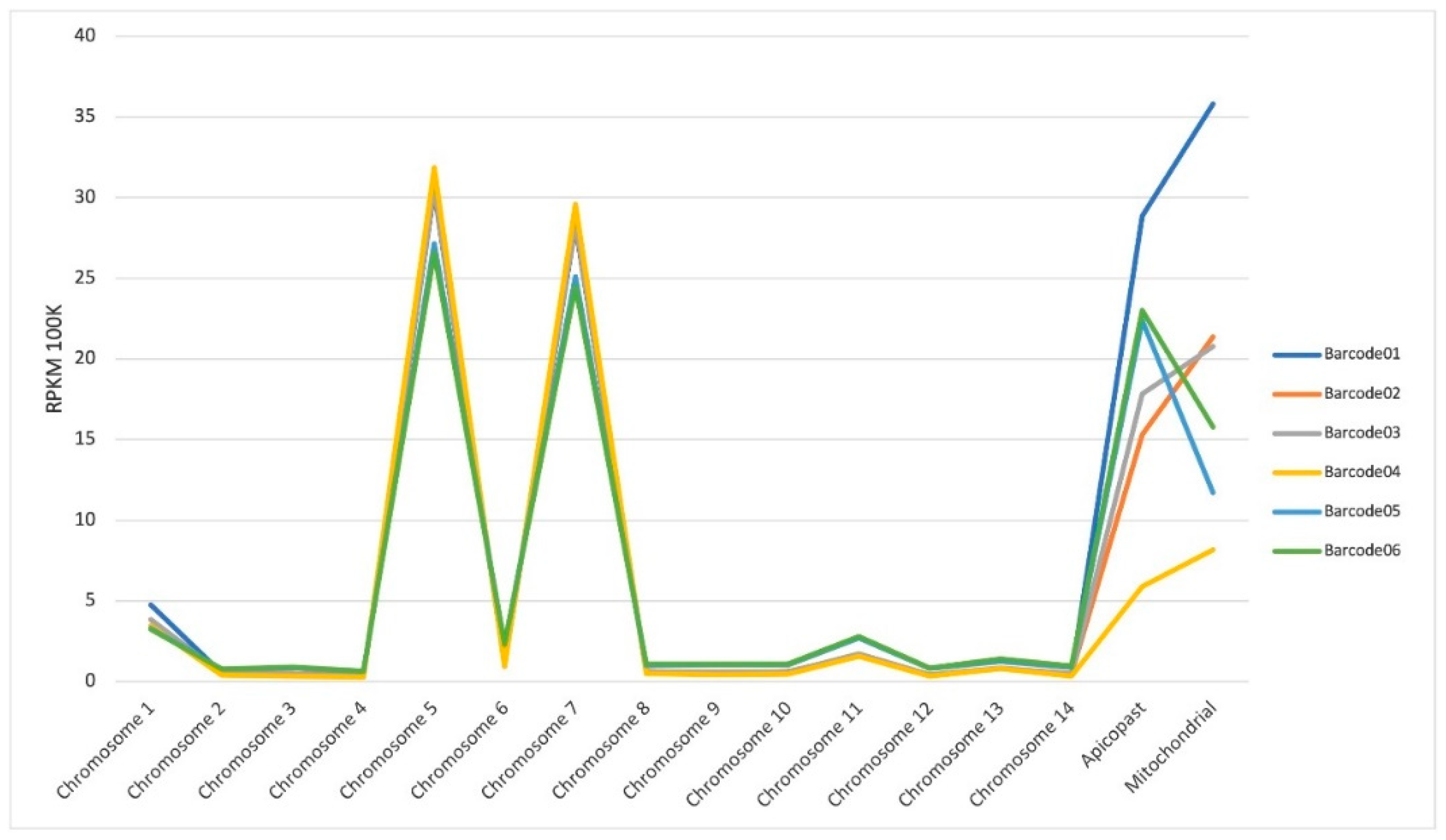
A.1.2. Alignment and mapping of reads against the 3D7 reference genome
| Control (untreated samples) | DPG (treated samples) | |||||
| Barcode01 | Barcode02 | Barcode03 | Barcode04 | Barcode05 | Barcode06 | |
| Number of Pass reads (%) | 124,497 (7.83%) | 262,680 (16.51%) | 165,905 (10.43% | 246,274 (15.48%) | 225,089 (14.15%) | 566,402 (35.60%) |
| 553,082 (34.77%) | 1,037,765 (65.23%) | |||||
| Total number of bases (Mbp) | 201.3 | 423.2 | 270.9 | 537.4 | 414.6 | 1000.0 |
| Sequence length | 128-46,300 | 117-23,141 | 110-21,960 | 104-29,694 | 141-29,207 | 127-30,804 |
| Total N. of Sequences | Mapped Reads | Unmapped Reads | Error rate | Average length | Average quality | ||
|
Control (untreated samples) |
Barcode01 | 124,497 | 122,745 (98.6%) | 1,752 (1.4%) | 9.63e-2 | 1,617 | 19.2 |
| Barcode02 | 262,680 | 257,908 (98.2%) | 4,772 (1.8%) | 9.65e-2 | 1,611 | 19.1 | |
| Barcode03 | 165,905 | 163,915 (98.8%) | 1,990 (1.2%) | 9.63e-2 | 1,633 | 19.3 | |
|
DPG (treated samples) |
Barcode04 | 246,274 | 242,957 (98.7%) | 3,317 (1.3%) | 9.49e-2 | 2,182 | 19.1 |
| Barcode05 | 225,089 | 222,821 (99%) |
2,268 (1%) |
1.04e-1 | 1,842 | 19.8 | |
| Barcode06 | 566,402 | 561,228 (99.1%) | 5,174 (0.9%) | 1.07e-1 | 1,826 | 20.3 |
References
- Prudêncio, M.; Costa, J.C. Research Funding after COVID-19. Nat. Microbiol. 2020, 5, 986–986. [Google Scholar] [CrossRef]
- World Health Organization World Malaria Report 2022; World Health Organization: Geneva, 2022; ISBN 978-92-4-006489-8.
- Andrade, M.V.; Noronha, K.; Diniz, B.P.C.; Guedes, G.; Carvalho, L.R.; Silva, V.A.; Calazans, J.A.; Santos, A.S.; Silva, D.N.; Castro, M.C. The Economic Burden of Malaria: A Systematic Review. Malar. J. 2022, 21, 283. [Google Scholar] [CrossRef] [PubMed]
- Mathews, E.S.; Odom John, A.R. Tackling Resistance: Emerging Antimalarials and New Parasite Targets in the Era of Elimination. F1000Research 2018, 7, 1170. [Google Scholar] [CrossRef] [PubMed]
- Lelliott, P.M.; McMorran, B.J.; Foote, S.J.; Burgio, G. The Influence of Host Genetics on Erythrocytes and Malaria Infection: Is There Therapeutic Potential? Malar. J. 2015, 14, 289. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Rao, M.; Wallis, R.S.; Kaufmann, S.H.E.; Rustomjee, R.; Mwaba, P.; Vilaplana, C.; Yeboah-Manu, D.; Chakaya, J.; Ippolito, G.; et al. Host-Directed Therapies for Infectious Diseases: Current Status, Recent Progress, and Future Prospects. Lancet Infect. Dis. 2016, 16, e47–e63. [Google Scholar] [CrossRef]
- Min-Oo, G.; Fortin, A.; Tam, M.-F.; Nantel, A.; Stevenson, M.M.; Gros, P. Pyruvate Kinase Deficiency in Mice Protects against Malaria. Nat. Genet. 2003, 35, 357–362. [Google Scholar] [CrossRef]
- Ayi, K.; Liles, W.C.; Gros, P.; Kain, K.C. Adenosine Triphosphate Depletion of Erythrocytes Simulates the Phenotype Associated with Pyruvate Kinase Deficiency and Confers Protection against Plasmodium Falciparum In Vitro. J. Infect. Dis. 2009, 200, 1289–1299. [Google Scholar] [CrossRef]
- Alves, J.; Machado, P.; Silva, J.; Gonçalves, N.; Ribeiro, L.; Faustino, P.; do Rosário, V.E.; Manco, L.; Gusmão, L.; Amorim, A.; et al. Analysis of Malaria Associated Genetic Traits in Cabo Verde, a Melting Pot of European and Sub Saharan Settlers. Blood Cells Mol Dis 2010, 44, 62–68. [Google Scholar] [CrossRef]
- Machado, P.; Pereira, R.; Rocha, A.M.; Manco, L.; Fernandes, N.; Miranda, J.; Ribeiro, L.; Do Rosário, V.E.; Amorim, A.; Gusmão, L.; et al. Malaria: Looking for Selection Signatures in the Human PKLR Gene Region. Br. J. Haematol. 2010, 149, 775–784. [Google Scholar] [CrossRef]
- Machado, P.; Manco, L.; Gomes, C.; Mendes, C.; Fernandes, N.; Salomé, G.; Sitoe, L.; Chibute, S.; Langa, J.; Ribeiro, L.; et al. Pyruvate Kinase Deficiency in Sub-Saharan Africa: Identification of a Highly Frequent Missense Mutation (G829A;Glu277Lys) and Association with Malaria. PLOS ONE 2012, 7, e47071. [Google Scholar] [CrossRef]
- van Bruggen, R.; Gualtieri, C.; Iliescu, A.; Louicharoen Cheepsunthorn, C.; Mungkalasut, P.; Trape, J.-F.; Modiano, D.; Sodiomon Sirima, B.; Singhasivanon, P.; Lathrop, M.; et al. Modulation of Malaria Phenotypes by Pyruvate Kinase (PKLR) Variants in a Thai Population. PLOS ONE 2015, 10, e0144555. [Google Scholar] [CrossRef]
- van Wijk, R.; van Solinge, W.W. The Energy-Less Red Blood Cell Is Lost: Erythrocyte Enzyme Abnormalities of Glycolysis. Blood 2005, 106, 4034–4042. [Google Scholar] [CrossRef]
- Roth, E. Plasmodium Falciparum Carbohydrate Metabolism: A Connection between Host Cell and Parasite. Blood Cells 1990, 16, 453–460, discussion 461-466. [Google Scholar]
- Morais, I.; Medeiros, M.M.; Carvalho, M.; Morello, J.; Teixeira, S.M.; Maciel, S.; Nhantumbo, J.; Balau, A.; Rosa, M.T.G.; Nogueira, F.; et al. Synthetic Red Blood Cell-Specific Glycolytic Intermediate 2,3-Diphosphoglycerate (2,3-DPG) Inhibits Plasmodium Falciparum Development In Vitro. Front. Cell. Infect. Microbiol. 2022, 12. [Google Scholar] [CrossRef]
- Carvalho, M.; Medeiros, M.M.; Morais, I.; Lopes, C.S.; Balau, A.; Santos, N.C.; Carvalho, F.A.; Arez, A.P. 2,3-Diphosphoglycerate and the Protective Effect of Pyruvate Kinase Deficiency against Malaria Infection—Exploring the Role of the Red Blood Cell Membrane. Int. J. Mol. Sci. 2023, 24, 1336. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, K.; Hentzschel, F.; Valkiūnas, G.; Marti, M. Plasmodium Asexual Growth and Sexual Development in the Haematopoietic Niche of the Host. Nat. Rev. Microbiol. 2020, 18, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Mancio-Silva, L.; Slavic, K.; Grilo Ruivo, M.T.; Grosso, A.R.; Modrzynska, K.K.; Vera, I.M.; Sales-Dias, J.; Gomes, A.R.; MacPherson, C.R.; Crozet, P.; et al. Nutrient Sensing Modulates Malaria Parasite Virulence. Nature 2017, 547, 213–216. [Google Scholar] [CrossRef]
- Simon, C.S.; Stürmer, V.S.; Guizetti, J. How Many Is Enough? - Challenges of Multinucleated Cell Division in Malaria Parasites. Front. Cell. Infect. Microbiol. 2021, 11, 658616. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, W.L.; Ishengoma, D.S.; Parr, J.B.; Bridges, D.J.; Barry, A.E. Nanopore Sequencing for Malaria Molecular Surveillance: Opportunities and Challenges. Trends Parasitol. 2023, S1471492223002350. [Google Scholar] [CrossRef]
- Imai, K.; Tarumoto, N.; Runtuwene, L.R.; Sakai, J.; Hayashida, K.; Eshita, Y.; Maeda, R.; Tuda, J.; Ohno, H.; Murakami, T.; et al. An Innovative Diagnostic Technology for the Codon Mutation C580Y in Kelch13 of Plasmodium Falciparum with MinION Nanopore Sequencer. Malar. J. 2018, 17, 217. [Google Scholar] [CrossRef]
- Runtuwene, L.R.; Tuda, J.S.B.; Mongan, A.E.; Makalowski, W.; Frith, M.C.; Imwong, M.; Srisutham, S.; Nguyen Thi, L.A.; Tuan, N.N.; Eshita, Y.; et al. Nanopore Sequencing of Drug-Resistance-Associated Genes in Malaria Parasites, Plasmodium Falciparum. Sci. Rep. 2018, 8, 8286. [Google Scholar] [CrossRef] [PubMed]
- Godin, M.J.; Sebastian, A.; Albert, I.; Lindner, S.E. Long-Read Genome Assembly and Gene Model Annotations for the Rodent Malaria Parasite Plasmodium Yoelii 17XNL. J. Biol. Chem. 2023, 299, 104871. [Google Scholar] [CrossRef] [PubMed]
- Oresegun, D.R.; Thorpe, P.; Benavente, E.D.; Campino, S.; Muh, F.; Moon, R.W.; Clark, T.G.; Cox-Singh, J. De Novo Assembly of Plasmodium Knowlesi Genomes From Clinical Samples Explains the Counterintuitive Intrachromosomal Organization of Variant SICAvar and Kir Multiple Gene Family Members. Front. Genet. 2022, 13, 855052. [Google Scholar] [CrossRef] [PubMed]
- Voß, Y.; Klaus, S.; Guizetti, J.; Ganter, M. Plasmodium Schizogony, a Chronology of the Parasite’s Cell Cycle in the Blood Stage. PLOS Pathog. 2023, 19, e1011157. [Google Scholar] [CrossRef] [PubMed]
- Van Niekerk, D.D.; Du Toit, F.; Green, K.; Palm, D.; Snoep, J.L. A Detailed Kinetic Model of Glycolysis in Plasmodium Falciparum-Infected Red Blood Cells for Antimalarial Drug Target Identification. J. Biol. Chem. 2023, 299, 105111. [Google Scholar] [CrossRef] [PubMed]
- van Schalkwyk, D.A.; Priebe, W.; Saliba, K.J. The Inhibitory Effect of 2-Halo Derivatives of d-Glucose on Glycolysis and on the Proliferation of the Human Malaria Parasite Plasmodium Falciparum. J. Pharmacol. Exp. Ther. 2008, 327, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Avery, V.M. Naturally Acquired Kelch13 Mutations in Plasmodium Falciparum Strains Modulate In Vitro Ring-Stage Artemisinin-Based Drug Tolerance and Parasite Survival in Response to Hyperoxia. Microbiol. Spectr. 2022, 10, e01282–21. [Google Scholar] [CrossRef] [PubMed]
- Torrentino-Madamet, M.; Almeras, L. ; Travaille; Sinou; Pophillat; Belghazi; Fourquet; Jammes; Parzy Proteomic Analysis Revealed Alterations of the Plasmodium Falciparum Metabolism Following Salicylhydroxamic Acid Exposure. Res. Rep. Trop. Med. 2011, 109. [Google Scholar] [CrossRef]
- Hoo, R.; Zhu, L.; Amaladoss, A.; Mok, S.; Natalang, O.; Lapp, S.A.; Hu, G.; Liew, K.; Galinski, M.R.; Bozdech, Z.; et al. Integrated Analysis of the Plasmodium Species Transcriptome. EBioMedicine 2016, 7, 255–266. [Google Scholar] [CrossRef]
- Lu, X.M.; Batugedara, G.; Lee, M.; Prudhomme, J.; Bunnik, E.M.; Le Roch, K.G. Nascent RNA Sequencing Reveals Mechanisms of Gene Regulation in the Human Malaria Parasite Plasmodium Falciparum. Nucleic Acids Res. 2017, 45, 7825–7840. [Google Scholar] [CrossRef]
- Bozdech, Z.; Llinás, M.; Pulliam, B.L.; Wong, E.D.; Zhu, J.; DeRisi, J.L. The Transcriptome of the Intraerythrocytic Developmental Cycle of Plasmodium Falciparum. PLoS Biol. 2003, 1, e5. [Google Scholar] [CrossRef] [PubMed]
- Viscardi, M.J.; Arribere, J.A. Poly(a) Selection Introduces Bias and Undue Noise in Direct RNA-Sequencing. BMC Genomics 2022, 23, 530. [Google Scholar] [CrossRef] [PubMed]
- Rang, F.J.; Kloosterman, W.P.; De Ridder, J. From Squiggle to Basepair: Computational Approaches for Improving Nanopore Sequencing Read Accuracy. Genome Biol. 2018, 19, 90. [Google Scholar] [CrossRef] [PubMed]
- Kengne-Ouafo, J.A.; Bah, S.Y.; Kemp, A.; Stewart, L.; Amenga-Etego, L.; Deitsch, K.W.; Rayner, J.C.; Billker, O.; Binka, F.N.; Sutherland, C.J.; et al. The Global Transcriptome of Plasmodium Falciparum Mid-Stage Gametocytes (Stages II–IV) Appears Largely Conserved and Gametocyte-Specific Gene Expression Patterns Vary in Clinical Isolates. Microbiol. Spectr. 2023, e03820–22. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.V.; Judd, L.M.; Jex, A.R.; Holt, K.E.; Tonkin, C.J.; Ralph, S.A. Direct Nanopore Sequencing of mRNA Reveals Landscape of Transcript Isoforms in Apicomplexan Parasites. mSystems 2021, 6, e01081–20. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, T.; Leger, A. Nanopore RNA Sequencing Analysis. In RNA Bioinformatics; Picardi, E., Ed.; Methods in Molecular Biology; Springer US: New York, NY, 2021; Vol. 2284, pp. 569–578; ISBN 978-1-07-161306-1. [Google Scholar]
- Batugedara, G.; Lu, X.M.; Hristov, B.; Abel, S.; Chahine, Z.; Hollin, T.; Williams, D.; Wang, T.; Cort, A.; Lenz, T.; et al. Novel Insights into the Role of Long Non-Coding RNA in the Human Malaria Parasite, Plasmodium Falciparum. Nat. Commun. 2023, 14, 5086. [Google Scholar] [CrossRef] [PubMed]
- Maróti, Z.; Tombácz, D.; Moldován, N.; Torma, G.; Jefferson, V.A.; Csabai, Z.; Gulyás, G.; Dörmő, Á.; Boldogkői, M.; Kalmár, T.; et al. Time Course Profiling of Host Cell Response to Herpesvirus Infection Using Nanopore and Synthetic Long-Read Transcriptome Sequencing. Sci. Rep. 2021, 11, 14219. [Google Scholar] [CrossRef] [PubMed]
- Trager, W.; Jensen, J.B. Human Malaria Parasites in Continuous Culture. Science 1976, 193, 673–675. [Google Scholar] [CrossRef] [PubMed]
- Radfar, A.; Méndez, D.; Moneriz, C.; Linares, M.; Marín-García, P.; Puyet, A.; Diez, A.; Bautista, J.M. Synchronous Culture of Plasmodium Falciparum at High Parasitemia Levels. Nat. Protoc. 2009, 4, 1899–1915. [Google Scholar] [CrossRef]
- Batut, B.; Freeberg, M.; Heydarian, M.; Erxleben, A.; Videm, P.; Blank, C.; Doyle, M.; Soranzo, N.; van Heusden, P.; Delisle, L. Reference-Based RNA-Seq Data Analysis (Galaxy Training Materials) 2023a.
- Lansink, L.I.M.; Skinner, O.P.; Engel, J.A.; Lee, H.J.; Soon, M.S.F.; Williams, C.G.; SheelaNair, A.; Pernold, C.P.S.; Laohamonthonkul, P.; Akter, J.; et al. Systemic Host Inflammation Induces Stage-Specific Transcriptomic Modification and Slower Maturation in Malaria Parasites. mBio 2023, e01129–23. [Google Scholar] [CrossRef]
| GO category | GO term | Name | N. of genes | Study Freq. | Pop. Freq. | p-value | q-value |
| Molecular Function | GO:0008201 | heparin binding | 6 | 25% | 0.68% | 1.48E-9 | 8.11E-8 |
| GO:0005488 | binding | 23 | 96% | 64% | 2.70E-4 | 2.97E-3 | |
| GO:0005253 | monoatomic anion channel activity | 2 | 8.3% | 0.15% | 3.15E-4 | 2.97E-3 | |
| GO:0046812 | host cell surface binding | 6 | 25% | 4.4% | 4.36E-4 | 3.00E-3 | |
| Biological Process | GO:0044409 | entry into host | 11 | 50% | 7.4% | 7.87E-8 | 9.05E-6 |
| Cellular Component | GO:0020008 | rhoptry | 7 | 18% | 1.3% | 3.22E-7 | 1.87E-5 |
| GO:1903561 | extracellular vesicle | 9 | 24% | 2.9% | 8.73E-7 | 2.53E-5 | |
| GO:0016459 | myosin complex | 2 | 5.3% | 0.10% | 4.35E-4 | 4.2E-3 | |
| GO:0020039 | pellicle | 2 | 5.3% | 0.10% | 4.35E-4 | 4.2E-3 | |
| GO:0070258 | inner membrane pellicle complex | 5 | 13% | 2.0% | 8.80E-4 | 6.38E-3 | |
| GO:0033643 | host cell part | 15 | 39% | 18% | 1.67E-3 | 9.89E-3 | |
| GO:0009986 | cell surface | 6 | 16% | 4.6% | 7.31E-3 | 0.0353 | |
| GO:0020009 | microneme | 3 | 7.9% | 1.2% | 9.88E-3 | 0.0441 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





