Submitted:
09 November 2023
Posted:
10 November 2023
You are already at the latest version
Abstract
Keywords:
Introduction
1. Genomic selection
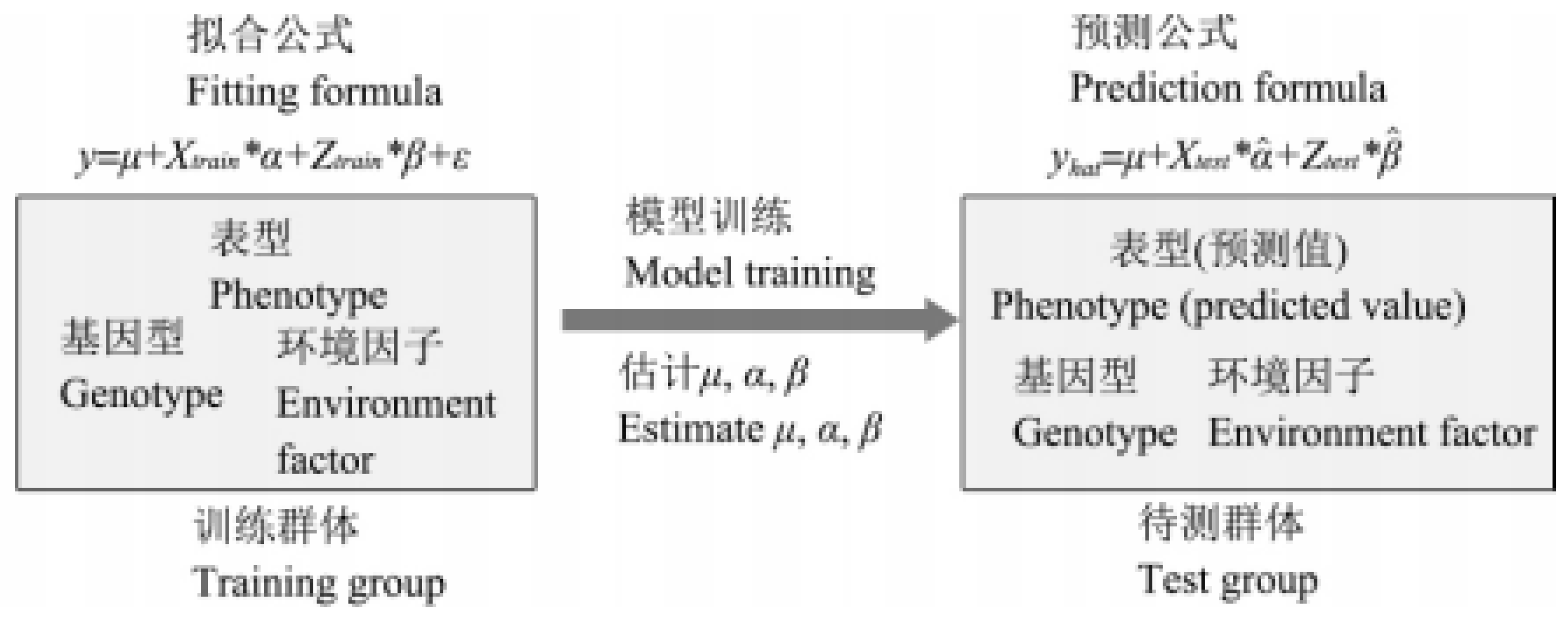
1.1. The principle of GS
1.1.1. Overview of GS
1.1.2. Reference group and candidate group
1.1.3. Breeding principle of GS
1.2. Research methods of GS
1.2.1. Bayesian method
1.2.2. Best linear unbiased prediction method
1.2.3. LASSO method
1.2.4. PLS method
1.2.5. SVM method
1.2.6. RKHS method
1.3. Application of GS in Breeding
2. Materials and methods
2.1. Data source
2.2. Research method
2.2.1. Research on different GS models
2.2.2. Study on the density of failing marks
3. Result
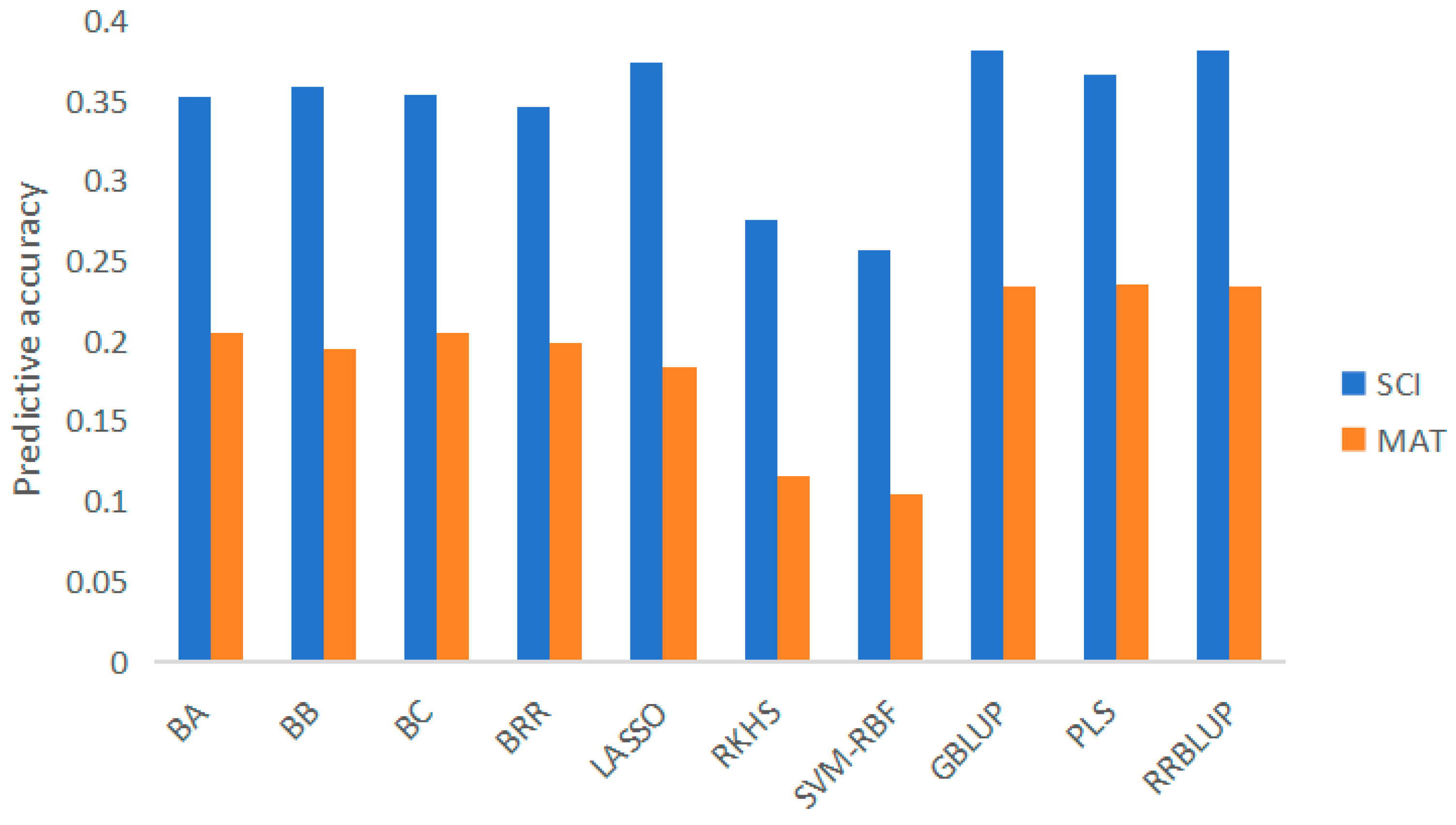
3.1. Prediction accuracy of different models
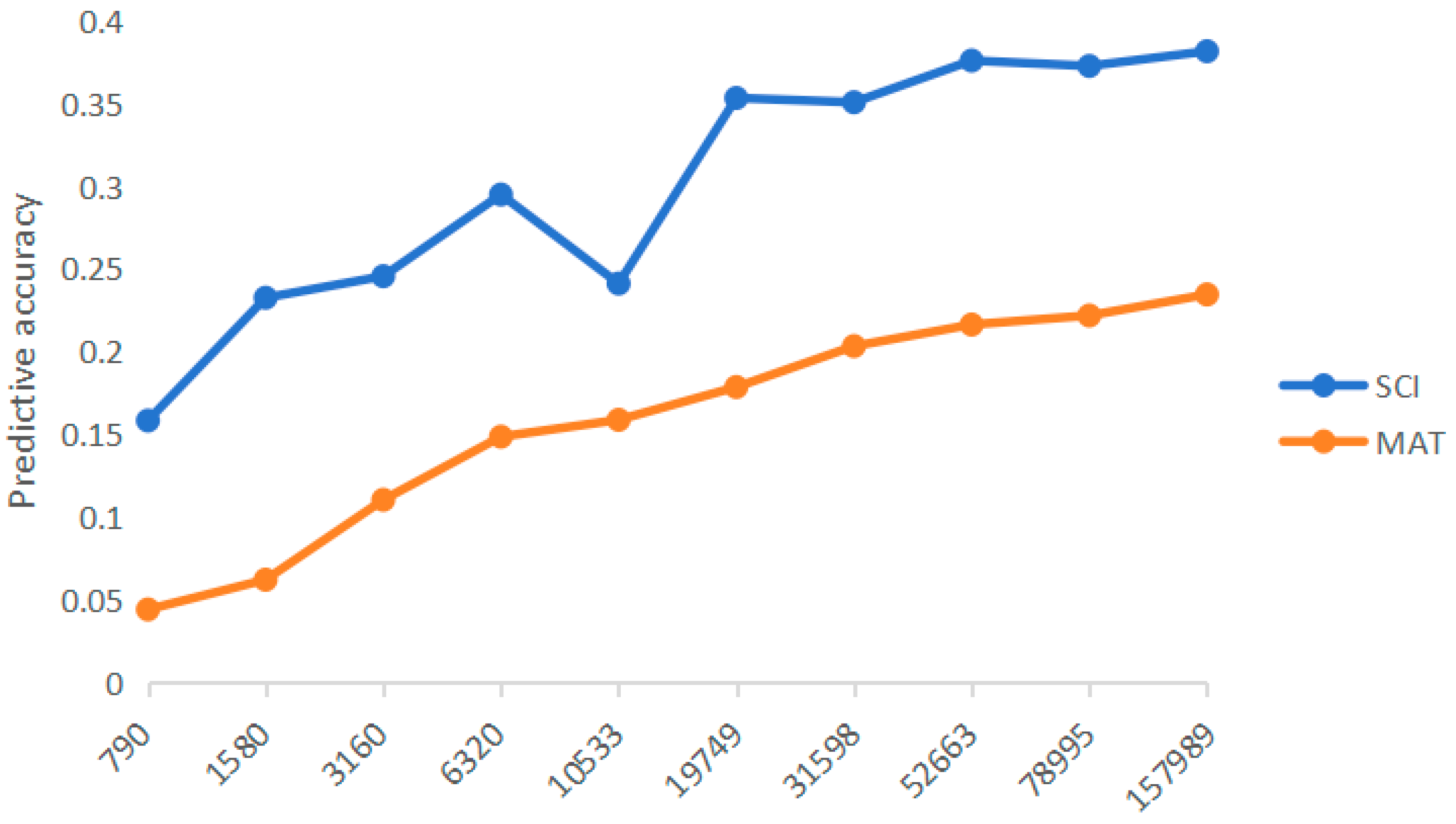
3.2. Comparison of prediction accuracy based on different marker densities
4. Discussion
4.1. Influence of different forecasting methods on forecasting accuracy
4.2. Influence of different marker densities on prediction accuracy
5. Conclusion
References
- Huang, G.; Zhu, Y.X. Breeding cotton with superior fiber quality: identification and utilization of multiple elite loci and exotic genetic resources. Science China Life sciences 2021, 64, 1197–1198. [Google Scholar] [CrossRef]
- Collard, B.C.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and markerassisted selection for crop improvement: the basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Xu, Y.; Crouch, J.H. Marker-Assisted Selection in Plant Breeding: From Publications to Practice. Crop. Sci. 2008, 48, 391–407. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, Y.; Xie, C.; Gao, S.; Wan, J.; Prasanna, B.M. Whole-genome strategies for marker-assisted plant breeding. Mol. Breed. 2012, 29, 833–854. [Google Scholar] [CrossRef]
- Patil, G.; Mian, R.; Vuong, T.; Pantalone, V.; Song, Q.; Chen, P.; Shannon, G.J.; Carter, T.C.; Nguyen, H.T. Molecular mapping and genomics of soybean seed protein: a review and perspective for the future. Theor. Appl. Genet. 2017, 130, 1975–1991. [Google Scholar] [CrossRef]
- Grevenhof, I.E.V.; Werf, J.H.V.D. Design of reference populations for genomic selection in crossbreeding programs. Genetics Selection Evolution 2015, 47, 1–9. [Google Scholar] [CrossRef]
- Goddard, M.E.; Hayes, B.J. Genomic selection. Journal of Animal Breeding & Genetics 2015, 124, 323–330. [Google Scholar]
- Heffner, E.L.; Sorrells, M.E.; Jannink, J. Genomic Selection for Crop Improvement. Crop. Sci. 2009, 49, 1–12. [Google Scholar] [CrossRef]
- Luan, T.; Woolliams, J.A.; Lien, S.; Kent, M.; Svendsen, M.; Meuwissen, T.H. . The accuracy of genomic selection in Norwegian red cattle assessed by cross-validation. Genetics 2009, 183, 1119–1126. [Google Scholar] [CrossRef]
- Jia, Z. Controlling the Overfitting of Heritability in Genomic Selection through Cross Validation. Sci. Rep. 2017, 7, 13678–13678. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient methods to compute genomic perdictions. J Dairy Sci 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Ricard, A.; Danvy, S.; Legarra, A. Computation of deregressed proofs for genomic selection when own phenotypes exist with an application in French show-jumping horses1. J. Anim. Sci. 2013, 91, 1076–1085. [Google Scholar] [CrossRef]
- Goddard, M. Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 2009, 136, 245–257. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression Shrinkage and Selection via the Lasso. Journal of the royal statistical society series b-methodological 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V.N. Support-Vector Networks. Machine Learning 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Cherkassky, V.; Ma, Y. Practical selection of SVM parameters and noise estimation for SVM regression. Neural networks : the official journal of the International Neural Network Society 2004, 17, 113–126. [Google Scholar] [CrossRef]
- Gianola, D.; Fernando, R.L.; Stella, A. Genomic-Assisted Prediction of Genetic Value With Semiparametric Procedures. Genetics 2006, 173, 1761–1776. [Google Scholar] [CrossRef]
- Beyene, Y.; Semagn, K.; Mugo, S.; Tarekegne, A.; Babu, R.; Meisel, B.; Sehabiague, P.; Makumbi, D.; Magorokosho, C.; Oikeh, S.; et al. Genetic Gains in Grain Yield Through Genomic Selection in Eight Bi-parental Maize Populations under Drought Stress. Crop. Sci. 2015, 55, 154–163. [Google Scholar] [CrossRef]
- Zhao, Y.; Gowda, M.; Liu, W.; Würschum, T.; Maurer, H.P.; Longin, F.H.; Ranc, N.; Reif, J.C. Accuracy of genomic selection in European maize elite breeding populations. Theor. Appl. Genet. 2011, 124, 769–776. [Google Scholar] [CrossRef]
- Gapare, W.; Liu, S.; Conaty, W.; Zhu, Q.H.; Gillespie, V.; Llewellyn, D.; Stiller, W.; Wilson, I. Historical Datasets Support Genomic Selection Models for the Prediction of Cotton Fiber Quality Phenotypes Across Multiple Environments. G3 (Bethesda, Md) 2018, 8, 1721–1732. [Google Scholar] [CrossRef]
- Islam, S.; Fang, D.D.; Jenkins, J.N.; Guo, J.; McCarty, J.C.; Jones, D.C. Evaluation of genomic selection methods for predicting fiber quality traits in Upland cotton. Mol. Genet. Genom. 2019, 295, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, S.; Conaty, W.; Zhu, Q.H.; Moncuquet, P.; Stiller, W.; Wilson, I. Genomic prediction of cotton fibre quality and yield traits using Bayesian regression methods. Heredity 2022, 129, 103–112. [Google Scholar] [CrossRef]
- Guo, T.; Li, H.; Yan, J.; Tang, J.; Li, J.; Zhang, Z.; Zhang, L.; Wang, J. Performance prediction of F1 hybrids between recombinant inbred lines derived from two elite maize inbred lines. Theoretical and Applied Genetics 2013, 126, 189–201. [Google Scholar] [CrossRef]
- Li, G.; Dong, Y.; Zhao, Y.; Tian, X.; Würschum, T.; Xue, J.; Chen, S.; Reif, J.C.; Xu, S.; Liu, W. Genome-wide prediction in a hybrid maize population adapted to Northwest China. The Crop Journal 2020, 8, 830–842. [Google Scholar] [CrossRef]
- Xiao, N.; Pan, C.; Li, Y.; Wu, Y.; Cai, Y.; Lu, Y.; Wang, R.; Yu, L.; Shi, W.; Kang, H.; et al. Genomic insight into balancing high yield, good quality, and blast resistance of japonica rice. Genome Biology 2021, 22, 283. [Google Scholar] [CrossRef]
- Qin, J.; Wang, F.; Zhao, Q.; Shi, A.; Zhao, T.; Song, Q.; Ravelombola, W.; An, H.; Yan, L.; Yang, C.; et al. Identification of Candidate Genes and Genomic Selection for Seed Protein in Soybean Breeding Pipeline. Front. Plant Sci. 2022, 13, 882732. [Google Scholar] [CrossRef]
- Liu, Y.-H.; Xu, Y.; Zhang, M.; Cui, Y.; Sze, S.-H.; Smith, C.W.; Xu, S.; Zhang, H.-B. Accurate Prediction of a Quantitative Trait Using the Genes Controlling the Trait for Gene-Based Breeding in Cotton. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Ma, Z.; He, S.; Wang, X.; Sun, J.; Zhang, Y.; Zhang, G.; Wu, L.; Li, Z.; Liu, Z.; Sun, G.; et al. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 2018, 50, 803–813. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genomics, Proteomics&Bioinformatics.
- Howard, R.; Carriquiry, A.L.; Beavis, W.D. Parametric and nonparametric statistical methods for genomic selection of traits with additive and epistatic genetic architectures. G3 (Bethesda, Md) 2014, 4, 1027–1046. [Google Scholar] [CrossRef]
- Budhlakoti, N.; Rai, A.; Mishra, D.C.; Jaggi, S.; Kumar, M.; Rao, A.R. Comparative study of different non-parametric genomic selection methods under diverse genetic architecture. Indian J. Genet. Plant Breed. (The) 2020, 80, 395–401. [Google Scholar] [CrossRef]
- Villumsen, T.; Janss, L.; Lund, M. The importance of haplotype length and heritability using genomic selection in dairy cattle. J. Anim. Breed. Genet. 2009, 126, 3–13. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



