1. Introduction
Candidemias represent a significant healthcare challenge, ranking among the foremost causes of morbidity and mortality, particularly in patients with healthcare-associated infections (HAI) [1–5]. Invasive candidiasis (IC) accounts for over 95% of these infections, with Candida albicans, Candida glabrata, Candida tropicalis, Candida parapsilosis, Candida krusei, and, in certain regions, Candida auris being the primary culprits. The Latin American region witnesses a significant burden of IC, with incidence rates ranging from 0.74 to 6.0 per 1000 hospital admissions and alarmingly high mortality rates ranging from 30% to 76% [6–9]. Notably, admission to intensive care units (ICUs) significantly escalates the risk, with at least half of ICU patients succumbing to this diagnosis [10]. The surge in infections by these Candida species is attributed to multiple factors, encompassing sustained exposure to antifungal agents, the utilization of catheters in hospitalized patients, underlying malignancies, advancing age, and geographic distribution. Furthermore, the development of antifungal resistance and the capacity to elude host immunity add to the intricate challenge of managing candidiasis in clinical practice [11–13].
Among the non-albicans Candida species, Candida tropicalis stands out for its remarkable relevance. It commands a high incidence in IC and a concerning resistance profile against commonly employed antifungal drugs [14,15]. This pathogen not only possesses the capability to infiltrate vital organs [16] but is also linked to elevated mortality rates when compared to C. albicans and other non-albicans Candida species. Candida tropicalis exhibits a propensity for dissemination, particularly among neutropenic individuals and those grappling with malignancies, notably in cases demanding prolonged catheterization, extensive use of broad-spectrum antibiotics, or underlying cancer [17]. Recent increases in antifungal resistance have been associated with mutations in the ERG11 gene, encoding ergosterol-synthase, and the overexpression of the transcriptional regulator UPC2 [18,19]. Considering these developments, Candida tropicalis has emerged as a prominent etiological agent in IC, notably in Latin America and certain Asian countries, further accentuating its clinical significance [21].
Regrettably, the three principal antifungal drug classes (polyenes, echinocandins, and azoles) have been unable to stem the surge in life-threatening fungal infections observed over the past few decades [22]. In response to this pressing clinical challenge, compounds derived from botanical sources have gained traction as a promising alternative [23–25]. In our previous investigations, we established that Isoespintanol (ISO), a monoterpene derived from Oxandra xylopioides, exhibits potent antifungal properties against Candida tropicalis [26,27]. Our findings elucidated ISO's ability to induce intracellular reactive oxygen species, disrupt cell membranes, and eradicate mature biofilms as part of its multifaceted antifungal action.
However, the precise mechanism by which ISO orchestrates genetic dysregulation in Candida tropicalis remains an enigma. Herein, we propose that ISO plays a pivotal role in reshaping the transcriptional landscape of Candida tropicalis. Our primary objective is to decipher the differential gene expression patterns underlying processes such as membrane biogenesis and biofilm formation, among other critical biological pathways impacted by ISO. By delving into the potential targets of this antifungal biomolecule and their distinctive gene expression profiles, our research offers essential insights. These insights are poised to unlock novel therapeutic avenues for the treatment of candidiasis, presenting a holistic understanding of the genetic perturbations provoked by ISO.
Our study endeavors to provide a perspective on potential therapeutic targets in the battle against candidiasis. Our transcriptomics-based approach offers a comprehensive view of the biological behavior of these pathogenic yeasts and their genetic adaptations when exposed to ISO. These findings hold the promise of paving the way for innovative therapies to combat these resilient pathogens within the realm of medical practice.
2. Materials and Methods
2.1. Isolation and Identification of Isoespintanol
Isoespintanol (ISO) was isolated from "yaya prieta" leaves (Oxandra xylopioides) collected in October 2019. The specimen was located at coordinates 08°48'17" north latitude and 75°42'07" west longitude in the Municipality of Montería, Department of Córdoba, Colombia. A herbarium specimen (collection number JAUM 037849) was deposited in the Joaquín Antonio Uribe Botanical Garden in Medellín, Colombia. To isolate ISO, 5 g of petroleum benzine extract was subjected to hydrodistillation and successive crystallizations with n-hexane, following a modified version of the methodology described in [28]. This process yielded 1.2 g of purified ISO. The purity of ISO was confirmed using gas chromatography coupled to a Thermo Scientific model Trace 1310 mass spectrometer equipped with an AB-5MS column (30 m × 0.25 mm id × 0.25 µm). The temperature gradient system initiated at 80 °C for 10 minutes and increased to 200 °C at a rate of 10 °C/min. Subsequently, the temperature was raised to 240 °C at 4 °C/min and finally to 290 °C for 10 minutes at 10 °C/min. The injection was split less, with a volume of 1 µL. Mass spectrometry was conducted in electron impact ionization mode at 70 eV. The structure of ISO was elucidated through a combination of spectroscopic techniques, including 1H-NMR, 13C-NMR, DEPT, 1H-1H COSY, HMQC, and HMBC spectra. These analyses were performed on a 400 MHz Bruker Advance DRX spectrometer using deuterated chloroform (CDCl3) as the solvent.
2.2. Yeast strain and culture conditions
The yeast strain utilized in this study, identified as Candida tropicalis (CLI007), was originally isolated from a blood culture sample obtained from a hospitalized patient at Salud Social S.A.S. in Sincelejo, Colombia. Standard methods for yeast identification, including Vitek 2 Compact, Biomerieux SA, Y.S.T. Vitek 2 Card, and AST-YS08 Vitek 2 Card (Ref 420739), were employed to initially identify the yeast strain. Further confirmation of its identification was established through a comprehensive genome-wide taxonomic study, as detailed in Contreras [26]. To maintain the yeast cultures, Sabouraud Dextrose Agar (SDA) and BBL CHROMagar Candida media were employed. Prior to conducting the experiments, a yeast suspension was meticulously adjusted to a concentration of 107 colony-forming units per milliliter (CFU/mL) in phosphate-buffered saline (PBS) with a pH of 7.4. This standardized yeast suspension served as the inoculum for the subsequent assays.
2.3. RNA-Sequencing and Read Count Data Acquisition
Total RNA was extracted from yeast samples following a 4-hour exposure to ISO at its minimum inhibitory concentration (MIC: 391.6 µg/mL) [26]. Additionally, control yeast samples without ISO treatment were processed in parallel. RNA extraction was carried out using the Trizol reagent, following the manufacturer's standard protocol (Trizol TM Reagent, Invitrogen; Total RNA extraction protocol). The quantification of the total RNA was performed using the ribogreen colorimetric method (Invitrogen). Furthermore, the integrity of the RNA was assessed by measuring the RNA Integrity Number (RIN) through capillary electrophoresis on an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). A RIN score exceeding 7.0 was established as the threshold for acceptable RNA quality. Subsequently, the extracted RNA was employed to construct RNA-Seq libraries for sequencing, utilizing the Illumina Novaseq platform with 150 bp paired-end sequencing (Illumina-Transcriptome, TruSeq mRNA/Illumina). For the generation of RNA counts, Bowtie 2 was utilized, aligning the sequencing data to the Ensemble GTF files corresponding to the reference genome Candida tropicalis (REF GCA000006335v3).
2.4. Bioinformatics Analyses
2.4.1. Differential Gene Expression Analysis
To identify differentially expressed genes (DEGs), we utilized the read counts generated from the RNA-sequencing data. The differential gene expression analysis was conducted using the EdgeR package within the RStudio environment. The dataset was submitted to the analysis pipeline to assess the directionality of gene expression changes between two stages: Stage A, which comprised control/untreated Candida tropicalis samples, and Stage B, which included ISO-treated Candida tropicalis samples. We considered genes with a log2-fold change (Log2FC) greater than 1 and a false discovery rate (FDR) less than 0.05 as significant DEGs. DEGs with positive log2-fold change and significant p-values were categorized as exhibiting increased expression (UP), while DEGs with negative log2-fold change represented decreased expression (DN). Genes with p-values exceeding 0.05 were categorized as showing no significant change in expression between the two stages (NC).
2.4.2. Functional Enrichment Analysis for DEGs
Functional enrichment analysis was conducted on the set of 186 differentially expressed genes (DEGs) identified in the
Candida tropicalis dataset. This analysis was performed using Gene Set Enrichment Analysis (GSEA), encompassing all Gene Ontology (GO) terms. To ensure robustness, GO terms with values exceeding 0.05 were filtered out from the analysis. Subsequently, the resulting
p-values underwent adjustment for multiple hypothesis testing using the Benjamini and Hochberg approach [29]. For the assessment of functional enrichment, two distinct gene sets were examined: The upregulated genes (UP) and the downregulated genes (DN). The background universe for these analyses was defined as the total number of DEGs. To facilitate data analysis and visualization, we employed ShinyGO 0.77, based on the
Candida tropicalis genome derived from STRING-db. This resource provided comprehensive insights into functional enrichment of GO terms (
http://bioinformatics.sdstate.edu/go, accessed on 10 May 2023).
2.5. Computational Prediction of DEGs and Primer Design
To enhance the functional annotation of the transcriptome, we obtained an orthologous gene pair list between
Candida tropicalis and
Candida albicans from the Ensembl database. This was achieved through the utilization of the BioMart package, facilitated by R scripts. From the pool of 186 differentially expressed genes (DEGs), we refined our selection to focus on genes enriched in key biological functions, namely mitochondria, cell wall, lipid metabolism, and general metabolism. For data visualization, we employed the RStudio environment to create plots featuring the selected genes. Subsequently, we designed primers for specific genes of interest, namely ERG6 (upregulated, associated with steroid metabolism), KRE1 (upregulated, linked to cell wall processes), and CTRG_03786 (downregulated, implicated in cell wall functions). These primers were designed using PrimerQuest (IDT, Integrated DNA Technologies,
https://www.idtdna.com/pages/tools/primerquest?returnurl=%2FPrimerQuest%2FHome%2FIndex accessed on June 16, 2023). For primer details, please refer to
Table S1 in Supplementary Materials.
2.6. Gene Expression Assessment by Quantitative RT-PCR
For experimental validation, RNA isolation was carried out using the TRIzol® reagent, following the manufacturer's instructions precisely (Trizol TM Reagent, Invitrogen; Total RNA extraction protocol), as previously detailed in
Section 2.3. For gene expression analysis, cDNA synthesis was conducted using SuperScript III (Invitrogen Cat No. 12574026) following the manufacturer's recommended protocols. Quantitative RT-PCR was performed using the StepOnePlus system (Applied Biosystems) with Power Up SYBR Master Mix (Applied Biosystems). This approach allowed us to determine the relative expression levels of mRNA transcripts for three target genes: ERG6, KRE1, and CTRG_03786. The housekeeping gene ACTIN-1 served as the reference for normalization. Each reaction mixture consisted of 100 ng of cDNA, 2X master mix, and 500 picomolar of each primer, resulting in a final volume of 10 μl. PCR cycling conditions encompassed an initial denaturation step at 95 °C for 2 minutes, followed by 40 cycles of 15 seconds at 95 °C, 15 seconds at 62 °C, and 60 seconds at 72 °C. The primer sequences for ERG6, KRE1, CTRG_03786, and ACTIN-1 are detailed in
Supplementary Table S1. Gene expression analysis was carried out employing the 2^(-ΔΔCT) method of relative quantification [32].
2.7. Determination of Total Ergosterol Content
The quantification of the total ergosterol content in C. tropicalis isolates treated with ISO was conducted in accordance with the protocol outlined in [33], with some modifications. Initially, the cells were exposed to ISO at their respective minimum inhibitory concentrations (MIC) and incubated at 35°C for a duration of 3 hours. Subsequently, the treated cells were subjected to centrifugation and washed with phosphate-buffered saline (PBS). To initiate ergosterol extraction, a wet weight of 0.5 g of cells was mixed with PBS. Saponification was achieved by adding 4 mL of a freshly prepared 30% (w/v) methanolic KOH solution and 8 mL of absolute ethanol. This mixture was maintained at 80°C for a period of 1 hour. The ensuing mixture was extracted using petroleum ether and then washed with a saturated NaCl solution. Following the extraction, the samples were concentrated under vacuum conditions at 60°C. The resulting residue was dissolved in 0.5 mL of methanol and subsequently filtered through a 0.45 µm micromembrane. The quantification of ergosterol content was determined by comparing the peak areas of the samples against a standard curve generated from ergosterol (95% Sigma-Aldrich). The concentrations used in the standard curve spanned 1, 10, 50, 100, 250, 500, 750, and 1000 mg/L of ergosterol. Ergosterol content analysis was performed using a UHPLC Ultimate 3000 system (Thermo Scientific, USA) equipped with a diode array detector (DAD). A C18 Hypersil Gold column (150 mm × 4.6 mm, 5 µm) was maintained at 30°C. Each injection had a volume of 30 µL, and the mobile phase consisted of methanol/water (97/3, 100% HPLC grade) at a flow rate of 0.6 mL/min. All measurements were conducted in triplicate, and ergosterol was detected at a wavelength of 205 nm.
The ergosterol inhibition ratio was calculated as follows:
Ergosterol Inhibition Ratio = (1 - Ergosterol Content of Treated Cells / Ergosterol Content of Untreated Cells) × 100%.
3. Results
3.1. Isolation and Purification of Isoespintanol Molecules
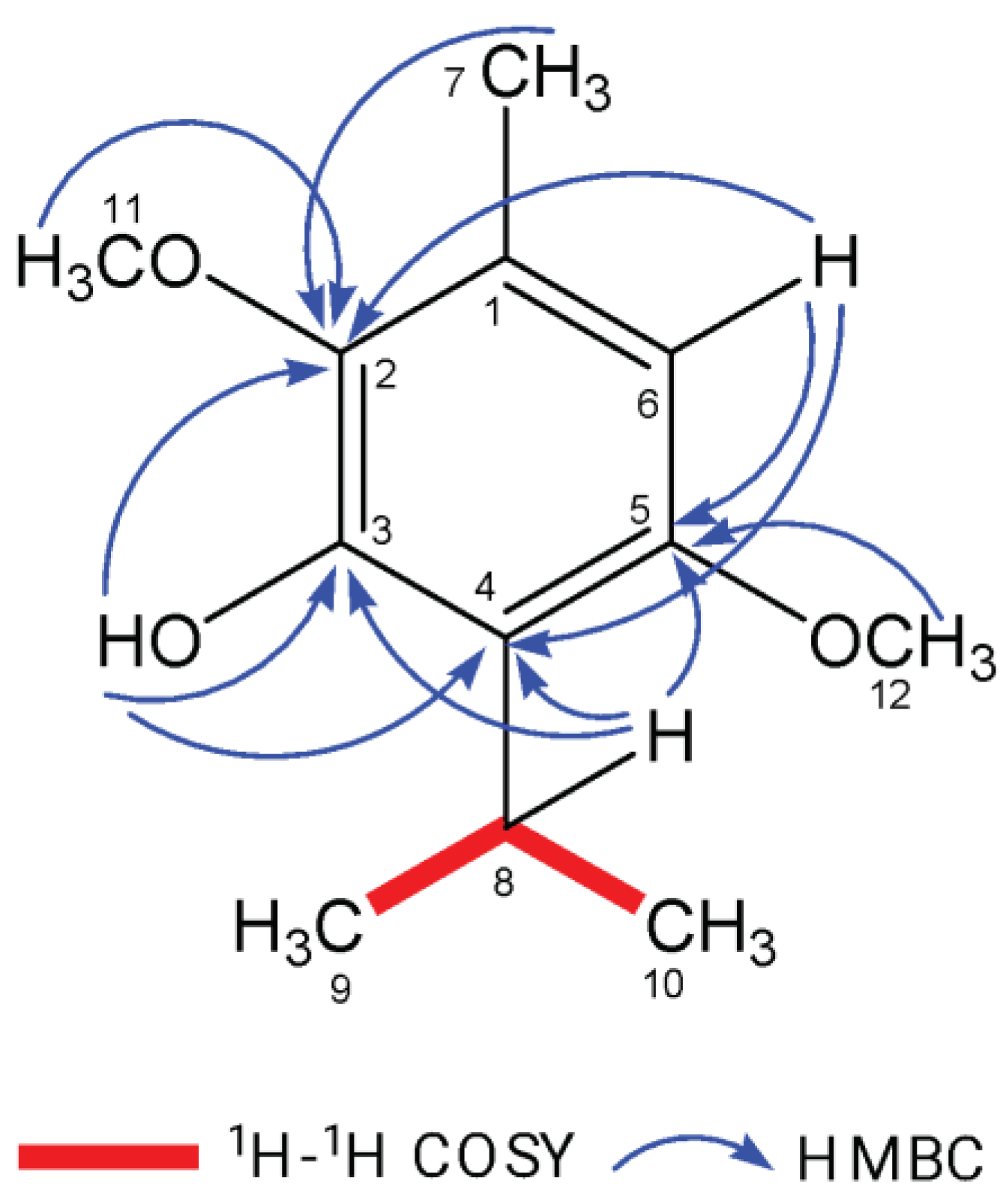
Isoespintanol (ISO) in the amount of 1.2 g was isolated and purified to a crystalline amorphous solid with a purity exceeding 99%. The high purity was verified using Gas Chromatography-Mass Spectrometry (GC-MS). Structural identification of ISO was accomplished through various spectroscopic techniques, including
1H-NMR,
13C-NMR, DEPT,
1H-
1H COSY, HMQC, and HMBC. This rigorous analysis led to the unequivocal proposal of the structure of 2,5-dimethoxy-3-hydroxy-
p-cymene isoespintanol, as illustrated in
Figure 1. Furthermore, Electron Impact Mass Spectrometry (EI-MS) provided essential data, with the observed molecular ion at m/z 210 (49%) and prominent fragment ions at m/z 195 (100%), 180, 165, 150, 135, and 91. Detailed nuclear magnetic resonance (NMR) spectra were recorded. In the
1H-NMR spectrum (400 MHz, CDCl
3), the chemical shifts were observed at δ 6.22 (singlet, 1H, H6), δ 5.85 (singlet, 1H, HO-3), δ 3.77 (singlet, 3H, H12), δ 3.76 (singlet, 3H, H11), δ 3.52 (heptet, J=7.1 Hz, 1H, H8), δ 2.29 (singlet, 3H, H7), and δ 1.33 (doublet, J=7.1 Hz, 6H, H9-H10). In the
13C-NMR spectrum (100 MHz, CDCl
3), the carbon chemical shifts were observed at δ 154.3 (C5), δ 147.4 (C3), δ 139.7 (C2), δ 126.8 (C1), δ 120.4 (C4), δ 104.4 (C6), δ 24.6 (C8), δ 60.8 (C11), δ 55.7 (C12), δ 20.6 (C9, C10), and δ 15.8 (C7).
3.2. RNA preparation, library construction, and sequence analysis
The results of RNA extraction from
Candida tropicalis, both with ISO treatment and without treatment (control), are summarized in
Table S2. Specifically, we obtained a total RNA yield of 0.797 µg for ISO-treated samples and 0.785 µg for untreated control cells. For a detailed overview of the sequencing statistics and transcriptome data, including the mapping of reads to the
Candida tropicalis reference genome (REF GCA000006335v3), please refer to
Tables S3 and S4, in Supplementary Materials.
Transcriptomes obtained by RNA-seq were mapped on the reference genome Candida_tropicalis REF GCA000006335v3.
3.3. Analysis of the Transcriptional Profile of Differentially Expressed Genes
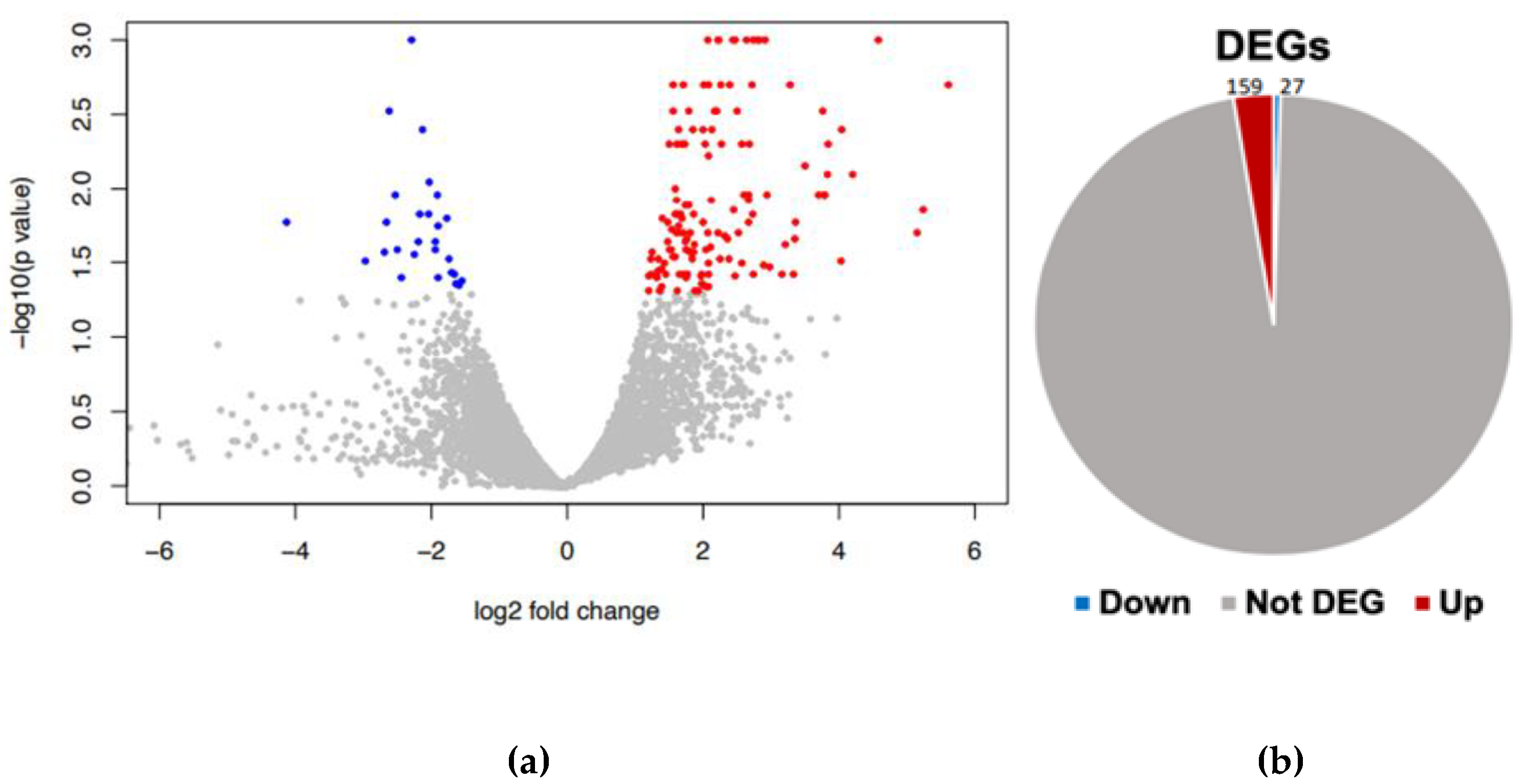
Our findings reveal distinct patterns of gene expression between yeast exposed to ISO and untreated yeast. A comprehensive analysis identified a total of 186 differentially expressed genes. Among these, 159 genes displayed induction in their expression levels, while 27 genes exhibited repression (
Figure 2).
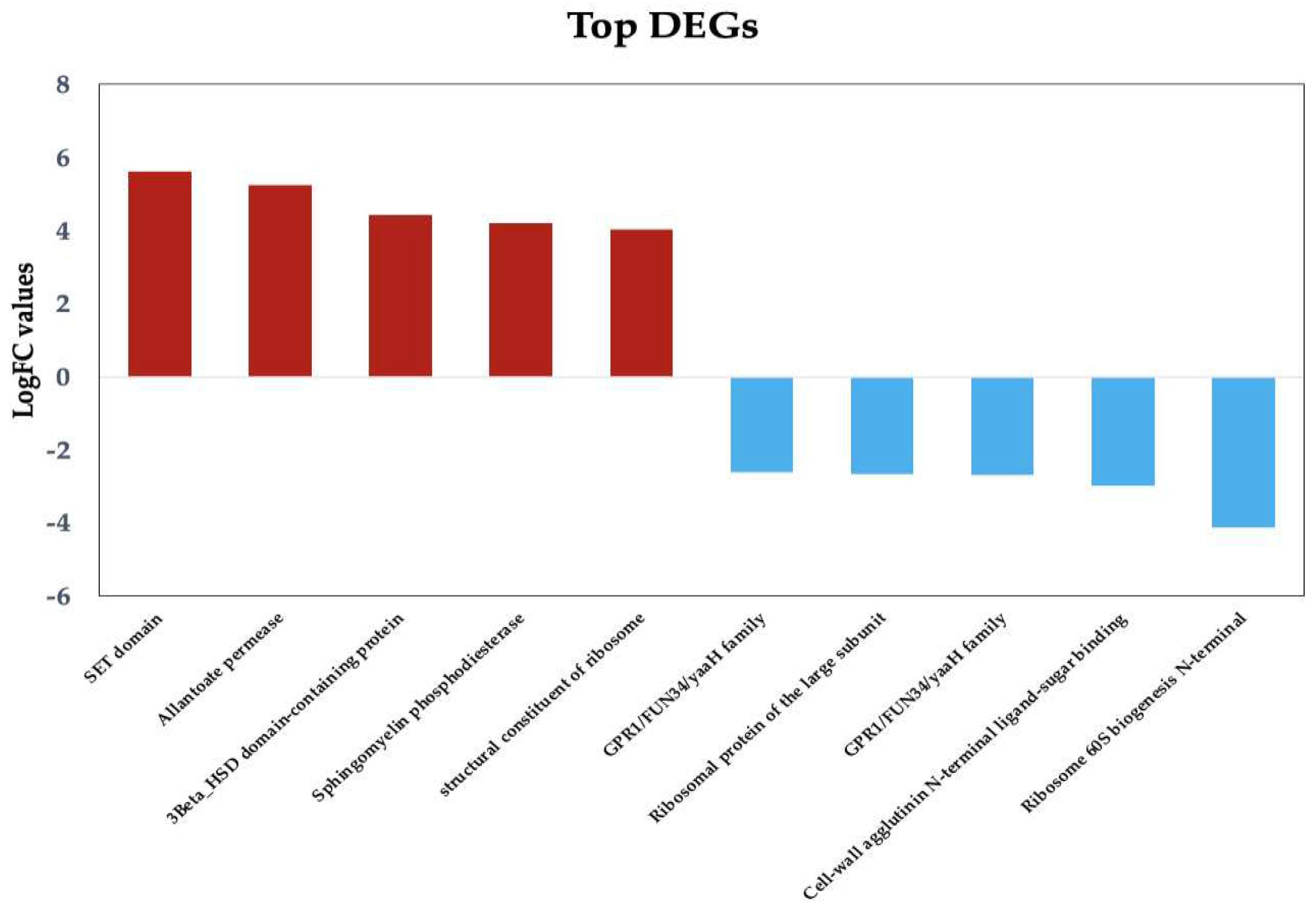
Figure 3 presents the key DEGs that are upregulated and downregulated in response to ISO treatment in the
C. tropicalis dataset. Noteworthy among the upregulated genes is the SET domain methyltransferase protein superfamily, encompassing proteins known for their histone methylation activity at lysine residues. Histone methylation is a pivotal process involved in chromatin regulation and gene expression [34]. Additionally, the upregulated genes encompass components of transmembrane transport systems, particularly those associated with the allenoate transport system [35]. Furthermore, genes related to the 3β-HSD enzyme system, crucial for the biosynthesis of various classes of steroid hormones, are observed [36,37]. The dataset also includes hydrolase enzyme genes involved in sphingolipid metabolism reactions [38,39], as well as genes contributing to the structural integrity of the ribosome.
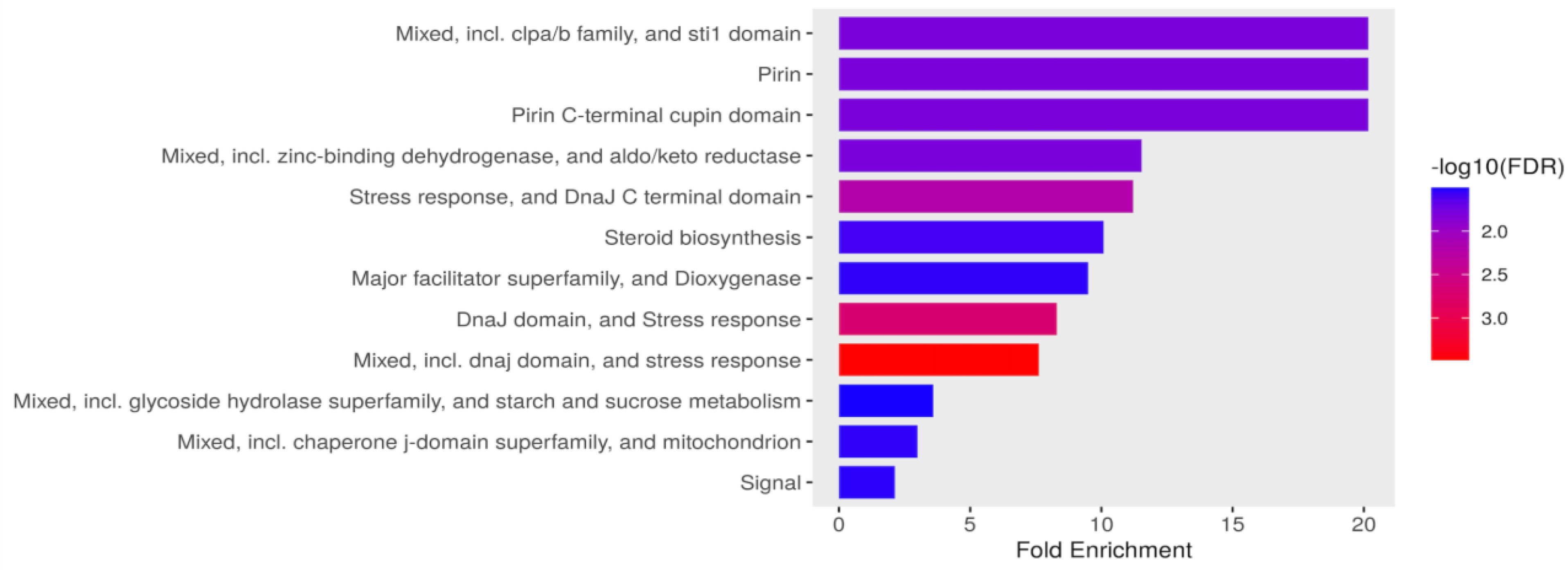
3.3.1. Stress response, metabolism, and mitochondrial functional processes are enriched in upregulated genes in C. tropicalis treated with ISO.
In response to ISO treatment, upregulated genes in
C. tropicalis were found to be associated with critical biological processes. Notably, these upregulated genes included ATP-dependent proteases of the Clpa/b family and the DnaJ domain (DnaJ/Hsp40: heat shock protein 40), which serve as essential chaperones involved in translation, protein folding, and protein translocation [40,41]. This suggests that ISO may impact protein synthesis in this yeast. Furthermore, the expression of the STI1 domain, known for its presence in the DNA damage response protein Rad23, was notable. This domain has a role in recognizing DNA damage, indicating that ISO's antifungal action might involve DNA damage to these pathogens. The upregulated genes also included those from the Aldo-Keto Reductase (AKR) superfamily, catalyzing redox transformations essential for biosynthesis, intermediary metabolism, and detoxification [43]. Additionally, overexpression of zinc-binding dehydrogenase family proteins, involved in oxidoreductase and catalytic activities, was observed, suggesting their role in responding to oxidative stress. Moreover, proteins related to the cellular response to organic substances, such as dioxygenases catalyzing critical reactions in aerobic microbial degradation of aromatic compounds, were upregulated.
Figure 4 illustrates the overexpression of genes associated with ergosterol biosynthesis, mitochondrial function, signaling pathways, and carbohydrate metabolism.
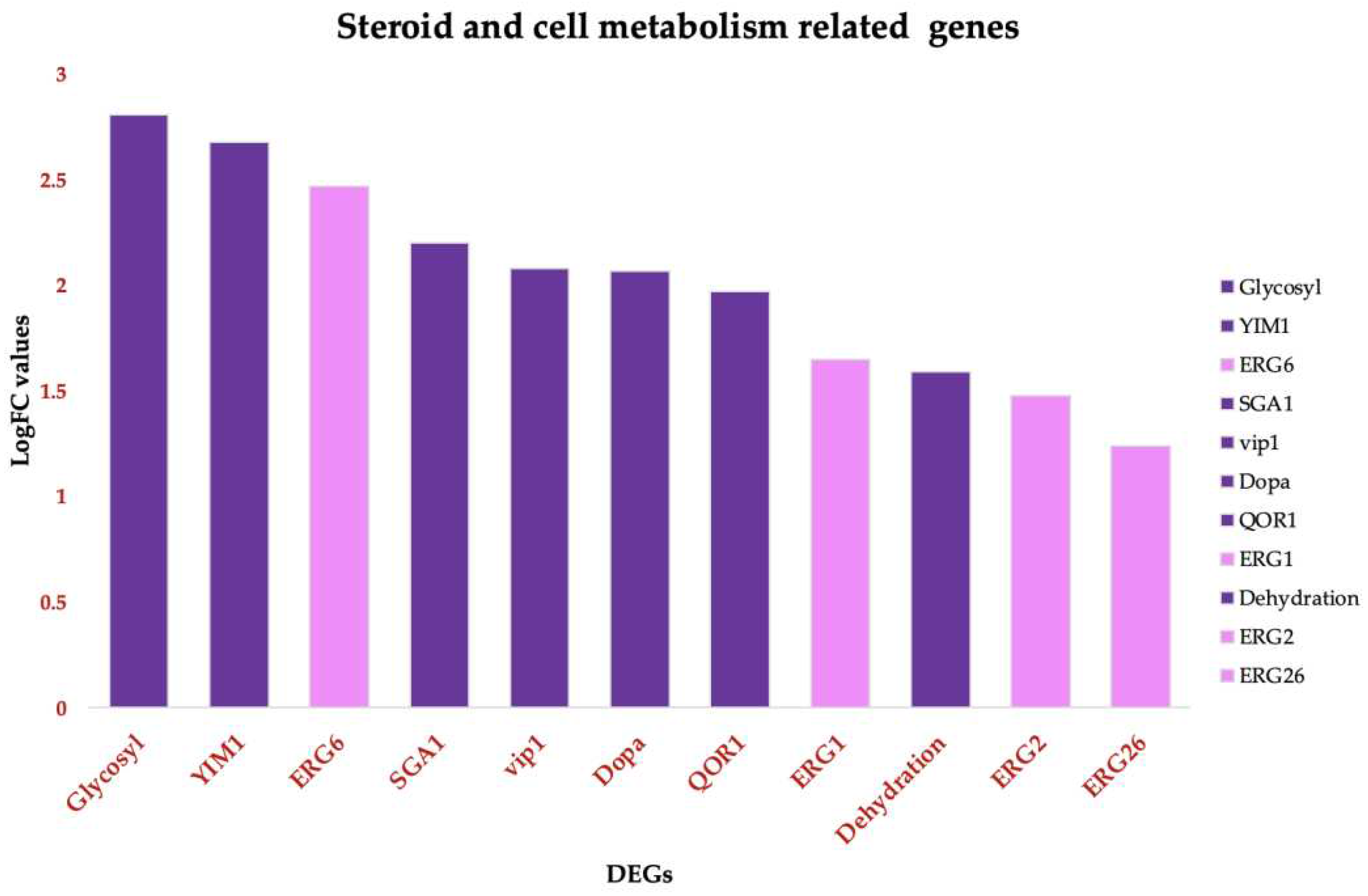
3.3.2. Upregulation of specific steroid and cell metabolism genes is an effect of C. tropicalis treated with ISO
Treatment of
Candida tropicalis with ISO at its Minimum Inhibitory Concentration (MIC) led to the notable upregulation of specific genes involved in ergosterol biosynthesis, including ERG1, ERG6, ERG2, and ERG26, (
Figure 5). Ergosterol is a fundamental component of the plasma membrane, and the increased expression of these genes suggests that ISO might impact its synthesis. Additionally, our results indicate the induction of genes associated with metabolism and the upregulation of Vip1, an inositol polyphosphate kinase known for its role as a signaling molecule. Vip1 regulates various essential biophysical processes, including autophagy and pathogenicity in
C. albicans [45].
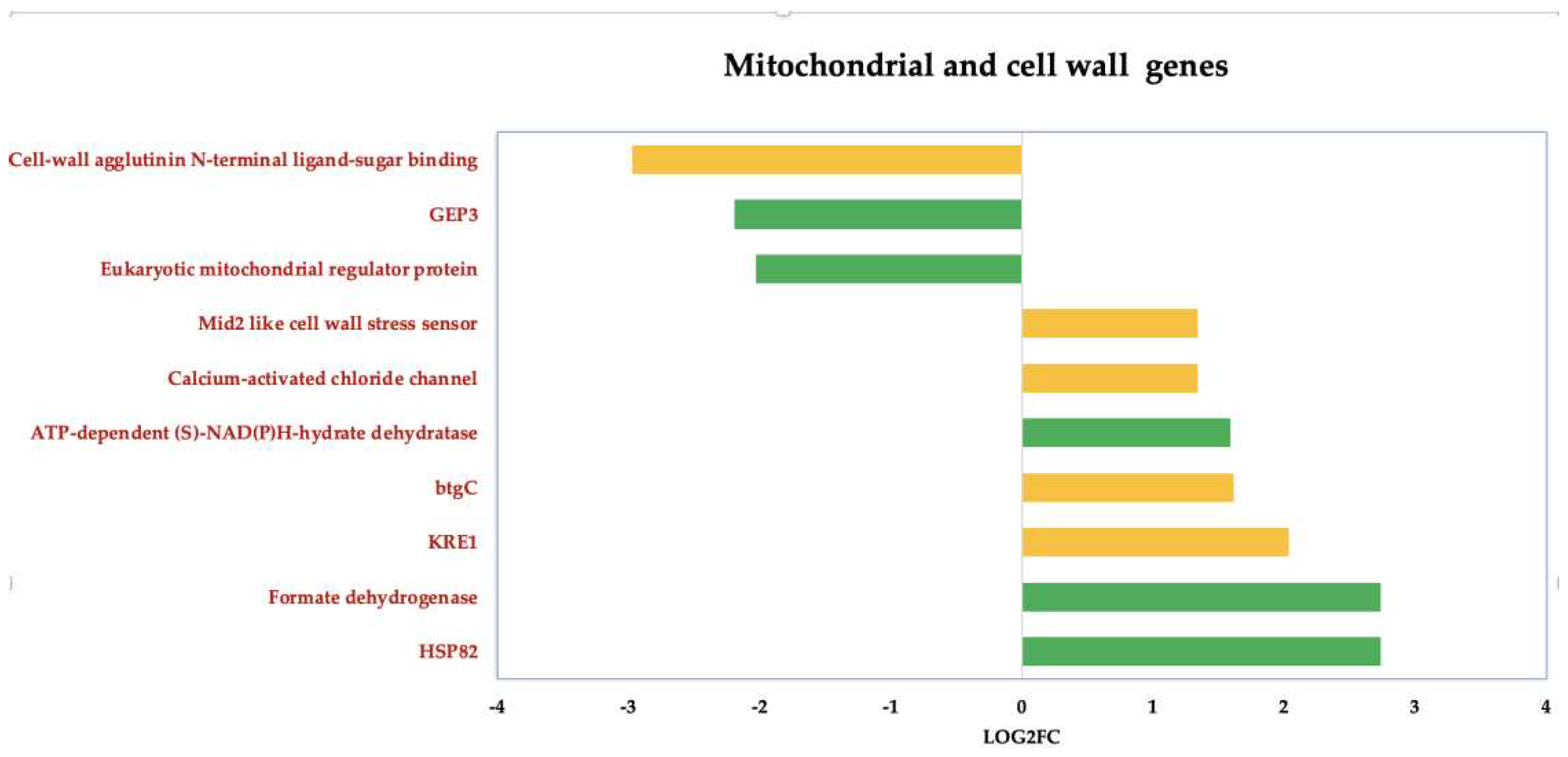
3.3.3. ISO-Induced Dysregulation of Mitochondrial and Cell Wall Genes in C. tropicalis
We also deepen insight into the genes that undergo dysregulation in response to ISO treatment, particularly those associated with mitochondrial function and cell wall pathways, (
Figure 6). Noteworthy among these genes is KRE1, which is involved in the synthesis of β-glucan in the cell wall of
C. albicans [46]. Additionally, the upregulation of the btgC gene, encoding a glucan endo-1,3-β-glucosidase, suggests potential involvement in β-glucan degradation. Furthermore, the overexpression of Mid2, cell surface sensors in cell wall integrity signaling [47], and calcium-activated chloride channels (CaCC), essential for cell physiology [48], was observed in
C. tropicalis. ISO treatment also induced the overexpression of HSP82 heat shock proteins, an essential gene family in yeast cells [49]. Additionally, enzymes such as formate dehydrogenase (FDH), crucial metalloenzymes catalyzing the reversible conversion of formate into carbon dioxide [50], were upregulated. These enzymes can utilize various electron donors, including ferredoxin, NAD, NADP, quinones, or F420, playing a vital role in mitochondrial metabolism.
Furthermore, the upregulation of ATP-dependent-(S)-NAD(P)H-hydrate dehydratase, catalyzing the dehydration of the S-form of NAD(P)HX at the expense of ATP, converting it to ADP, was also observed.
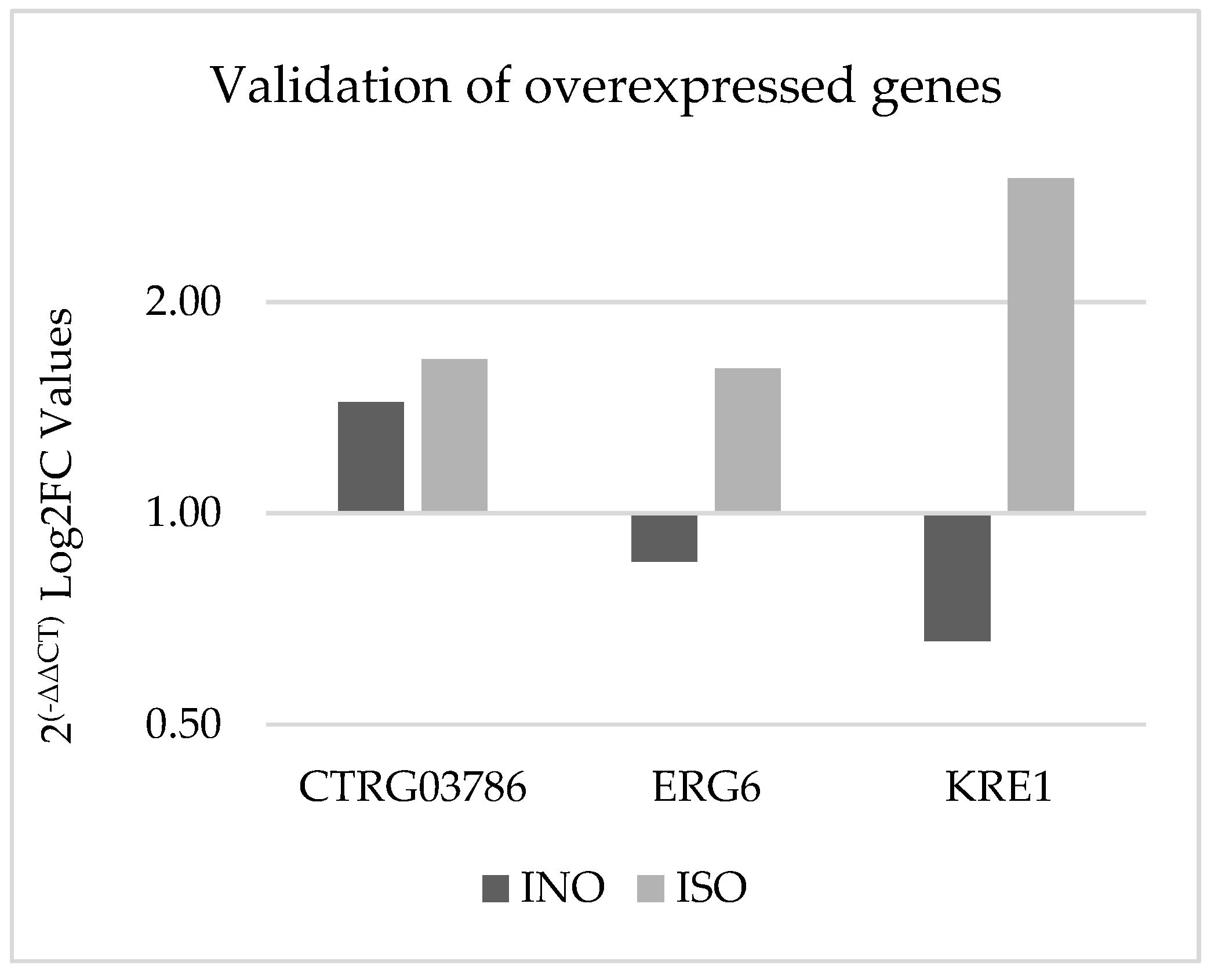
3.4. Gene expression validation using qPCR
Experimental validation was conducted using quantitative PCR (qPCR) to confirm the differential expression of specific genes in
C. tropicalis treated with ISO, as depicted in
Figure 7. Notably, the CTRG_03786 and ERG6 genes exhibited 1.14- and 1.8-fold higher expression, respectively, compared to the housekeeping gene in the case group. In contrast, the KRE1 gene demonstrated an eightfold higher expression in yeasts treated with isoespintanol compared to ACTIN-1. These qPCR results align with the findings obtained from the biostatistical analysis of RNA-seq.
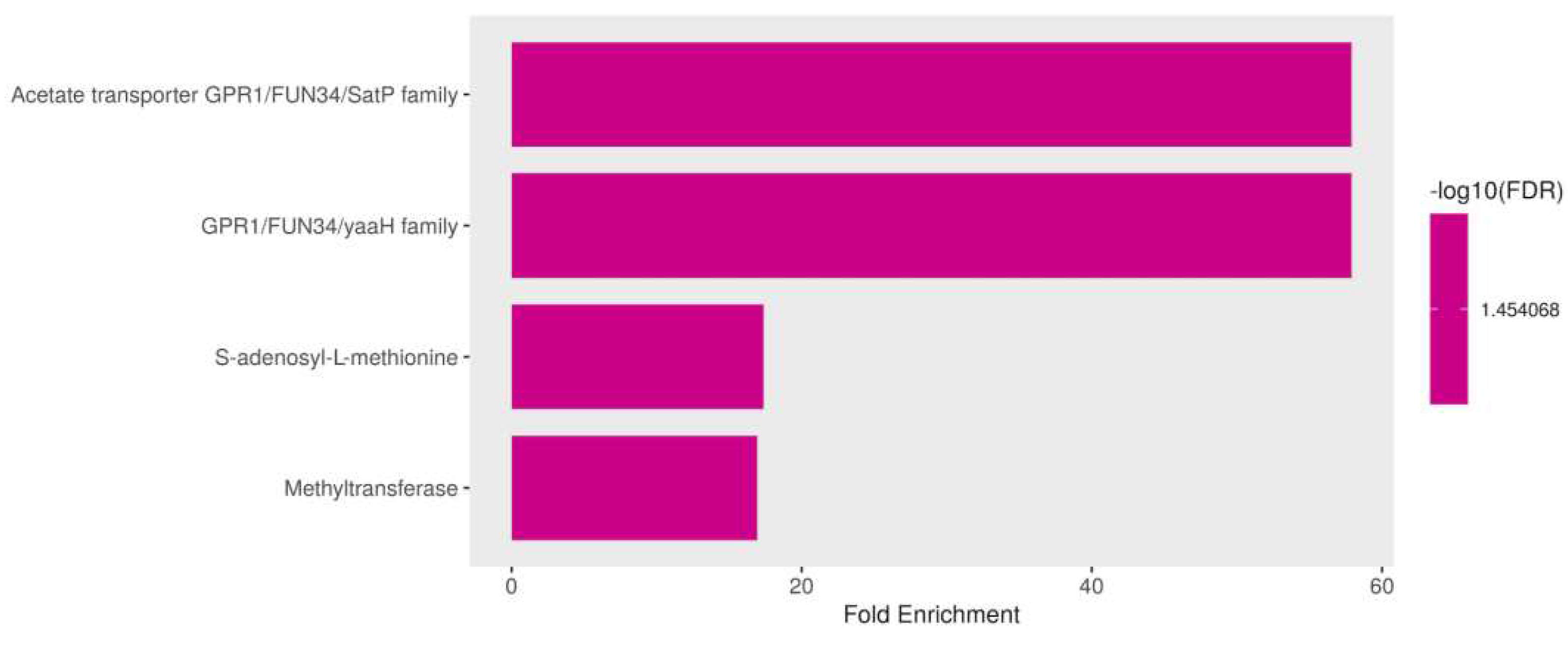
3.3.5. Down Regulation of Membrane Proteins and S-adenosyl-L-methionine (SAM) in Response to ISO Treatment
In addition to the previously mentioned down regulated genes in C. tropicalis exposed to ISO treatment, several genes exhibited down regulation, shedding light on the multifaceted impact of ISO on the yeast's transcriptome. These down regulated genes encompass membrane proteins and SAM-related processes, each with unique implications for the cellular functions and metabolism of the yeast. Among the down regulated genes, members of the membrane protein family, including mug84GPR1/FUN34/YaaH and mug86GPR1/FUN34/Satp, drew attention. These genes encode membrane proteins that are involved in acetate permease activity [59], a process responsible for transporting acetate across the fungal cell membrane. The down regulation of these genes implies that ISO treatment may disrupt acetate transport in C. tropicalis, potentially affecting various metabolic pathways. Furthermore, these membrane proteins are known to play a role in the regulation of morphogenesis and hyphal formation in C. albicans [60,61]. Consequently, their down regulation by ISO treatment may contribute to the inhibition of hyphal growth and morphological changes observed in C. tropicalis. Moreover, the down regulation of S-adenosyl-L-methionine (SAM) was observed. SAM is a critical nucleoside that serves as a central molecule in various cellular processes. It acts as a methyl donor for trans-methylation reactions, playing essential roles in epigenetic modifications, gene regulation, and protein methylation [62,63]. Additionally, SAM is involved in trans-sulfuration and trans-propylamine reactions, contributing to the synthesis of crucial molecules such as cysteine and polyamines. The down regulation of SAM in response to ISO treatment suggests that this monoterpene disrupts these essential metabolic pathways. Consequently, this disruption may affect gene regulation, protein function, and the overall cellular physiology of C. tropicalis.
In summary, the down regulation of specific membrane protein genes and SAM-related processes highlights the diverse and profound effects of ISO on the yeast's cellular processes. ISO's impact extends beyond the previously discussed deregulated genes and encompasses disruptions in acetate transport, morphogenesis regulation, and critical metabolic pathways like trans-methylation and trans-sulfuration. These findings underscore the intricate and multifaceted nature of ISO's antifungal mechanism, providing valuable targets for further exploration into the molecular mechanisms underlying ISO's antifungal activity and its potential as a therapeutic agent against
C. tropicalis infections, (
Figure 9).
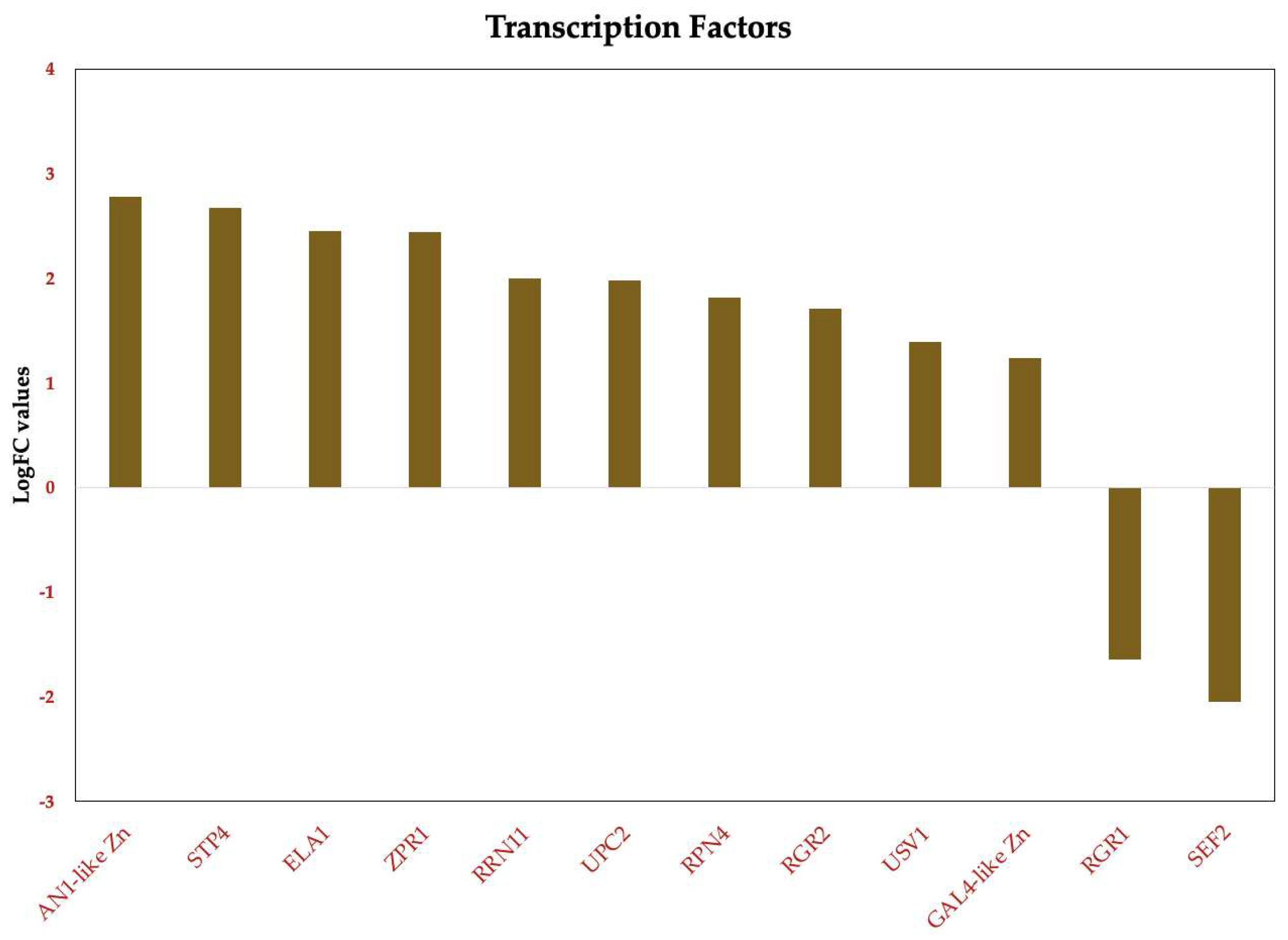
3.3.6. Overexpression of Zinc-Regulated Transcription Factors (TFs) in Candida tropicalis Induced by ISO Treatment
In this study, we observed a notable overexpression of genes encoding transcription factors (TFs) in
C. tropicalis in response to ISO treatment, shedding light on the intricate transcriptional changes elicited by this monoterpene. The upregulation of these TFs indicates their pivotal role in orchestrating various cellular responses and adaptations to ISO exposure. The observed overexpression of zinc-regulated TFs in
C. tropicalis subjected to ISO treatment sheds light on the intricate transcriptional landscape influenced by this monoterpene. The upregulation of GAL4-like Zn, a DNA binding TF, suggests its involvement in regulating the transcription of specific genes. GAL4-like Zn is known for its role in recognizing the palindromic DNA sequence of galactose metabolizing enzyme genes [64]. The overexpression of this TF may signify a shift in metabolic processes in response to ISO treatment, potentially influencing galactose metabolism or related pathways. Another induced TF is RRN11, a DNA-binding TF for RNA polymerase I, it plays a crucial role in the transcription of large nuclear rRNA transcripts [65]. Its overexpression suggests potential alterations in rRNA transcription in
C. tropicalis following ISO exposure, which may impact ribosomal biogenesis and protein synthesis. The upregulated GAL4-like Zn TF hints at potential alterations in metabolic pathways, possibly affecting galactose metabolism or related processes [64]. Concurrently, the upregulation of RRN11 TF points toward changes in rRNA transcription, impacting ribosomal biogenesis and protein synthesis [65]. Moreover, the overexpression of RPN4 TF suggests ISO-induced stress responses, potentially affecting protein turnover and DNA repair mechanisms [66,67]. The upregulated USV1 TF implies shifts in respiratory pathways, impacting energy metabolism and cellular respiration [68,69]. Additionally, the upregulation of ZPR1 suggests potential disruptions in the cell cycle progression [70]. ELA1 overexpression indicates ISO-induced DNA damage responses [71]. Furthermore, the overexpression of UPC2 TF implies ISO's impact on ergosterol biosynthesis pathways, influencing membrane composition and integrity [72,73]. Lastly, the overexpression of genes related to environmental stress responses, such as STP4, suggests that ISO triggers stress-related adaptations in
C. tropicalis [74], (
Figure 10).
Overall, the overexpression of these zinc-regulated TFs underscores the complex and multifaceted transcriptional responses of C. tropicalis to ISO treatment. These TFs play pivotal roles in regulating various cellular processes, including metabolism, stress responses, DNA damage repair, and cell cycle progression. Understanding the regulatory networks governed by these TFs is crucial for unraveling the mechanisms underlying ISO's antifungal activity and its potential as a therapeutic agent.
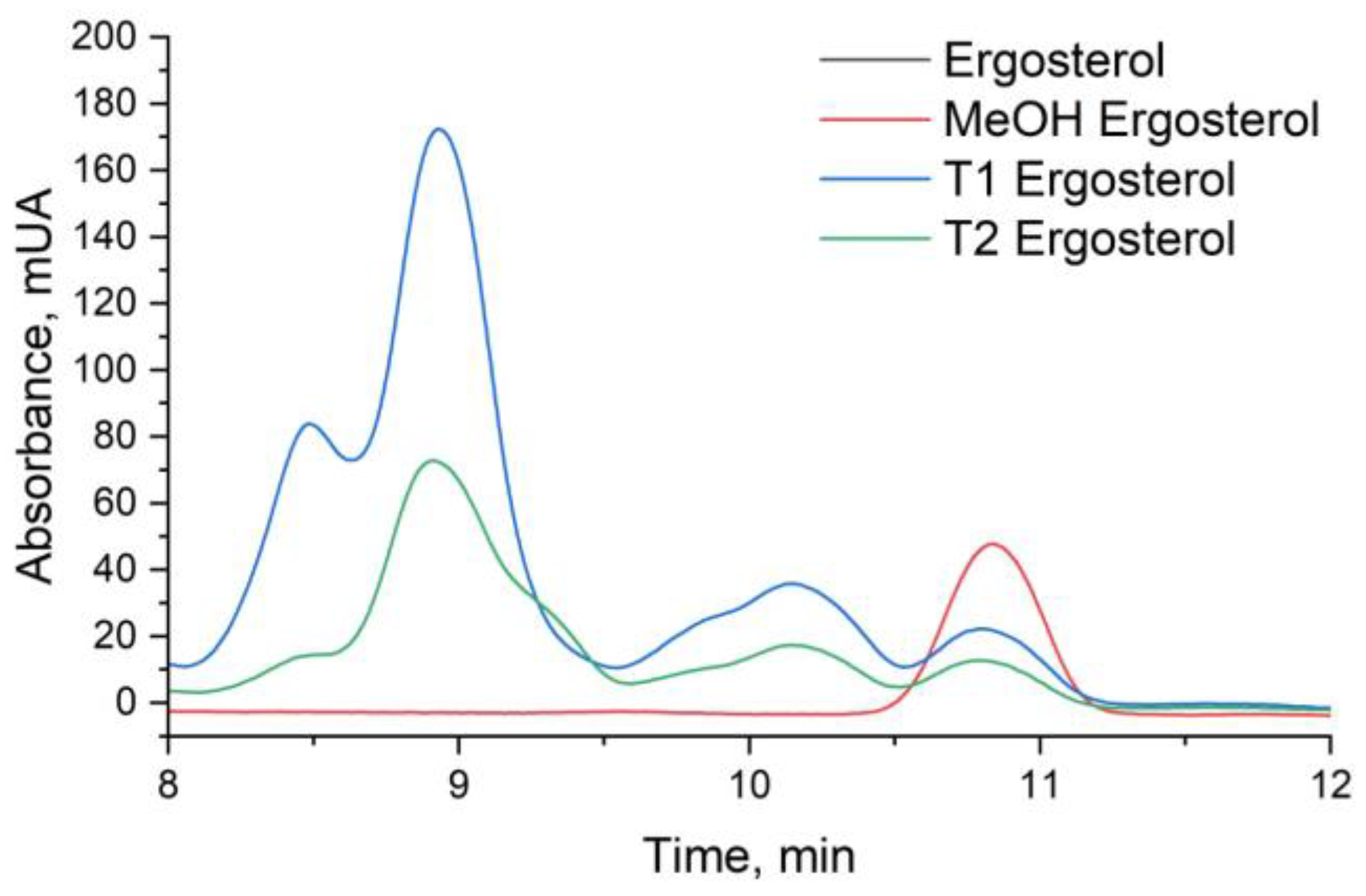
3.4. Disruption of Fungal Membrane Integrity: Ergosterol Content Analysis Reveals ISO's Antifungal Mechanism
The analysis of ergosterol content was conducted using UHPLC-DAD, revealing pertinent insights into the impact of ISO treatment on
C. tropicalis. The retention time for ergosterol, as determined by UHPLC-DAD analysis, was consistently observed at approximately 10.8 minutes (
Figure 11). The ergosterol content was quantified in both control (T1) and ISO-treated (T2) samples following a 3-hour incubation period. Notably, the ergosterol content in the control samples (T1) was measured at 28.40 ± 1.48 mg/L, while ISO-treated samples (T2) exhibited a notably reduced ergosterol content of 15.58 ± 0.37 mg/L (p < 0.05), as detailed in
Table 1. Furthermore, the standard curve employed in the analysis demonstrated excellent linearity, as evidenced by an R
2 value of 0.9965. These findings unequivocally indicate that ISO treatment resulted in a substantial reduction of ergosterol content in the membrane of
C. tropicalis, underscoring the pronounced effect of ISO on the fungal membrane composition. The significant reduction in ergosterol content in ISO-treated samples compared to control samples is a compelling finding. Ergosterol is essential for maintaining the integrity and fluidity of the fungal cell membrane. Therefore, a decrease in ergosterol levels can lead to membrane dysfunction, compromising the ability of
C. tropicalis to adapt and survive in different environments. This reduction in ergosterol content is consistent with the observed upregulation of genes associated with ergosterol biosynthesis in the transcriptomic analysis, suggesting that ISO may disrupt the synthesis of this critical membrane component.
The linearity of the standard curve, as indicated by an R2 value of 0.9965, further validates the accuracy of the ergosterol quantification method, strengthening the credibility of the results. These findings collectively emphasize the pronounced effect of ISO on the fungal membrane composition, which is crucial for the pathogenicity and survival of C. tropicalis. Understanding the impact of ISO on ergosterol content provides valuable information for future research and the development of antifungal therapies targeting membrane integrity and function.
4. Discussion.
Candida tropicalis has become a critical species within the Candida genus due to its increasing incidence, drug resistance, and high mortality rates, particularly among immunocompromised individuals [14,16,18,75,76]. Despite the efficacy of existing antifungals, the evolution of drug resistance in Candida spp. poses a significant therapeutic challenge [77]. Moreover, the limited treatment options and potential toxicity associated with current therapies necessitate the search for novel antifungal compounds, with natural plant-based compounds emerging as promising alternatives [81–83]. In previous studies, we established ISO's activity against clinical Candida spp. isolates, including C. tropicalis, and characterized its targets [26,27,84]. Here, we employ a transcriptomic approach to identify gene expression profiles, shedding light on the intricate mechanisms underlying ISO's antifungal activity.
Ergosterol, an essential component of fungal cell membranes, is distributed throughout various cellular components, including the cell membrane, intracellular endomembranes, and mitochondrial membranes [85]. Due to its absence in mammals, ergosterol serves as an attractive target for antifungal agents. These lipids house a multitude of biologically essential proteins involved in signaling, stress responses, nutrient transport, and other vital processes [77]. Ergosterol biosynthesis is a complex process regulated by numerous ERG genes [86]. Our results demonstrate the upregulation of ERG1, ERG6, ERG2, and ERG26 genes, all involved in ergosterol biosynthesis. The exact mechanism responsible for the global upregulation of ERG genes in response to azoles remains unclear. However, it has been proposed that disruptions in ergosterol or other sterol pathway components lead to global ERG gene expression increases. Alternatively, accumulation of early substrates or toxic sterol by products may trigger ERG expression [33]. We hypothesize that ISO treatment may disrupt ergosterol synthesis in C. tropicalis' plasma membrane, reducing its levels and subsequently increasing the expression of ERG genes. These results align with previous findings where C. albicans treated with azoles exhibited similar responses [90,91]. Notably, the ERG6 gene, which is not involved in cholesterol biosynthesis, may serve as a potential target for a new generation of antifungal drugs [92]. This supports previous observations that cell membrane damage is one of the mechanisms through which ISO exerts its antifungal action.
Fungal cell walls are vital for viability, morphogenesis, and pathogenesis [93]. Candida albicans' cell wall consists of an inner skeletal layer rich in β-(1,3)- and β-(1,6)-glucan and chitin, as well as an outer fibrillar layer primarily composed of highly mannosylated cell wall proteins [94,95]. Since the cell wall plays a crucial role in maintaining cellular osmotic integrity, damage to it triggers various responses [96]. Previous studies have shown that the inhibition of β-(1,3)-glucan synthesis leads to the compensatory synthesis of chitin and alterations in wall macromolecule arrangements in C. albicans. Similarly, C. tropicalis has been shown to activate a chitin compensatory response upon exposure to D-limonene [95] and ISO [84].
Of particular note is the upregulation of the KRE1 gene, which is implicated in β-glucan synthesis and assembly in yeast cell walls such as Saccharomyces cerevisiae and C. albicans. Mutations in this gene are associated with abnormal (1,6)-β-glucan production in yeast cell walls [46,97,98]. This suggests that ISO may indeed alter the cell wall structure in C. tropicalis. As cell wall β-glucans are common in fungi and absent in mammals, enzymes involved in β-glucan biosynthesis are potential targets for specific antifungal agents [93,99].
Monoterpenes have been reported to interact with various intracellular targets, including DNA and RNA, leading to disruptions in protein synthesis, metabolic processes, biosynthetic pathways, mitochondrial membrane potential, and more [100,101]. Our results demonstrate that ISO can penetrate C. tropicalis cells, interacting with intracellular structures, thereby disrupting the pathogens' normal metabolic processes, enzymatic activity, mitochondrial function, and signaling pathways. Notably, we observe the upregulation of genes related to protein translation and folding, suggesting that protein synthesis may be a crucial target in ISO's antifungal activity. Furthermore, the upregulation of the STI1 domain, known for its role in various cellular processes, including the transfer of hydrophobic substrates and the DNA damage response, suggests that ISO's antifungal action also involves DNA damage in C. tropicalis. Our results align with previous studies in C. tropicalis and C. parapsilosis, where terpenes like D-limonene have exhibited multiple action targets [24,25,100].
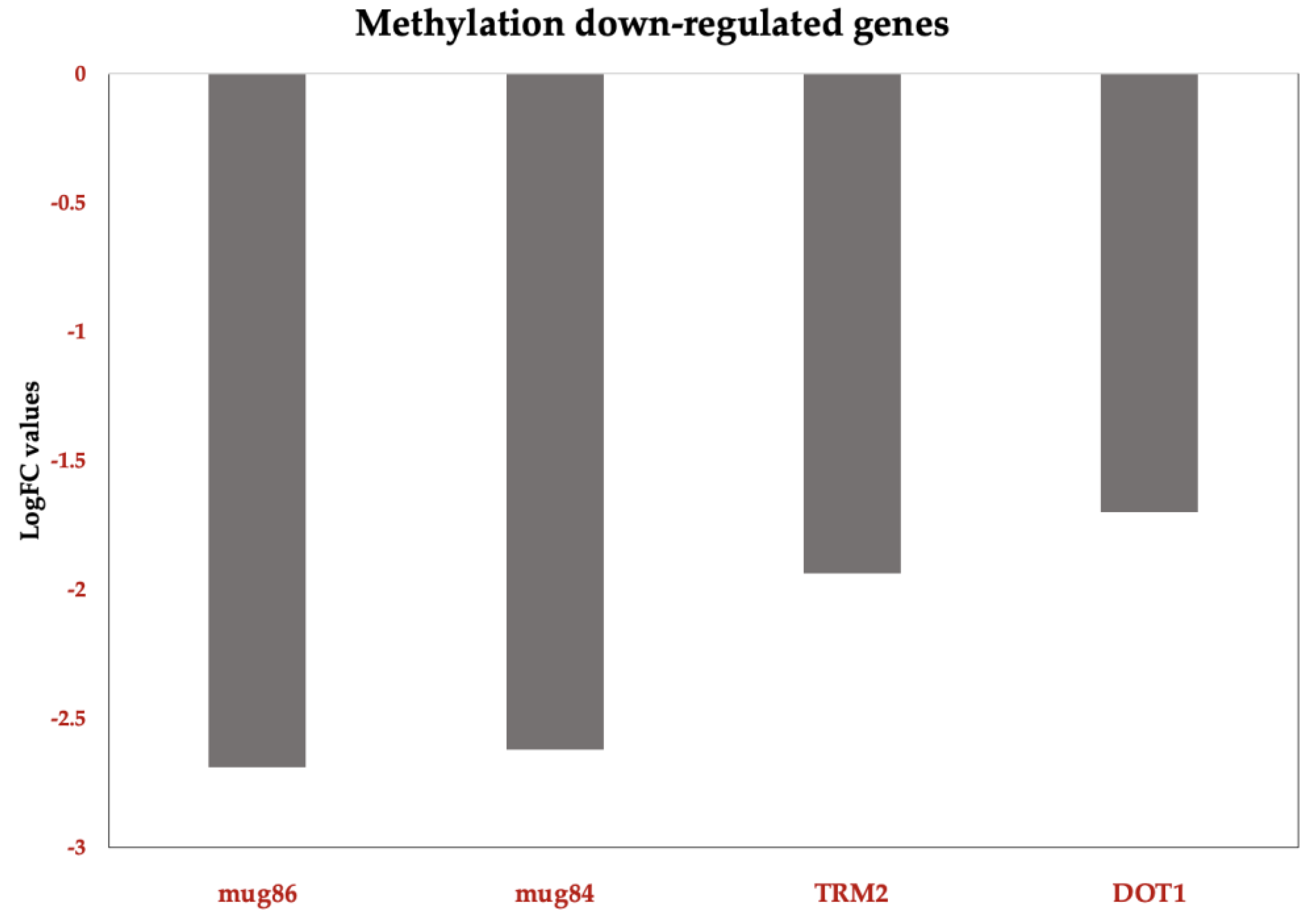
We also observe the downregulation of methylation-related genes in C. tropicalis following ISO treatment. Genes such as TRM7, TRM2, and DOT1, known for their roles in tRNA modification and histone methylation, were among those downregulated. Additionally, genes involved in acetate permease activity and the expression of S-adenosyl-L-methionine (SAM), a vital nucleoside serving as a methyl donor in various processes including biosynthesis of choline, carnitine, creatine, DNA and protein methylation, were found to be downregulated. These findings suggest that ISO's antifungal action involves complex regulation processes, potentially including post-transcriptional events.
Interestingly, our results reveal the upregulation of genes encoding transcription factors (TFs) in response to ISO treatment. These include GAL4-like Zn, RRN11, RPN4, USV1, ZPR1, ELA1, and UPC2. These TFs play various roles, from binding to specific DNA sequences to regulating DNA repair and stress responses. This upregulation highlights the complexity of ISO's antifungal mechanism, suggesting that it may involve multiple regulatory processes, including post-transcriptional events. These findings underscore the multi-faceted nature of ISO's action against C. tropicalis.
In conclusion, ISO's antifungal activity against C. tropicalis involves a multi-targeted approach that disrupts ergosterol biosynthesis, alters cell wall structure, affects multiple cellular processes, and potentially induces DNA damage. The complexity of ISO's mechanism highlights its potential as a promising antifungal agent against drug-resistant Candida species.
5. Conclusions
In this study, we conducted a comprehensive transcriptomic analysis of C. tropicalis exposed to ISO, a natural monoterpene isolated from O. xylopioides. Our findings not only reaffirm the antifungal properties of ISO, as previously reported, but also shed light on the intricate mechanisms underlying its antifungal activity. The complexity of ISO's action, as revealed by our results, underscores its potential as a promising antifungal agent against C. tropicalis, a clinically significant pathogenic yeast known for its increasing drug resistance and mortality rates among immunocompromised individuals. These results provide valuable insights into potential clinical molecular targets within C. tropicalis that may be explored in the future for the development of novel treatments against this pathogenic yeast. This research contributes to our understanding of the molecular basis of ISO's antifungal activity and opens new avenues for the development of effective therapies for combating drug-resistant Candida species in clinical settings.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.. Table S1: List of real-time qPCR primers. Table S2. RNA extraction of C. tropicalis. Quality control total RNA extracted from Zoea-type larvae of Callinectus sapidus. Table S3. NGS sequencing by RNA-seq of C. tropicalis. Table S4. Mapping of transcriptomes obtained by RNA-seq.
Author Contributions
Conceptualization, O.I.C.M., K.A.P. and A.A.O.; methodology, O.I.C.M., M.C.V.P., K.A.P. and A.A.O.; software, O.I.C.M. and K.A.P.; validation, O.I.C.M., M.C.V.P., M.F.Y. and K.A.P.; formal analysis, O.I.C.M., K.A.P., M.C.V.P. and A.A.O.; investigation, O.I.C.M., M.C.V.P., and K.A.P.; resources, A.A.O.; M.F.Y. and G.S.P.; writing—original draft preparation, O.I.C.M., K.A.P., M.C.V.P. and A.A.O.; writing—review and editing, O.I.C.M., A.A.O., K.A.P. and G.S.P.; visualization, O.I.C.M. and K.A.P.; supervision, O.I.C.M., A.A.O. and G.S.P.; funding acquisition, A.A.O., M.F.Y. and G.S.P. All authors have read and agreed to the published version of the manuscript.
Funding
O.C.M. thanks the scholarship program of the Ministry of Science, Technology and Innovation of Colombia for the granting of the doctoral scholarship. This research was funded with resources from the FCB-02-19 project of the University of Córdoba, Montería, Colombia. K.A.P (CVU:227919) received financial support from CONACyT and a fellowship from the Fulbright García-Robles Foundation.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Code and Supplementary Material have been deposited in https://github.com/kap8416/transcriptomicsofcandidatropicalis_isoespintanol.
Acknowledgments
The authors thank Loranda Calderón Zamora for her technical assistance in data visualization.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schils, R.; Rampat, R.; Rakic, J.-M.; Crahay, F.-X. Candida Chorioretinitis in Renal Transplant Recipient with Candidemia Related to Contaminated Organ Preservation Fluid : A Role for Dilated Fundus Examination in Its Management. IDCases 2023, 32 (March), 0–1. [CrossRef]
- Ohashi, Y.; Matono, T.; Suzuki, S.; Yoshino, S.; Alshahni, M. M.; Komori, A.; Makimura, K. The First Case of Clade I Candida auris Candidemia in a Patient with COVID-19 in Japan. J. Infect. Chemother. 2023, 29 (7), 713–717. [CrossRef]
- Papadimitriou-Olivgeris, M.; Kolonitsiou, F.; Kefala, S.; Spiliopoulou, A.; Aretha, D.; Bartzavali, C.; Siapika, A.; Marangos, M.; Fligou, F. Increased Incidence of Candidemia in Critically Ill Patients during the Coronavirus Disease 2019 (COVID-19) Pandemic. Brazilian J. Infect. Dis. 2022, 26 (2), 102353. [CrossRef]
- Kimura, S.; Kameda, K.; Harada, K.; Saburi, M.; Okinaka, K.; Shinohara, A.; Uchida, N.; Nishijima, A.; Ozawa, Y.; Tanaka, M.; Keriyama, T.; Katayama, Y.; Sawa, M.; Ikegame, K.; Kawakita, T.; Kanda, Y.; Nakamae, H.; Ara, T.; Kimura, T.; Sato, A.; Fukuda, T.; Atsuta, Y.; Nakasone, H. Risk and Predictive Factors for Candidemia After Allogeneic Hematopoietic Cell Transplantation: JSTCT Transplant Complications Working Group. Transplant. Cell. Ther. 2022, 28 (4), 209.e1-209.e9. [CrossRef]
- Lin, J.; Zhou, M.; Chen, J.; Zhang, L.; Lu, M.; Liu, Z. De-Escalation from Echinocandins to Azole Treatment in Critically Ill Patients with Candidemia. Int. J. Infect. Dis. 2022, 121, 69–74. [CrossRef]
- Saiprom, N.; Wongsuk, T.; Oonanant, W.; Sukphopetch, P.; Chantratita, N.; Boonsilp, S. Characterization of Virulence Factors in Candida Species Causing Candidemia in a Tertiary Care Hospital in Bangkok, Thailand. J. Fungi 2023, 9, 353. [CrossRef]
- Vargas-Espíndola, L. A.; Cuervo-Maldonado, S. I.; Enciso-Olivera, J.; Gómez-Rincón, J.; Jiménez-Cetina, L.; Sánchez-Pedraza, R.; García-Guzmán, K.; López-Mora, M. J.; Álvarez-Moreno, C.; Cortés, J. A.; Garzón-Herazo, J.; Martínez-Vernaza, S.; Sierra-Parada, C.; Murillo-Sarmiento, B. Fungemia in Hospitalized Adult Patients with Hematological Malignancies : Epidemiology and Risk Factors. J. Fungi 2023, 9, 400. [CrossRef]
- Alvarez-Moreno, C. A.; Morales-López, S.; Rodriguez, G. J.; Rodriguez, J. Y.; Robert, E.; Picot, C.; Ceballos-Garzon, A.; Parra-Giraldo, C. M.; Le Pape, P. The Mortality Attributable to Candidemia in C. auris Is Higher than That in Other Candida Species: Myth or Reality? J. Fungi 2023, 9 (4), 430. [CrossRef]
- Pappas, P.; Lionakis, M.; Cavling, M.; Ostrosky-Zeichner, L.; Kullberg, B. Invasive Candidiasis. Dis. Prim. 2018, 4. [CrossRef]
- McCarty, T. P.; White, C. M.; Pappas, P. G. Candidemia and Invasive Candidiasis. Infect. Dis. Clin. North Am. 2021, 35 (2), 389–413. [CrossRef]
- Gómez-Gaviria, M.; Ramírez-Sotelo, U.; Mora-Montes, H. M. Non-Albicans Candida Species: Immune Response, Evasion Mechanisms, and New Plant-Derived Alternative Therapies. J. Fungi 2023, 9 (1). [CrossRef]
- Ceballos-Garzon, A.; Peñuela, A.; Valderrama-Beltrán, S.; Vargas-Casanova, Y.; Ariza, B.; Parra-Giraldo, C. M. Emergence and Circulation of Azole-Resistant C. albicans, C. auris and C. parapsilosis Bloodstream Isolates Carrying Y132F, K143R or T220L Erg11p Substitutions in Colombia. Front. Cell. Infect. Microbiol. 2023, 13 (March), 1–11. [CrossRef]
- Osset-Trénor, P.; Pascual-Ahuir, A.; Proft, M. Fungal Drug Response and Antimicrobial Resistance. J. Fungi 2023, 9 (5). [CrossRef]
- El-kholy, M. A.; Helaly, G. F.; El Ghazzawi, E. F.; El-sawaf, G.; Shawky, S. M. Virulence Factors and Antifungal Susceptibility Profile of C. tropicalis Isolated from Various Clinical Specimens in Alexandria, Egypt. J. Fungi 2021, 7 (5), 351. [CrossRef]
- De Oliveira, J. S.; Pereira, V. S.; Castelo-Branco, D. de S. C. M.; Cordeiro, R. de A.; Sidrim, J. J. C.; Brilhante, R. S. N.; Rocha, M. F. G. The Yeast, the Antifungal, and the Wardrobe: A Journey into Antifungal Resistance Mechanisms of Candida tropicalis. Can. J. Microbiol. 2020, 66 (6), 377–388. [CrossRef]
- Munhoz-Alves, N.; Nishiyama Mimura, L. A.; Viero, R. M.; Bagagli, E.; Schatzmann, J.; Sartori, A.; de Campos. Candida tropicalis Systemic Infection Redirects Leukocyte Infiltration to the Kidneys Attenuating Encephalomyelitis. J. Fungi 2021, 7, 757. [CrossRef]
- Silva, S.; Negri, M.; Henriques, M.; Oliveira, R.; Williams, D. W.; Azeredo, J. Candida glabrata, Candida parapsilosis and Candida tropicalis: Biology, Epidemiology, Pathogenicity and Antifungal Resistance. FEMS Microbiol. Rev. 2012, 36 (2), 288–305. [CrossRef]
- Wang, D.; An, N.; Yang, Y.; Yang, X.; Fan, Y.; Feng, J. Candida tropicalis Distribution and Drug Resistance Is Correlated with ERG11 and UPC2 Expression. 2021, 1–9. [CrossRef]
- Fan, X.; Xiao, M.; Liao, K.; Kudinha, T.; Wang, H.; Zhang, L.; Hou, X.; Kong, F.; Xu, Y. C. Notable Increasing Trend in Azole Non-Susceptible Candida tropicalis Causing Invasive Candidiasis in China (August 2009 to July 2014): Molecular Epidemiology and Clinical Azole Consumption. Front. Microbiol. 2017, 8 (MAR), 1–9. [CrossRef]
- Spruijtenburg, B.; Baqueiro, C. C. S. Z.; Colombo, A. L.; Meijer, E. F. J.; de Almeida, J.; Berrio, I.; Fernández, N. B.; Chaves, G. M.; Meis, J.; de Groot, T. Short Tandem Repeat Genotyping and Antifungal Susceptibility Testing of Latin American Candida tropicalis Isolates. J. Fungi 2023, 9, 1–11. [CrossRef]
- Zuza-Alves, D. L.; Sila-Rocha, W. P.; Chaves, G. An Update on Candida tropicalis Based on Basic and Clinical Approaches. Front. Microbiol. 2017, 8. [CrossRef]
- Argüelles, A.; Sánchez-Fresneda, R.; Guirao-Abad, J. P.; Lozano, J. A.; Solano, F.; Argüelles, J. C. Insight into the Antifungal Effects of Propolis and Carnosic Acid—Extension to the Pathogenic Yeast Candida glabrata: New Propolis Fractionation and Potential Synergistic Applications. J. Fungi 2023, 9 (4). [CrossRef]
- Iraji, A.; Yazdanpanah, S.; Alizadeh, F.; Mirzamohammadi, S.; Ghasemi, Y.; Pakshir, K.; Yang, Y.; Zomorodian, K. Screening the Antifungal Activities of Monoterpenes and Their Isomers against Candida Species. J. Appl. Microbiol. 2020, 129 (6), 1541–1551. [CrossRef]
- Leite-Andrade, M. C.; de Araújo Neto, L. N.; Buonafina-Paz, M. D. S.; de Assis Graciano dos Santos, F.; da Silva Alves, A. I.; de Castro, M. C. A. B.; Mori, E.; de Lacerda, B. C. G. V.; Araújo, I. M.; Coutinho, H. D. M.; Kowalska, G.; Kowalski, R.; Baj, T.; Neves, R. P. Antifungal Effect and Inhibition of the Virulence Mechanism of D-Limonene against Candida parapsilosis. Molecules 2022, 27 (24), 1–12. [CrossRef]
- Yu, H.; Lin, Z.; Xiang, W.; Huang, M.; Tang, J.; Lu, Y.; Zhao, Q.; Zhang, Q.; Rao, Y.; Liu, L. Antifungal Activity and Mechanism of D -Limonene against Foodborne Opportunistic Pathogen Candida tropicalis. LWT-Food Sci. Technol. 2022, 159, 113144. [CrossRef]
- Contreras Martínez, O. I.; Angulo Ortíz, A. A.; Santafé Patiño, G. Mechanism of Antifungal Action of Monoterpene Isoespintanol against Clinical Isolates of Candida tropicalis. Molecules 2022, 27, 5808. [CrossRef]
- Contreras Martínez, O. I.; Angulo Ortíz, A. A.; Santafé Patiño, G. Antifungal Potential of Isoespintanol Extracted from Oxandra xylopioides Diels (Annonaceae) against Intrahospital Isolations of Candida SPP. Heliyon 2022, 8 (10). [CrossRef]
- Ramírez, R. D.; Páez, M. S.; Angulo, A. A. Obtención de Isoespintanol Por Hidrodestilación y Cristalización a Partir Del Extracto Bencínico de Oxandra xylopioides. Inf. Tecnol. 2015, 26 (6), 13–18. [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57 (1), 289–300. [CrossRef]
- Ge, S. X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36 (8), 2628–2629. [CrossRef]
- Silva, M.; Cardozo, D.; Fernando, P.; Freitas, F.; Fuzo, C.; Santos, C.; Rodrigues, M.; Santana, J. Modulation of ERG Genes Expression in Clinical Isolates of Candida tropicalis Susceptible and Resistant to Fluconazole and Itraconazole. Mycopathologia 2020, 6, 675–684. [CrossRef]
- Livak, K. J.; Schmittgen, T. D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25 (4), 402–408. [CrossRef]
- Zhang, X.; Zhang, T.; Guo, S.; Zhang, Y.; Sheng, R.; Sun, R.; Chen, L.; Lv, R.; Qi, Y. In Vitro Antifungal Activity and Mechanism of Ag3PW12O40 Composites against Candida Species. Molecules 2020, 25, 6012. [CrossRef]
- Dillon, S. C.; Zhang, X.; Trievel, R. C.; Cheng, X. The SET-Domain Protein Superfamily: Protein Lysine Methyltransferases. Genome Biol. 2005, 6 (8), 227. [CrossRef]
- Rai, R.; Genbauffe, F.; Cooper, T. Structure and Transcription of the Allantoate Permease Gene (DAL5) from Saccharomyces cerevisiae. J. Bacteriol. 1988, 170 (1), 266–271. [CrossRef]
- Simard, J.; Ricketts, M.; Gingras, S.; Soucy, P.; Feltus, A.; Melner, M. Molecular Biology of the 3β-Hydroxysteroid Dehydrogenase/Δ5-Δ4 Isomerase Gene Family. Endocr. Rev. 2005, 26 (4), 525–582. [CrossRef]
- Wang, L.; Salavaggione, E.; Pelleymounter, L.; Eckloff, B.; Wieben, E.; Weinshilboum, R. Human 3β-Hydroxysteroid Dehydrogenase Types 1 and 2: Gene Sequence Variation and Functional Genomics. J Steroid Biochem Mol Biol 2007, 107, 88–99. [CrossRef]
- Han, L.; Ding, G.; Liu, Y.; Huang, J.; Wu, J. Characterization of Sphingomyelin Phosphodiesterase Expression in Bumblebee (Bombus lantschouensis). J. Insect Sci. 2018, 18 (5), 1–7. [CrossRef]
- Ueda, S.; Manabe, Y.; Kubo, N.; Morino, N.; Yuasa, H.; Shiotsu, M.; Tsuji, T.; Sugawara, T.; Kambe, T. Early Secretory Pathway-Resident Zn Transporter Proteins Contribute to Cellular Sphingolipid Metabolism through Activation of Sphingomyelin Phosphodiesterase 1. Am. J. Physiol. Cell Physiol. 2022, 322. [CrossRef]
- Sherman, M. Y. S.; Goldberg, A. L. Involvement of Molecular Chaperones in Intracellular Protein Breakdown. Stress. Cell. Responses 1996. [CrossRef]
- Qiu, X. B.; Shao, Y. M.; Miao, S.; Wang, L. The Diversity of the DnaJ/Hsp40 Family, the Crucial Partners for Hsp70 Chaperones. Cell. Mol. Life Sci. 2006, 63 (22), 2560–2570. [CrossRef]
- Fry, M. Y.; Saladi, S. M.; Clemons, W. M. The STI1-Domain Is a Flexible Alpha-Helical Fold with a Hydrophobic Groove. Protein Sci. 2021, 30 (4), 882–898. [CrossRef]
- Barski, O. A.; Tipparaju, S. M.; Bhatnagar, A. The Aldo-Keto Reductase Superfamily and Its Role in Drug Metabolism and Detoxification. Drug Metab Rev. 2008, 40 (4), 553-624. [CrossRef]
- Fetzner, S. Ring-Cleaving Dioxygenases with a Cupin Fold. Appl. Environ. Microbiol. 2012, 2505–2514. [CrossRef]
- Ma, T.; Yu, Q.; Ma, C.; Mao, X.; Liu, Y.; Peng, X.; Li, M. Role of the Inositol Polyphosphate Kinase Vip1 in Autophagy and Pathogenesis in Candida albicans. Future Microbiol. 2020, 15 (14), 1363–1377. [CrossRef]
- Boone, C.; Sdicu, A. M.; Laroche, M.; Bussey, H. Isolation from Candida albicans of a Functional Homolog of the Saccharomyces cerevisiae KRE1 Gene, Which Is Involved in Cell Wall β-Glucan Synthesis. J. Bacteriol. 1991, 173 (21), 6859–6864. [CrossRef]
- Philip, B.; Levin, D. E. Wsc1 and Mid2 Are Cell Surface Sensors for Cell Wall Integrity Signaling That Act through Rom2, a Guanine Nucleotide Exchange Factor for Rho1. Mol. Cell. Biol. 2001, 21 (1), 271–280. [CrossRef]
- Hartzell, C.; Putzier, I.; Arreola, J. Calcium-Activated Chloride Channels. Annu. Rev. Physiol. 2005, 67, 719–758. [CrossRef]
- Borkovich, K. A.; Farrelly, F. W.; Finkelstein, D. B.; Taulien, J.; Lindquist, S. Hsp82 Is an Essential Protein That Is Required in Higher Concentrations for Growth of Cells at Higher Temperatures. Mol. Cell. Biol. 1989, 9 (9), 3919–3930. [CrossRef]
- Dong, G.; Ryde, U. Reaction Mechanism of Formate Dehydrogenase Studied by Computational Methods. J. Biol. Inorg. Chem. 2018, 23 (8), 1243–1254. [CrossRef]
- Ljungdahl, L. G. Formate Dehydrogenases: Role of Molybdenum, Tungsten and Selenium. Pergamon Press Ltd, 1980; pp 463–486. [CrossRef]
- Hirata, A.; Okada, K.; Yoshii, K.; Shiraishi, H.; Saijo, S.; Yonezawa, K.; Shimizu, N.; Hori, H. Structure of TRNA Methyltransferase Complex of Trm7 and Trm734 Reveals a Novel Binding Interface for TRNA Recognition. Nucleic Acids Res. 2019, 47 (20), 10942–10955. [CrossRef]
- Pintard, L.; Lecointe, F.; Bujnicki, J. M.; Bonnerot, C.; Grosjean, H.; Lapeyre, B. Trm7p Catalyses the Formation of Two 2¢-O-Methylriboses in Yeast TRNA Anticodon Loop Lionel. EMBO J. Vol. 2002, 21 (7), 1811–1820. [CrossRef]
- Nordlund, M. E.; Johansson, J. O. M.; Von Pawel-Rammingen, U.; Byström, A. S. Identification of the TRM2 Gene Encoding the TRNA(M5U54) Methyltransferase of Saccharomyces cerevisiae. RNA 2000, 6 (6), 844–860. [CrossRef]
- Leeuwen, F. Van; Gafken, P. R.; Gottschling, D. E. Dot1p Modulates Silencing in Yeast by Methylation of the Nucleosome Core. Cell 2002, 109, 745–756. [CrossRef]
- San-segundo, P. A.; Roeder, G. S. Role for the Silencing Protein Dot1 in Meiotic Checkpoint Control. Mol. Biol. Cell 2000, 11 (October), 3601–3615.
- Wysocki, R.; Javaheri, A.; Sha, F.; Co, J. Role of Dot1-Dependent Histone H3 Methylation in G 1 and S Phase DNA Damage Checkpoint Functions of Rad9. Mol. Cell. Biol. 2005, 25 (19), 8430–8443. [CrossRef]
- Frederiks, F.; Heynen, G. J. J. E.; Van Deventer, S. J.; Janssen, H.; Van Leeuwen, F. Two Dot1 Isoforms in Saccharomyces cerevisiae as a Result of Leaky Scanning by the Ribosome. Nucleic Acids Res. 2009, 37 (21), 7047–7058. [CrossRef]
- Robellet, X.; Flipphi, M.; Pégot, S.; MacCabe, A.; Vélot, C. AcpA , a Member of the GPR1 / FUN34 / YaaH Membrane Protein Family , Is Essential for Acetate Permease Activity in the Hyphal Fungus Aspergillus nidulans. Biochem J. 2008, 493, 485–493. [CrossRef]
- Miwa, T.; Takagi, Y.; Shinozaki, M.; Yun, C. W.; Schell, W. A.; Perfect, J. R.; Kumagai, H.; Tamaki, H. Gpr1, a Putative G-Protein-Coupled Receptor, Regulates Morphogenesis and Hypha Formation in the Pathogenic Fungus Candida albicans. Eukaryot. Cell 2004, 3 (4), 919–931. [CrossRef]
- Maidan, M. M.; De Rop, L.; Serneels, J.; Exler, S.; Rupp, S.; Tournu, H.; Thevelein, J.; Van Dijck, P. The G Protein-Coupled Receptor Gpr1 and the Gα Protein Gpa2 Act through the CAMP-Protein Kinase A Pathway to Induce Morphogenesis in Candida albicans. Mol. Biol. Cell 2005, 16, 1971–1986. [CrossRef]
- Fontecave, M.; Atta, M.; Mulliez, E. S-Adenosylmethionine: Nothing Goes to Waste. Trends Biochem. Sci. 2004, 29 (5), 243–249. [CrossRef]
- Chen, H.; Wang, Z.; Cai, H.; Zhou, C. Progress in the Microbial Production of S-Adenosyl-l-Methionine. World J. Microbiol. Biotechnol. 2016, 32 (9), 1–8. [CrossRef]
- Pan, T.; Coleman, J. E. GAL4 Transcription Factor Is Not a “Zinc Finger” but Forms a Zn(II)2Cys6 Binuclear Cluster. Proc. Natl. Acad. Sci. U. S. A. 1990, 87 (6), 2077–2081. [CrossRef]
- Lalo, D.; Steffan, J. S.; Dodd, J. A.; Nomura, M. RRN11 Encodes the Third Subunit of the Complex Containing Rrn6p and Rrn7p That Is Essential for the Initiation of RDNA Transcription by Yeast RNA Polymerase I. J. Biol. Chem. 1996, 271 (35), 21062–21067. [CrossRef]
- Xie, Y.; Varshavsky, A. RPN4 Is a Ligand , Substrate , and Transcriptional Regulator of the 26S Proteasome : A Negative Feedback Circuit. PNAS 2001, 98 (6), 3056–3061. [CrossRef]
- Mannhaupt, G.; Schnall, R.; Karpov, V.; Vetter, I.; Feldmann, H. Rpn4p Acts as a Transcription Factor by Binding to PACE, a Nonamer Box Found Upstream of 26S Proteasomal and Other Genes in Yeast. FEBS Lett. 1999, 450 (1–2), 27–34. [CrossRef]
- Böhm, S.; Frishman, D.; Werner Mewes, H. Variations of the C2H2 Zinc Finger Motif in the Yeast Genome and Classification of Yeast Zinc Finger Proteins. Nucleic Acids Res. 1997, 25 (12), 2464–2469. [CrossRef]
- Hlynialuk, C.; Schierholtz, R.; Vernooy, A.; van der Merwe, G. Nsf1/Ypl230w Participates in Transcriptional Activation during Non-Fermentative Growth and in Response to Salt Stress in Saccharomyces cerevisiae. Microbiology 2008, 154 (8), 2482–2491. [CrossRef]
- Gangwani, L.; Mikrut, M.; Galcheva-Gargova, Z.; Davis, R. J. Interaction of ZPR1 with Translation Elongation Factor-1α in Proliferating Cells. J. Cell Biol. 1998, 143 (6), 1471–1484. [CrossRef]
- Ribar, B.; Prakash, L.; Prakash, S. ELA1 and CUL3 Are Required Along with ELC1 for RNA Polymerase II Polyubiquitylation and Degradation in DNA-Damaged Yeast Cells . Mol. Cell. Biol. 2007, 27 (8), 3211–3216. [CrossRef]
- Marie, C.; Leyde, S.; White, T. C. Cytoplasmic Localization of Sterol Transcription Factors Upc2p and Ecm22p in S. cerevisiae. Fungal Genet. Biol. 2008, 45 (10), 1430–1438. [CrossRef]
- Vik, A.; Rine, J. Upc2p and Ecm22p , Dual Regulators of Sterol Biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21 (19), 6395–6405. [CrossRef]
- Truernit, E.; Schmid, J.; Epple, P.; Illig, J.; Sauer, N. The Sink-Specific and Stress-Regulated Arabidopsis SPP4 Gene : Enhanced Expression of a Gene Encoding a Monosaccharide Transporter by Wounding , Elicitors , and Pathogen Challenge. Plant Cell 1996, 8, 2169–2182. [CrossRef]
- Chen, P.; Chuang, Y.; Wu, U.; Sun, H.; Wang, J.; Sheng, W.; Chen, Y.; Chang, S. Mechanisms of Azole Resistance and Trailing in Candida tropicalis Bloodstream Isolates. Journal of Fungi 2021, 7 (8). [CrossRef]
- Maikan, H. K.; Jabbar, S.; Al-Haishawi, H. Isolation and Identification of Candida tropicalis as a Cause of Cutaneous Candidiasis in Kalar District, Iraq. Arch. Razi Inst. 2022, 77 (4), 1377–1382. [CrossRef]
- Bhattacharya, S.; Sae-Tia, S.; Fries, B. C. Candidiasis and Mechanisms of Antifungal Resistance. Antibiotics 2020, 9 (6), 1–19. [CrossRef]
- Beattie, S. R.; Krysan, D. J. Antifungal Drug Screening: Thinking Outside the Box to Identify Novel Antifungal Scaffolds. Curr. Opin. Microbiol. 2020, 57, 1–6. [CrossRef]
- Houšť, J.; Spížek, J.; Havlíček, V. Antifungal Drugs. Metabolites 2020, 10 (3). [CrossRef]
- Scorneaux, B.; Angulo, D.; Borroto-Esoda, K.; Ghannoum, M.; Peel, M.; Wring, S. SCY-078 Is Fungicidal against Candida Species in Time-Kill Studies. Antimicrob. Agents Chemother. 2017, 61 (3), 1–10. [CrossRef]
- Avato, P. Editorial to the Special Issue –“ Natural Products and Drug Discovery". Molecules 2020, 25, 1128. [CrossRef]
- Atanasov, A.; Zotchev, S.; Dirsch, V.; Taskforce, T. I. N. P. S.; Supuran, C. Natural Products in Drug Discovery: Advances and Opportunities. Nat. Rev. 2021, 20, 200–216. [CrossRef]
- Aylate, A.; Agize, M.; Ekero, D.; Kiros, A.; Ayledo, G.; Gendiche, K. In-Vitro and In-Vivo Antibacterial Activities of Croton macrostachyus Methanol Extract against E. coli and S. aureus. Adv. Anim. Vet. Sci. 2017, 5, 107–114. [CrossRef]
- Contreras Martínez, O. I.; Angulo Ortíz, A.; Santafé Patiño, G.; Peñata-Taborda, A.; Berrio Soto, R. Isoespintanol Antifungal Activity Involves Mitochondrial Dysfunction, Inhibition of Biofilm Formation, and Damage to Cell Wall Integrity in Candida tropicalis. Int. J. Mol. Sci. 2023, 24 (12), 10187. [CrossRef]
- Sun, L.; Liao, K. The Effect of Honokiol on Ergosterol Biosynthesis and Vacuole Function in Candida albicans. J. Microbiol. Biotechnol. 2020, 30 (12), 1835–1842. [CrossRef]
- Liu, J. F.; Xia, J. J.; Nie, K. L.; Wang, F.; Deng, L. Outline of the Biosynthesis and Regulation of Ergosterol in Yeast. World J. Microbiol. Biotechnol. 2019, 35 (7), 1–8. [CrossRef]
- Bhattacharya, S.; Esquivel, B. D.; White, T. C. Overexpression or Deletion of Ergosterol Biosynthesis Genes Alters Doubling Time, Response to Stress Agents, and Drug Susceptibility in Saccharomyces cerevisiae. MBio 2018, 9 (4). [CrossRef]
- Jordá, T.; Puig, S. Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae. Genes (Basel). 2020, 11 (7), 1–18. [CrossRef]
- Cheng, R.; Xu, Q.; Hu, F.; Li, H.; Yang, B.; Duan, Z.; Zhang, K.; Wu, J.; Li, W.; Luo, Z. Antifungal Activity of MAF-1A Peptide against Candida albicans. Int. Microbiol. 2021, 24, 233–242. [CrossRef]
- De Backer, M. D.; Ilyina, T.; Ma, X. J.; Vandoninck, S.; Luyten, W. H. M. L.; Bossche, H. Vanden. Genomic Profiling of the Response of Candida albicans to Itraconazole Treatment Using a DNA Microarray. Antimicrob. Agents Chemother. 2001, 45 (6), 1660–1670. [CrossRef]
- Florio, A. R.; Ferrari, S.; De Carolis, E.; Torelli, R.; Fadda, G.; Sanguinetti, M.; Sanglard, D.; Posteraro, B. Genome-Wide Expression Profiling of the Response to Short-Term Exposure to Fluconazole in Cryptococcus neoformans Serotype A. BMC Microbiol. 2011, 11, 1–13. [CrossRef]
- Kodedová, M.; Sychrová, H. Changes in the Sterol Composition of the Plasma Membrane Affect Membrane Potential, Salt Tolerance and the Activity of Multidrug Resistance Pumps in Saccharomyces cerevisiae. PLoS One 2015, 10 (9), 1–19. [CrossRef]
- Ahmadipour, S.; Field, R. A.; Miller, G. J. Prospects for Anti-Candida Therapy through Targeting the Cell Wall: A Mini-Review. Cell Surf. 2021, 7, 100063. [CrossRef]
- Lenardon, M. D.; Sood, P.; Dorfmueller, H. C.; Brown, A. J. P.; Gow, N. A. R. Scalar Nanostructure of the Candida albicans Cell Wall; a Molecular, Cellular and Ultrastructural Analysis and Interpretation. Cell Surf. 2020, 6, 100047. [CrossRef]
- Ibe, C.; Munro, C. A. Fungal Cell Wall Proteins and Signaling Pathways Form a Cytoprotective Network to Combat Stresses. J. Fungi 2021, 7 (9). [CrossRef]
- Popolo, L.; Gualtieri, T.; Ragni, E. The Yeast Cell-Wall Salvage Pathway. Med. Mycol. Suppl. 2001, 39 (1), 111–121. [CrossRef]
- Boone, C.; Sommer, S. S.; Hensel, A.; Bussey, H. Yeast KRE Genes Provide Evidence for a Pathway of Cell Wall β-Glucan Assembly. J. Cell Biol. 1990, 110 (5), 1833–1843. [CrossRef]
- Nagahash, S.; Lussier, M.; Bussey, H. Isolation of Candida glabrata Homologs of the Saccharomyces cerevisiae KRE9 and KNH1 Genes and Their Involvement in Cell Wall β-1,6-Glucan Synthesis. J. Bacteriol. 1998, 180 (19), 5020–5029. [CrossRef]
- García, R.; Botet, J.; Rodríguez-Peña, J. M.; Bermejo, C.; Ribas, J. C.; Revuelta, J. L.; Nombela, C.; Arroyo, J. Genomic Profiling of Fungal Cell Wall-Interfering Compounds: Identification of a Common Gene Signature. BMC Genomics 2015, 16 (1), 1–20. [CrossRef]
- Xiong, H.; Zhou, X.; Xiang, W.; Huang, M.; Lin, Z.; Tang, J.; Cai, T.; Zhang, Q. Integrated Transcriptome Reveals That D -Limonene Inhibits Candida tropicalis by Disrupting Metabolism. LWT- Food Sci. Technol. 2023, 176, 114535. [CrossRef]
- un, F.-J.; Li, M.; Gu, L.; Wang, M.; Yang, M. Recent Progress on Anti-Candida Natural Products. Chin. J. Nat. Med. 2021, 19 (8), 561–579. [CrossRef]
- Gentsch, M.; Kuschel, M.; Schlegel, S.; Barth, G. Mutations at Different Sites in Members of the Gpr1/Fun34/YaaH Protein Family Cause Hypersensitivity to Acetic Acid in Saccharomyces cerevisiae as Well as in Yarrowia lipolytica. FEMS Yeast Res. 2007, 7 (3), 380–390. [CrossRef]
- Hymbaugh Bergman, S. J.; Comstock, L. R. N-Mustard Analogs of S-Adenosyl-l-Methionine as Biochemical Probes of Protein Arginine Methylation. Bioorganic Med. Chem. 2015, 23 (15), 5050–5055. [CrossRef]
Figure 1.
Key 1H–1H COSY and HMBC correlations of ISO.
Figure 1.
Key 1H–1H COSY and HMBC correlations of ISO.
Figure 2.
Differential gene expression analysis. (a) volcano plot of DEG, red (up) and blue (down) colored dots indicate the DEGs, while the grey dots represent genes without expression changes (NC) among Candida tropicalis and control samples. (b) Pie chart distribution of red (up) and blue (down) DEGs among treatments.
Figure 2.
Differential gene expression analysis. (a) volcano plot of DEG, red (up) and blue (down) colored dots indicate the DEGs, while the grey dots represent genes without expression changes (NC) among Candida tropicalis and control samples. (b) Pie chart distribution of red (up) and blue (down) DEGs among treatments.
Figure 3.
Top upregulated and downregulated DEGs with known function in Candida tropicalis dataset. Bars show the top upregulated (red) and downregulated (blue) genes, according to their Log2FC values.
Figure 3.
Top upregulated and downregulated DEGs with known function in Candida tropicalis dataset. Bars show the top upregulated (red) and downregulated (blue) genes, according to their Log2FC values.
Figure 4.
Functional enrichment of DEG in Candida tropicalis treatment. FDR is calculated based on a nominal p-value from the hypergeometric test. Fold Enrichment is defined as the percentage of differentially expressed genes belonging to a pathway, divided by the corresponding percentage in the background. FDR reports how likely the enrichment is by chance, higher values are colored on a scale of red to blue. In the x-axis, Fold Enrichment indicates how drastically genes of a certain pathway are overrepresented. .
Figure 4.
Functional enrichment of DEG in Candida tropicalis treatment. FDR is calculated based on a nominal p-value from the hypergeometric test. Fold Enrichment is defined as the percentage of differentially expressed genes belonging to a pathway, divided by the corresponding percentage in the background. FDR reports how likely the enrichment is by chance, higher values are colored on a scale of red to blue. In the x-axis, Fold Enrichment indicates how drastically genes of a certain pathway are overrepresented. .
Figure 5.
Functional annotated Up-DEGs in Candida tropicalis treatment enriched in steroid and cell metabolism functions. Steroid biosynthesis: ERG6, ERG26, ERG2, ERG1. Pink bars depict the upregulated genes according to their Log2FC values. Glycoside hydrolase superfamily, and starch and sucrose metabolism. Purple Bars show the upregulated enriched genes according to their corresponding Log2FC values. .
Figure 5.
Functional annotated Up-DEGs in Candida tropicalis treatment enriched in steroid and cell metabolism functions. Steroid biosynthesis: ERG6, ERG26, ERG2, ERG1. Pink bars depict the upregulated genes according to their Log2FC values. Glycoside hydrolase superfamily, and starch and sucrose metabolism. Purple Bars show the upregulated enriched genes according to their corresponding Log2FC values. .
Figure 6.
Deregulated genes in Candida tropicalis treatment enriched in mitochondria and cell wall processes. Green bars depict the deregulated transcripts involved in mitochondrial processes according to their Log2FC values. While cell- wall-related transcripts are depicted by yellow bars according to their corresponding Log2FC values. .
Figure 6.
Deregulated genes in Candida tropicalis treatment enriched in mitochondria and cell wall processes. Green bars depict the deregulated transcripts involved in mitochondrial processes according to their Log2FC values. While cell- wall-related transcripts are depicted by yellow bars according to their corresponding Log2FC values. .
Figure 7.
Validation of gene overexpression by qPCR using the 2(-ΔΔCT) method. The dark bars represent the genetic expression of C. tropicalis without treatment (control group) and the light bars represent C. tropicalis treated with ISO. The data are presented by calculating the logarithm to base 2 (Log2FC).
Figure 7.
Validation of gene overexpression by qPCR using the 2(-ΔΔCT) method. The dark bars represent the genetic expression of C. tropicalis without treatment (control group) and the light bars represent C. tropicalis treated with ISO. The data are presented by calculating the logarithm to base 2 (Log2FC).
Figure 8.
Functional Down-DEG in Candida tropicalis treatment enriched in methylation. methylation-related TRM7: Methylates the 2'-O-ribose of nucleotides at positions 32 and 34 of the tRNA anticodon loop of substrate tRNAs. TRM2; Belongs to the class I-like SAM-binding methyltransferase superfamily. RNA M5U methyltransferase family. DOT1: Histone methyltransferase that specifically methylates histone H3 to form H3K79me. mug84GPR1/FUN34/yaaH family; mug86 GPR1/FUN34/yaaH family. Bars show the top upregulated (red) genes according to Log2FC values. .
Figure 8.
Functional Down-DEG in Candida tropicalis treatment enriched in methylation. methylation-related TRM7: Methylates the 2'-O-ribose of nucleotides at positions 32 and 34 of the tRNA anticodon loop of substrate tRNAs. TRM2; Belongs to the class I-like SAM-binding methyltransferase superfamily. RNA M5U methyltransferase family. DOT1: Histone methyltransferase that specifically methylates histone H3 to form H3K79me. mug84GPR1/FUN34/yaaH family; mug86 GPR1/FUN34/yaaH family. Bars show the top upregulated (red) genes according to Log2FC values. .
Figure 9.
Functional enrichment of down regulated genes in Candida tropicalis treated with ISO. FDR is calculated based on a nominal p-value from the hypergeometric test. Fold Enrichment is defined as the percentage of differentially expressed genes belonging to a pathway divided by the corresponding percentage in the background. FDR reports how likely the enrichment is by chance. In the x-axis, Fold Enrichment indicates how drastically genes of a certain pathway are overrepresented. .
Figure 9.
Functional enrichment of down regulated genes in Candida tropicalis treated with ISO. FDR is calculated based on a nominal p-value from the hypergeometric test. Fold Enrichment is defined as the percentage of differentially expressed genes belonging to a pathway divided by the corresponding percentage in the background. FDR reports how likely the enrichment is by chance. In the x-axis, Fold Enrichment indicates how drastically genes of a certain pathway are overrepresented. .
Figure 10.
Deregulated transcription factors in Candida tropicalis treatment. Brown bars depict the deregulated transcripts according to their Log2FC values.
Figure 10.
Deregulated transcription factors in Candida tropicalis treatment. Brown bars depict the deregulated transcripts according to their Log2FC values.
Figure 11.
HPLC Chromatograms of Ergosterol in Candida tropicalis with and without ISO Treatment Ergosterol extractions from the negative control and the ISO-treated group were dissolved in methanol for analysis. Each graph represents the results of three repeated experiments.
Figure 11.
HPLC Chromatograms of Ergosterol in Candida tropicalis with and without ISO Treatment Ergosterol extractions from the negative control and the ISO-treated group were dissolved in methanol for analysis. Each graph represents the results of three repeated experiments.
Table 1.
Ergosterol content of Candida tropicalis treated with or without ISO. Data are presented as the mean ± SD of three independent experiments. p < 0.05 for control and ISO.
Table 1.
Ergosterol content of Candida tropicalis treated with or without ISO. Data are presented as the mean ± SD of three independent experiments. p < 0.05 for control and ISO.
| Experimental groups |
Ergosterol Content (mg/L) |
| Control |
28,40 ± 1.48 |
| ISO |
15,58 ± 0.37 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).