Submitted:
04 November 2023
Posted:
06 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
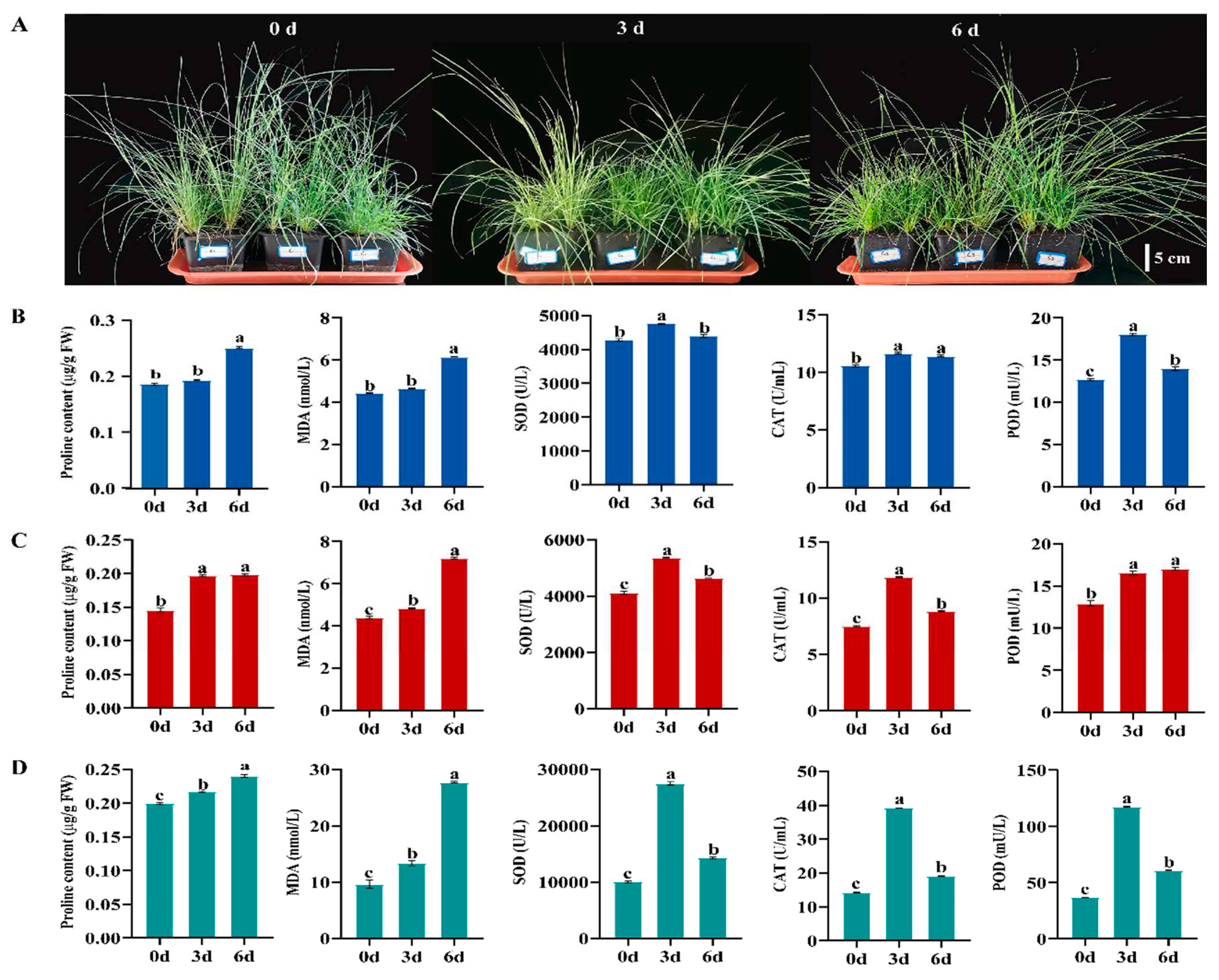
2.1. Physiological Responses of P. crymophila under Cold Stress
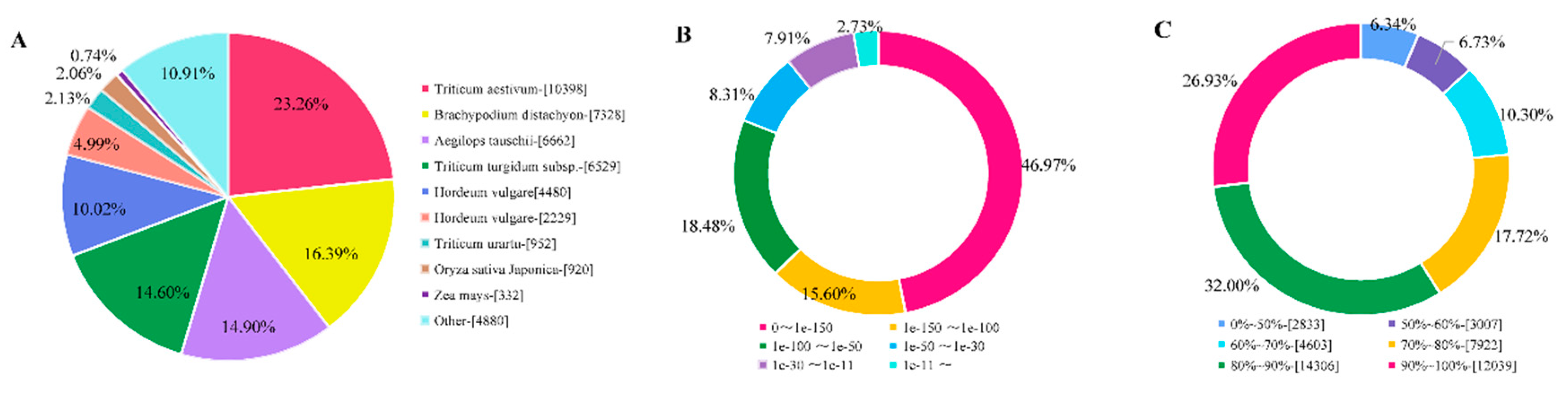
2.2. Transcriptome Assembly and Annotation
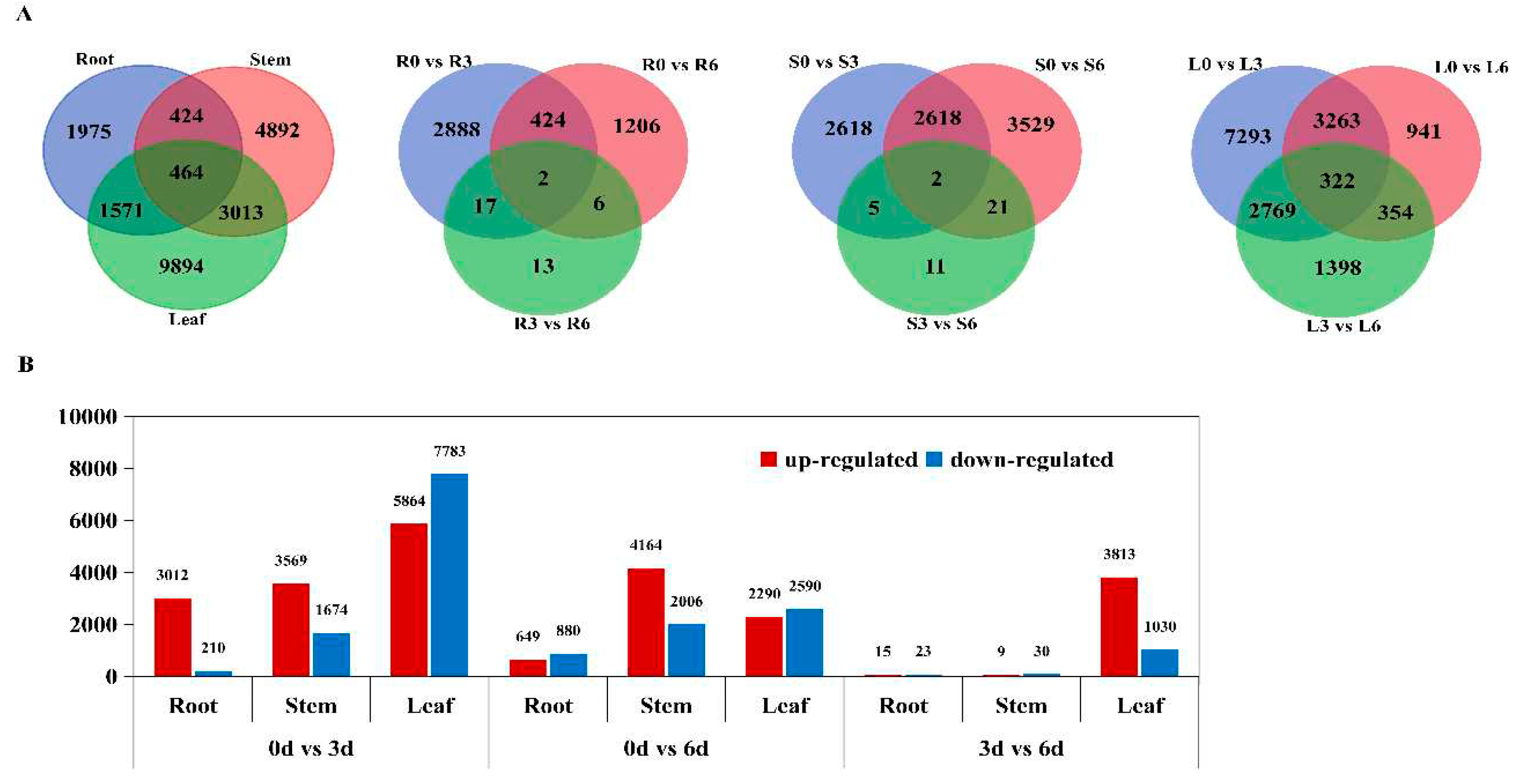
2.3. Identification and Analysis of DEGs
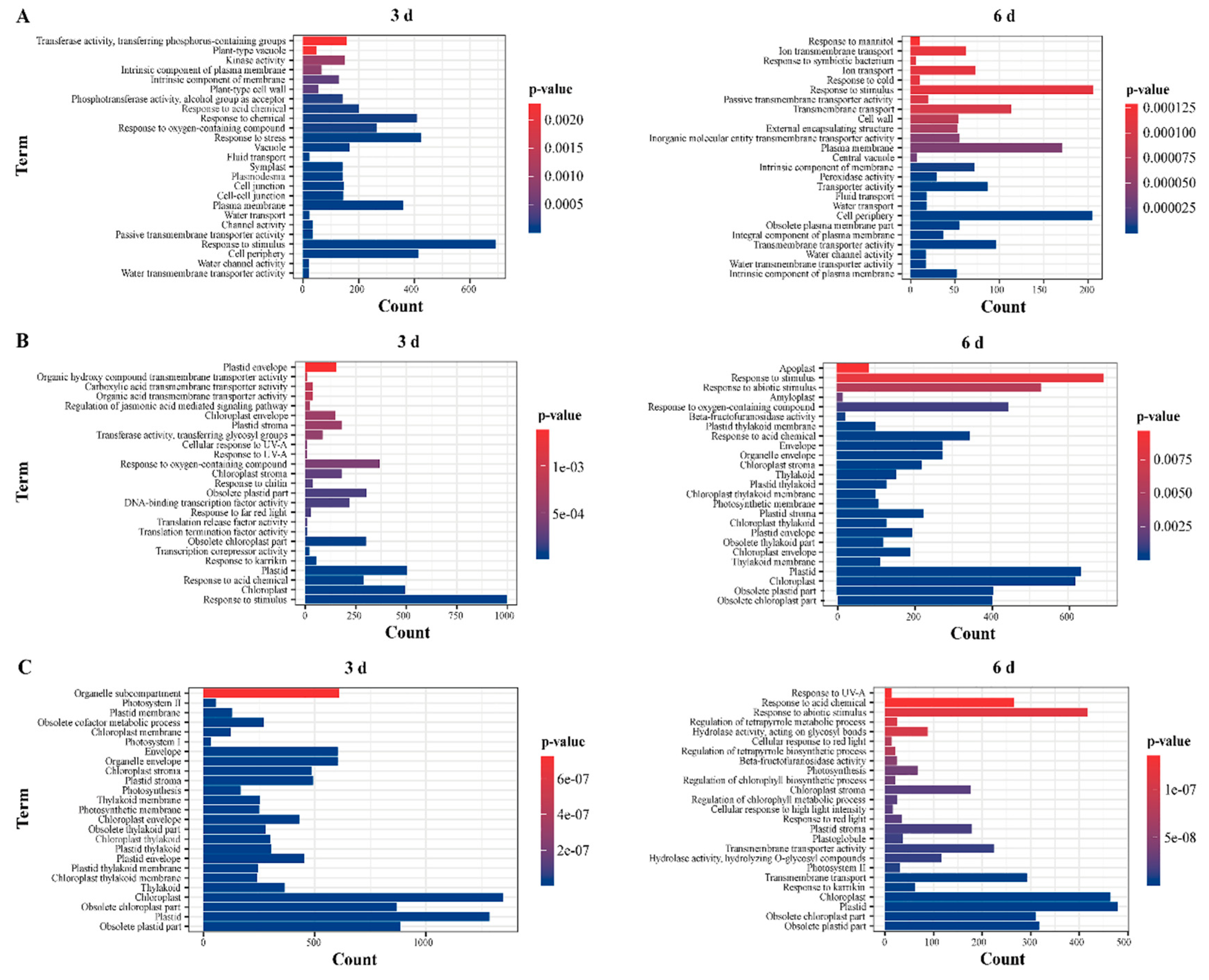
2.4. GO Enrichment Analysis of DEGs
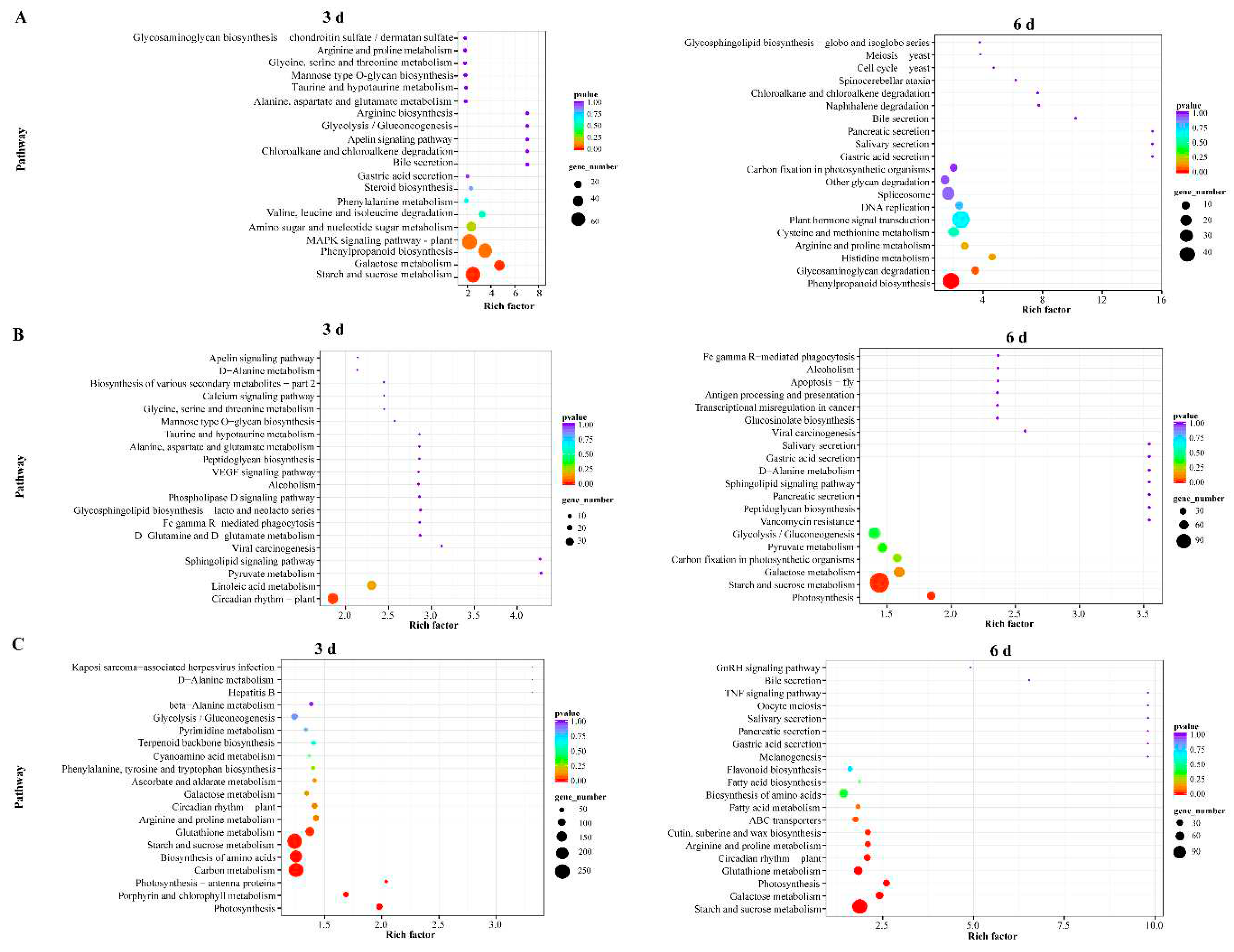
2.5. KEGG Enrichment Analysis of DEGs
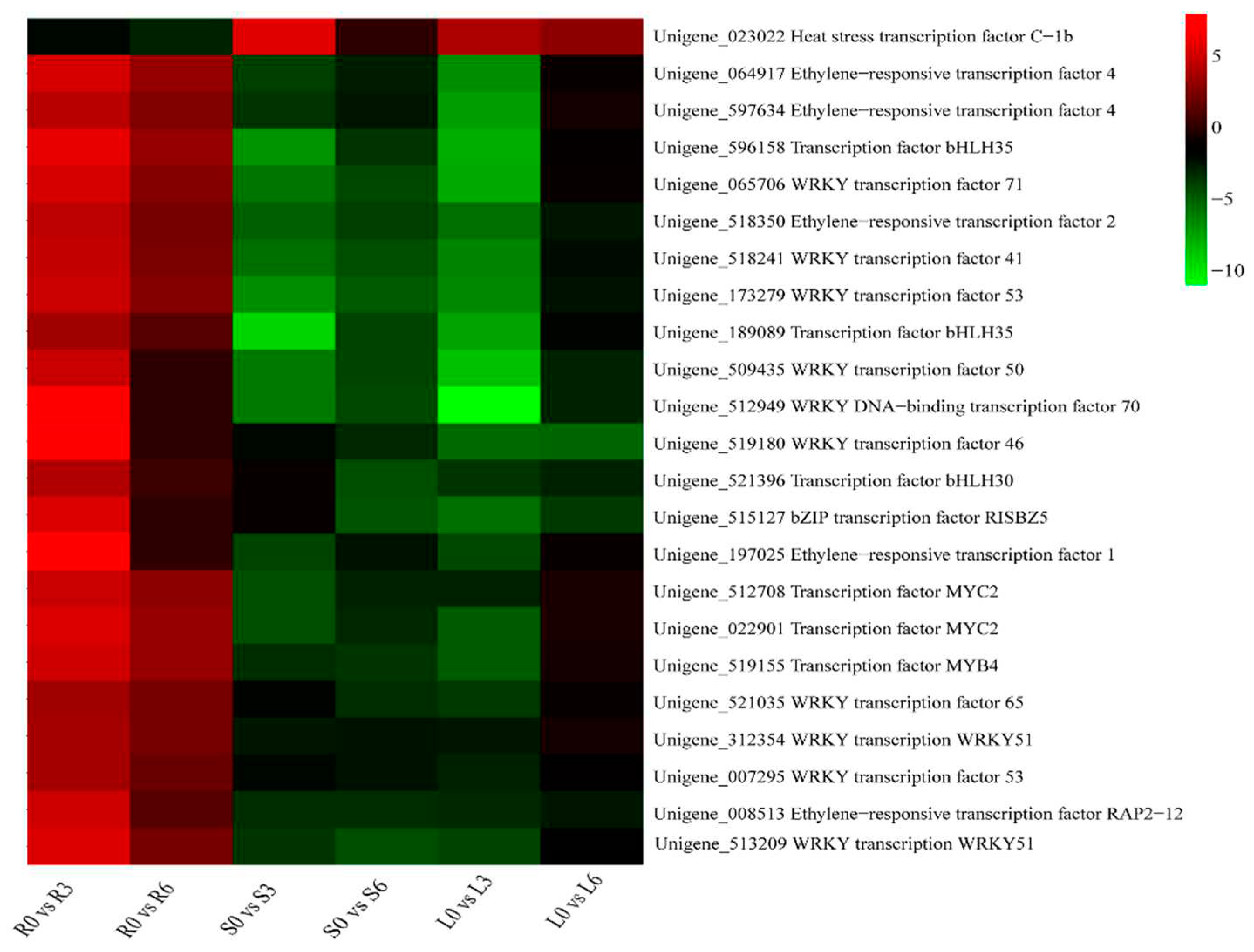
2.6. Identification of DEGs Related to Transcription Factors (TFs)
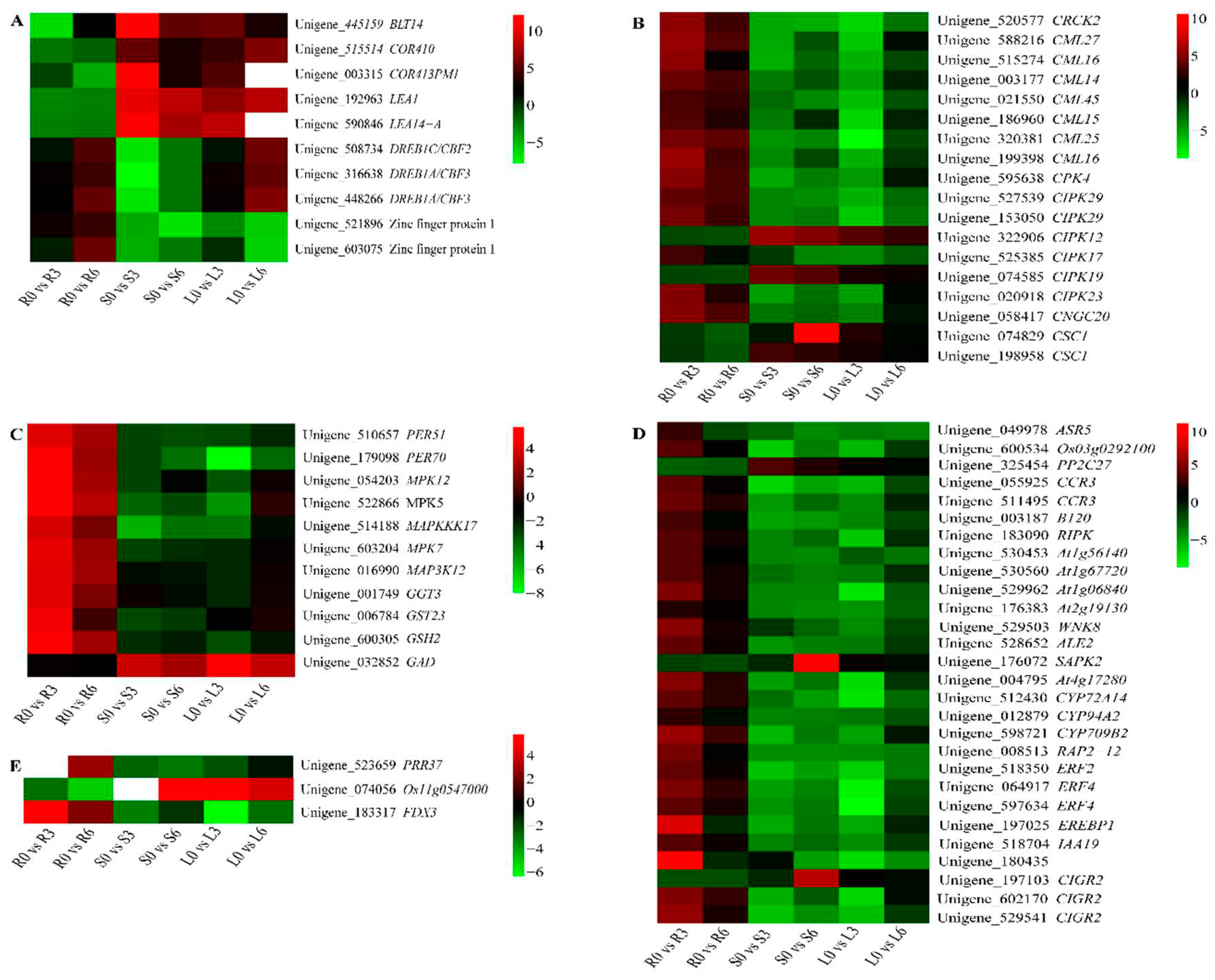
2.7. Identification of Key DEGs Involved in Cold Responses
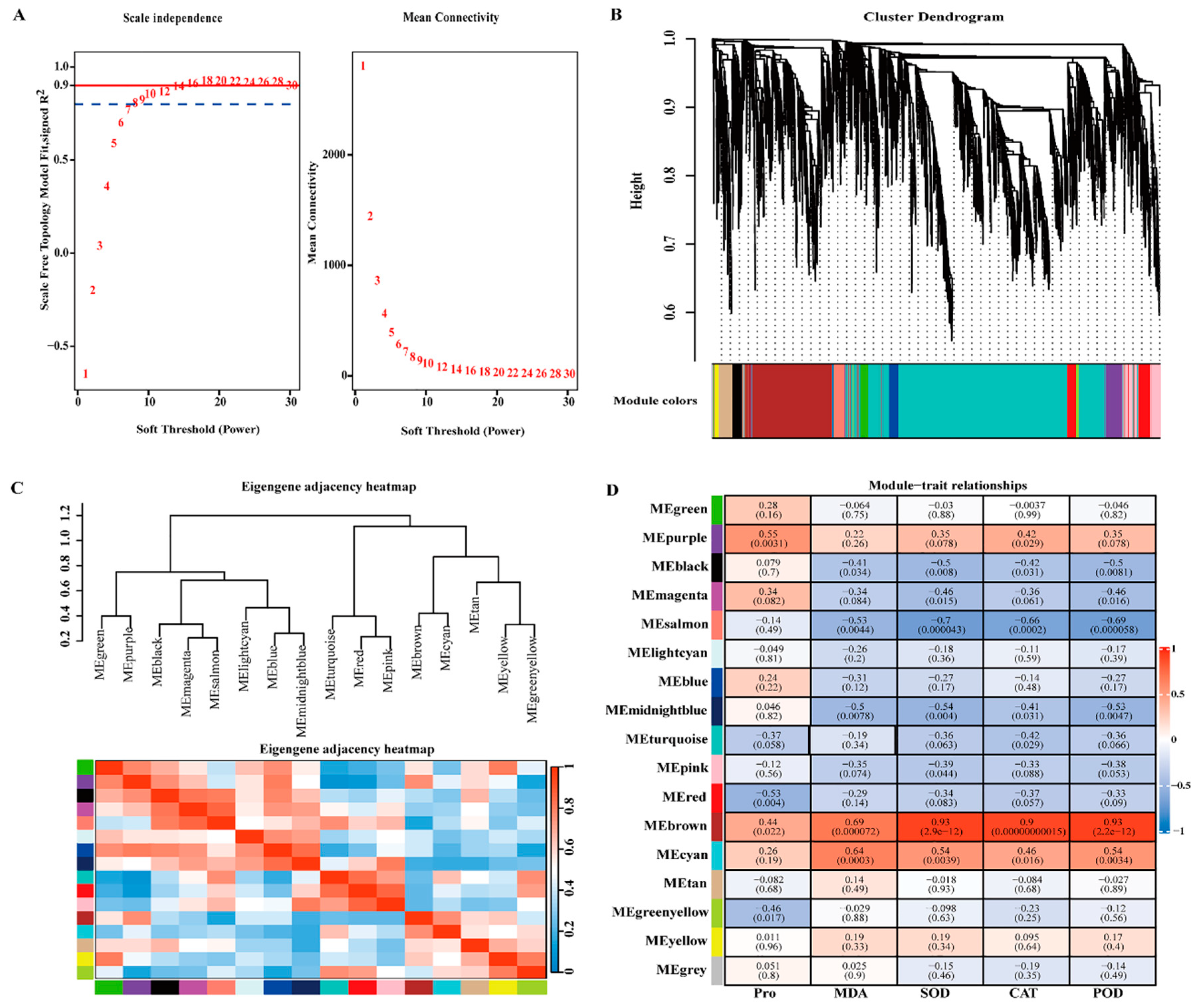
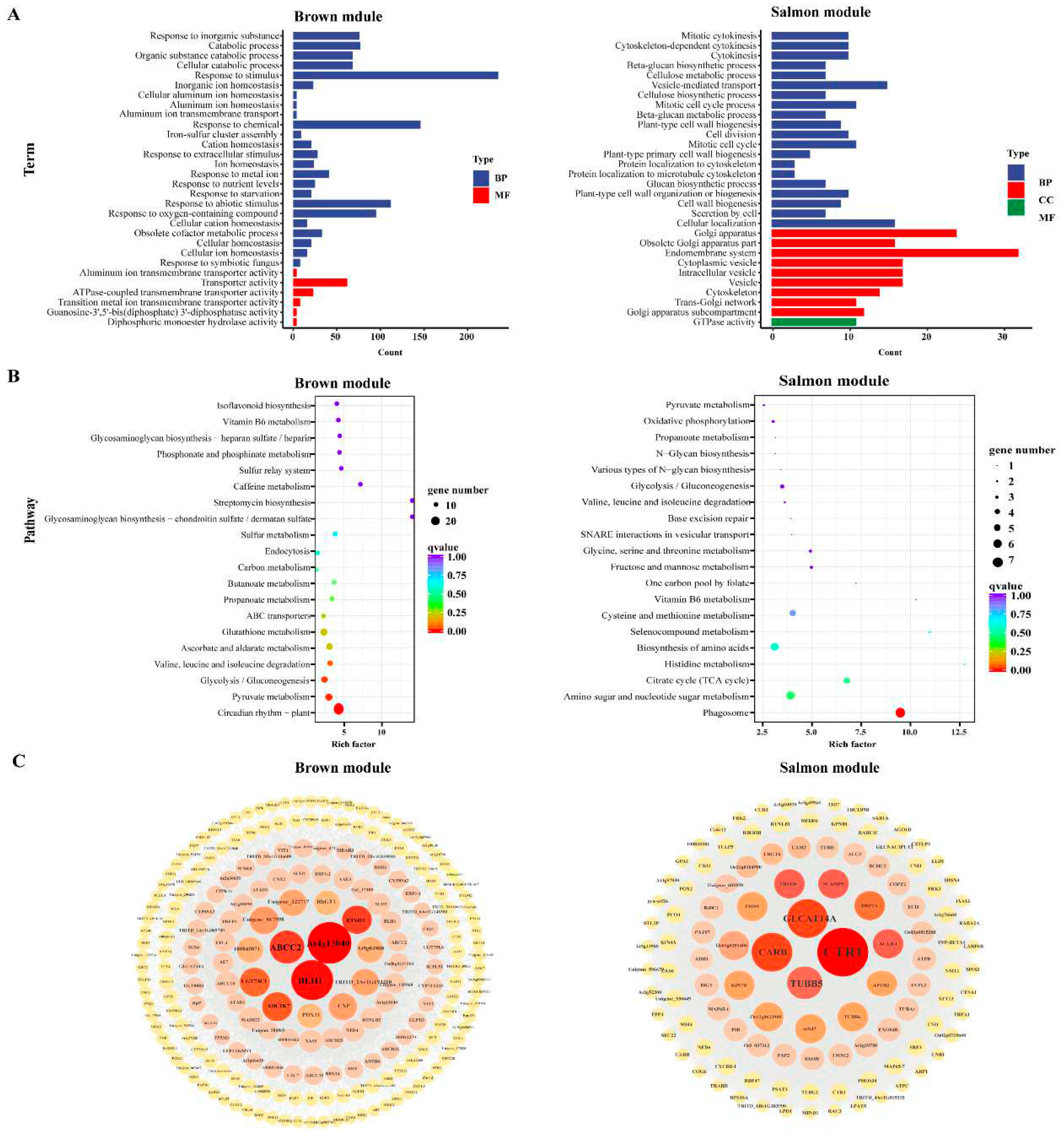
2.8. WGCNA and Identification of Key Genes
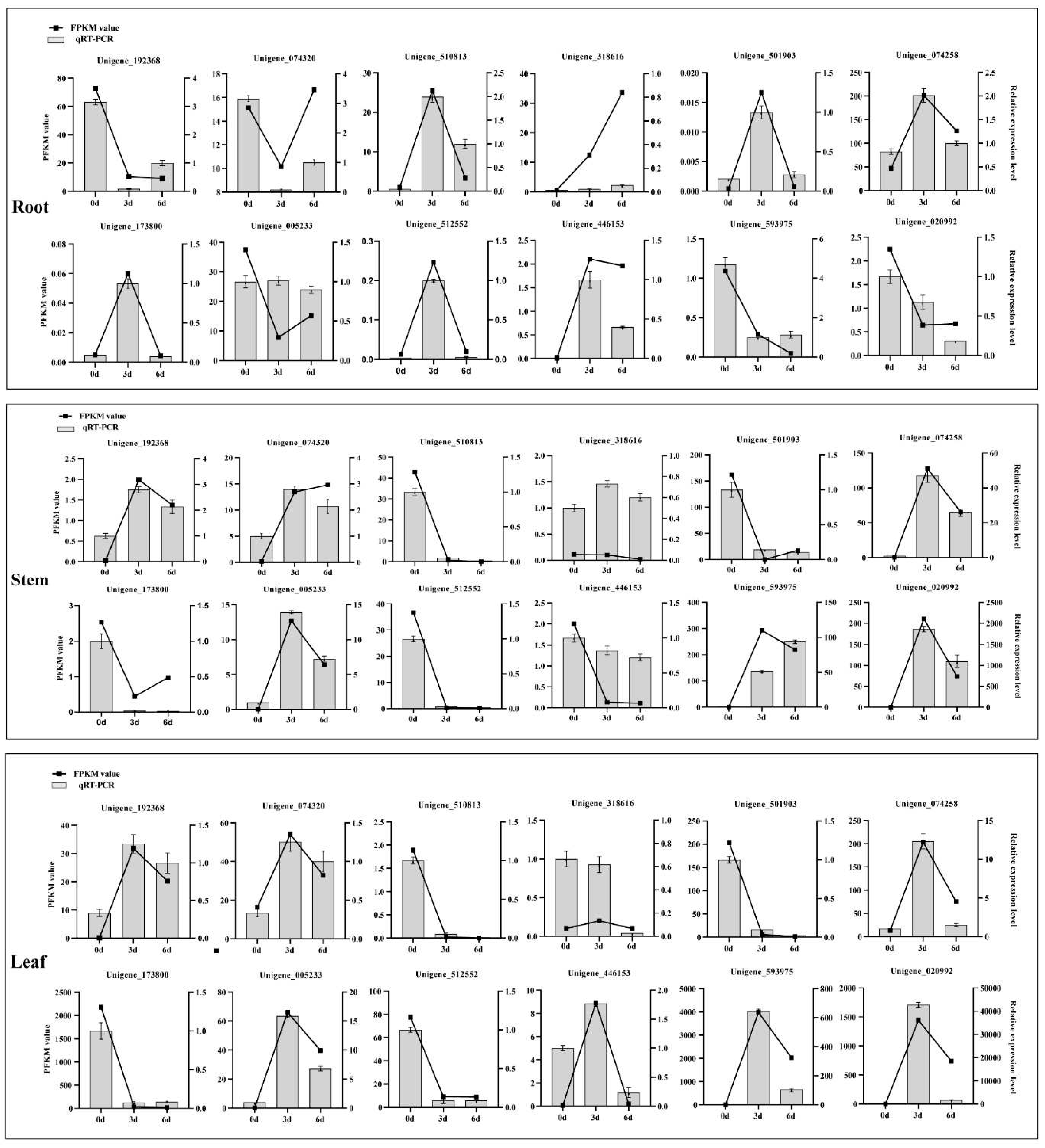
2.9. Experimental Validation
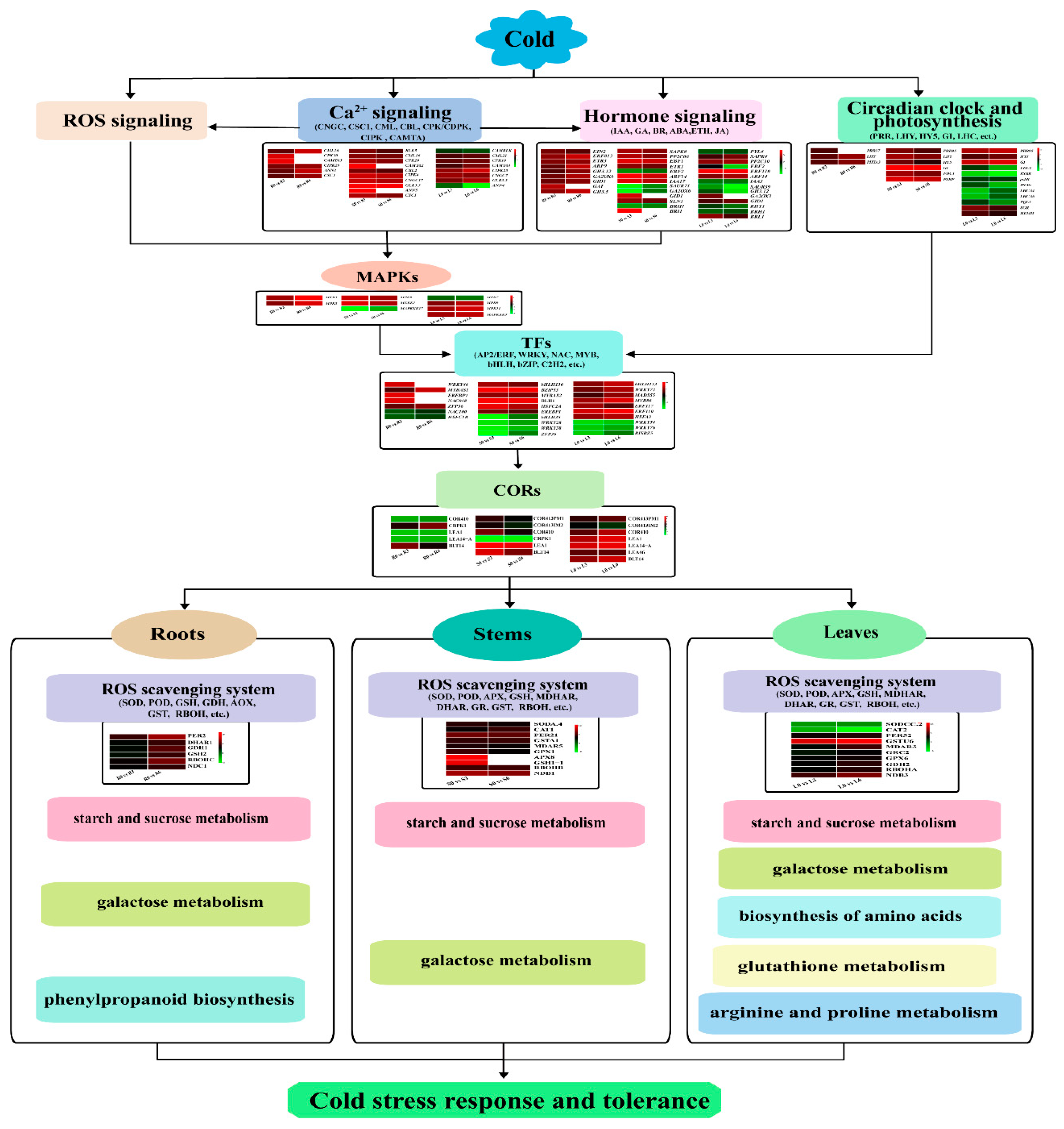
3. Discussion
3.1. Physiological Changes in P. crymophila Responding to Cold Stress
3.2. Tissue-specific RNA-seq Strategy Provides More Comprehensive Information
3.3. TFs involved in the cold-stress response
3.4. Role of ICE-CBF-COR in the Cold Response
3.5. Role of Ca2+ Signaling, Hormone Signaling, and ROS Scavenging System in the Cold Response
3.6. Role of the Circadian Clock and Photosynthesis in the Cold Response
3.7. Cold Tolerance Hub genes Selected by WGCNA
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Determination of MDA, Proline Contents, and Antioxidant Enzymatic Activities
4.3. Total RNA Extraction and Sequencing
4.4. Sequence Data Processing, De novo Assembly, and Annotation
4.5. Analysis of Differentially Expressed Genes
4.6. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.7. Validation Analysis of Transcriptome Data by qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, L.; Jian, Z.; Wang, P.; Zhao, L.; Chen, K.; Chaves, M. Combined physiological responses and differential expression of drought-responsive genes preliminarily explain the drought resistance mechanism of Lotus corniculatus. Functional Plant Biology 2022, 50, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Yang, S. Surviving and thriving: How plants perceive and respond to temperature stress. Developmental Cell 2022, 57, 947–958. [Google Scholar] [CrossRef]
- Dong, R.; Luo, B.; Tang, L.; Wang, Q.-x.; Lu, Z.-J.; Chen, C.; Yang, F.; Wang, S.; He, J. A comparative transcriptomic analysis reveals a coordinated mechanism activated in response to cold acclimation in common vetch (Vicia sativa L.). BMC Genomics 2022, 23, 814. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Sun, M.; Su, R.; Wang, X.; Lu, X.; Xiao, Y.; Deng, H.; Liu, X.; Tang, W.; Zhang, G. Transcriptomic profiling of cold stress-induced differentially expressed genes in seedling stage of Indica rice. Plants 2023, 12, 2675. [Google Scholar] [CrossRef]
- Gusain, S.; Joshi, S.; Joshi, R. Sensing, signalling, and regulatory mechanism of cold-stress tolerance in plants. Plant Physiology and Biochemistry 2023, 197, 107646. [Google Scholar] [CrossRef] [PubMed]
- Nouri, M.-Z.; Komatsu, S. Subcellular protein overexpression to develop abiotic stress tolerant plants. Frontiers in Plant Science 2013, 4, 2. [Google Scholar] [CrossRef]
- Cheng, M.; Pan, Z.; Cui, K.; Zheng, J.; Luo, X.; Chen, Y.; Yang, T.; Wang, H.; Li, X.; Zhou, Y.; Lei, X.; Li, Y.; Zhang, R.; Iqbal, M.Z.; He, R. RNA sequencing and weighted gene co-expression network analysis uncover the hub genes controlling cold tolerance in Helictotrichon virescens seedlings. Frontiers in Plant Science 2022, 13, 938859. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nature Reviews Genetics 2021, 23, 104–119. [Google Scholar] [CrossRef]
- Fursova, O.V.; Pogorelko, G.V.; Tarasov, V.A. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 2009, 429, 98–103. [Google Scholar] [CrossRef]
- Shu, Y.; Li, W.; Zhao, J.; Zhang, S.; Xu, H.; Liu, Y.; Guo, C. Transcriptome sequencing analysis of alfalfa reveals CBF genes potentially playing important roles in response to freezing stress. Genetics and Molecular Biology 2017, 40, 824–833. [Google Scholar] [CrossRef]
- Zhuo, C.; Liang, L.; Zhao, Y.; Guo, Z.; Lu, S. A cold responsive ethylene responsive factor from Medicago falcata confers cold tolerance by up-regulation of polyamine turnover, antioxidant protection, and proline accumulation. Plant, Cell & Environment 2018, 41, 2021–2032. [Google Scholar]
- Ritonga, F.N.; Chen, S. Physiological and molecular mechanism involved in cold Stress tolerance in plants. Plants 2020, 9, 560. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Cui, Y.; Dong, S.; Luo, D.; Fang, L.; Shi, Z.; Liu, W.; Wang, Z.; Nan, Z.; Liu, Z. Integrative physiological, transcriptome, and metabolome analyses reveal the associated genes and metabolites involved in cold stress response in common vetch (Vicia sativa L.). Food and Energy Security 2023, 12, e484. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, C.; Ren, J.; Dong, J.; Shi, X.; Zhao, X.; Wang, X.; Wang, J.; Zhong, C.; Zhao, S.; Liu, X.; Gao, S.; Yu, H. An advanced lipid metabolism system revealed by transcriptomic and lipidomic analyses plays a central role in peanut cold tolerance. Frontiers in Plant Science 2020, 11, 1110. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Shi, Y.; Terzaghi, W.; Yang, S.; Li, J. Integration of light and temperature signaling pathways in plants. Journal of Integrative Plant Biology 2022, 64, 393–411. [Google Scholar] [CrossRef]
- Min, X.; Liu, Z.; Wang, Y.; Liu, W. Comparative transcriptomic analysis provides insights into the coordinated mechanisms of leaves and roots response to cold stress in Common Vetch. Industrial Crops and Products 2020, 158, 112949. [Google Scholar] [CrossRef]
- Li, L.; Han, C.; Yang, J.; Tian, Z.; Jiang, R.; Yang, F.; Jiao, K.; Qi, M.; Liu, L.; Zhang, B.; Niu, J.; Jiang, Y.; Li, Y.; Yin, J. comprehensive transcriptome analysis of responses during cold Stress in Wheat (Triticum aestivum L.). Genes 2023, 14, 844. [Google Scholar] [CrossRef]
- Zou, F.; Hu, Q.; Li, H.; Lin, J.; Liu, Y.; Sun, F. Dynamic disturbance analysis of grasslands using nural networks and spatiotemporal indices fusion on the Qinghai-Tibet Plateau. Frontiers in Plant Science 2022, 12, 760551. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Hou, X.; Chen, Y.; Li, C.; Ma, X. Flexible response and rapid recovery strategies of the plateau forage Poa crymophila to cold and drought. Frontiers in Plant Science 2022, 13, 970496. [Google Scholar] [CrossRef]
- Hua, Q.; Yu, Y.; Dong, S.; Li, S.; Shen, H.; Han, Y.; Zhang, J.; Xiao, J.; Liu, S.; Dong, Q.; Zhou, H.; Wessell, K. Leaf spectral responses of Poa crymophila to nitrogen deposition and climate change on Qinghai-Tibetan Plateau. Agriculture, Ecosystems & Environment 2019, 284, 106598. [Google Scholar]
- Fu, J.; Miao, Y.; Shao, L.; Hu, T.; Yang, P. De novo transcriptome sequencing and gene expression profiling of Elymus nutans under cold stress. BMC Genomics 2016, 17, 870. [Google Scholar] [CrossRef]
- Dong, W.; Ma, X.; Jiang, H.; Zhao, C.; Ma, H. Physiological and transcriptome analysis of Poa pratensis var. anceps cv. Qinghai in response to cold stress. BMC Plant Biology 2020, 20, 362. [Google Scholar] [CrossRef]
- Qi, W.; Wang, F.; Ma, L.; Qi, Z.; Liu, S.; Chen, C.; Wu, J.; Wang, P.; Yang, C.; Wu, Y.; Sun, W. Physiological and biochemical mechanisms and cytology of cold tolerance in Brassica napus. Frontiers in Plant Science 2020, 11, 1241. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.-Y.; Li, C.-X.; He, Y.; Hou, X.-Y.; Ma, X.-R. The regulation of adaptation to cold and drought stresses in Poa crymophila Keng revealed by integrative transcriptomics and metabolomics analysis. Frontiers in Plant Science 2021, 12, 631117. [Google Scholar] [CrossRef]
- Tian, Y.; Peng, K.; Lou, G.; Ren, Z.; Sun, X.; Wang, Z.; Xing, J.; Song, C.; Cang, J. Transcriptome analysis of the winter wheat Dn1 in response to cold stress. BMC Plant Biology 2022, 22, 277. [Google Scholar] [CrossRef]
- Waititu, J.K.; Cai, Q.; Sun, Y.; Sun, Y.; Li, C.; Zhang, C.; Liu, J.; Wang, H. Transcriptome profiling of maize (Zea mays L.) leaves reveals key cold-responsive genes, transcription factors, and metabolic pathways regulating cold stress tolerance at the seedling stage. Genes 2021, 12, 1638. [Google Scholar] [CrossRef]
- Cai, X.; Chen, Y.; Wang, Y.; Shen, Y.; Yang, J.; Jia, B.; Sun, X.; Sun, M. A comprehensive investigation of the regulatory roles of OsERF096, an AP2/ERF transcription factor, in rice cold stress response. Plant Cell Reports 2023. Advance online publication. [Google Scholar] [CrossRef]
- Liu, C.; Ou, S.; Mao, B.; Tang, J.; Wang, W.; Wang, H.; Cao, S.; Schläppi, M.R.; Zhao, B.; Xiao, G.; Wang, X.; Chu, C. Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nature Communications 2018, 9, 3302. [Google Scholar] [CrossRef]
- Cheng, M.; Cui, K.; Zheng, M.; Yang, T.; Zheng, J.; Li, X.; Luo, X.; Zhou, Y.; Zhang, R.; Yan, D.; Yao, M.; Iqbal, M.Z.; Zhou, Q.; He, R. Physiological attributes and transcriptomics analyses reveal the mechanism response of Helictotrichon virescens to low temperature stress. BMC Genomics 2022, 23, 280. [Google Scholar] [CrossRef]
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 Have a Different Function than CBF2 in Cold Acclimation and Define Different Gene Classes in the CBF Regulon. Proceedings of the National Academy of Sciences of the United States of America 2007, 104, 21002–21007. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, L.; Baral, R.; Paudel, D.; Min, D.; Makaju, S.O.; Poudel, H.P.; Acharya, J.P.; Missaoui, A.M. Cold stress in plants: Strategies to improve cold tolerance in forage species. Plant Stress 2022, 4, 100081. [Google Scholar] [CrossRef]
- Shah, F.A.; Ni, J.; Yao, Y.; Hu, H.; Wei, R.; Wu, L. Overexpression of karrikins receptor gene sapium sebiferum KAI2 promotes the cold stress tolerance via regulating the redox homeostasis in Arabidopsis thaliana. Frontiers in Plant Science 2021, 12, 657960. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nature Reviews Molecular Cell Biology 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Eremina, M.; Rozhon, W.; Poppenberger, B. Hormonal control of cold stress responses in plants. Cellular and Molecular Life Sciences 2015, 73, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Li, H.; Chen, X.; Xiang, X.; Guo, Z.; Yu, J.; Zhou, Y. The role of calcium-dependent protein kinase in hydrogen peroxide, nitric oxide and ABA-dependent cold acclimation. Journal of Experimental Botany 2018, 69, 4127–4139. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wu, N.; Fu, J.; Wang, S.; Li, X.; Xiao, J.; Xiong, L. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. Journal of Experimental Botany 2012, 63, 6467–6480. [Google Scholar] [CrossRef]
- Shi, Q.; Zhang, Y.; To, V.-T.; Shi, J.; Zhang, D.; Cai, W. Genome-wide characterization and expression analyses of the auxin/indole-3-acetic acid (Aux/IAA) gene family in barley (Hordeum vulgare L.). Scientific Reports 2020, 10, 10242. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 2006, 225, 353–364. [Google Scholar] [CrossRef]
- Deng, S.; Ma, J.; Zhang, L.; Chen, F.; Sang, Z.; Jia, Z.; Ma, L. De novo transcriptome sequencing and gene expression profiling of Magnolia wufengensis in response to cold stress. BMC Plant Biology 2019, 19, 321. [Google Scholar] [CrossRef]
- Xiang, N.; Hu, J.-g.; Yan, S.; Guo, X. Plant hormones and volatiles response to temperature stress in sweet corn (Zea mays L.) seedlings. Journal of Agricultural and Food Chemistry 2021, 69, 6779–6790. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.; Jiang, C.; Chen, L.; Paul, A.; Chatterjee, A.; Shen, G. Achieving abiotic stress tolerance in plants through antioxidative defense mechanisms. Frontiers in Plant Science 2023, 14, 1110622. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nature Reviews Molecular Cell Biology 2022, 23, 663–679. [Google Scholar] [CrossRef]
- Kantakhoo, J.; Imahori, Y. Antioxidative responses to pre-storage hot water treatment of red sweet pepper (Capsicum annuum L.) fruit during cold storage. Foods 2021, 10, 3031. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Wu, T.; Li, S.; He, Q.; Yang, Z.; Zhang, W.; Gan, Y.; Sun, P.; Xiang, G.; Zhang, H.; Deng, H. The methylation patterns and transcriptional responses to chilling stress at the seedling stage in rice. International Journal of Molecular Sciences 2019, 20, 5089. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Zhang, L.; Wang, H.; Lu, J.; Wei, H.; Yu, S. Transcriptomic profiling of young cotyledons response to chilling stress in two contrasting cotton (Gossypium hirsutum L.) genotypes at the seedling stage. International Journal of Molecular Sciences 2020, 21, 5095. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Kato, H.; Imai, R. Biochemical identification of the OsMKK6–OsMPK3 signalling pathway for chilling stress tolerance in rice. Biochemical Journal 2012, 443, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, P.; Si, T.; Hsu, C.-C.; Wang, L.; Zayed, O.; Yu, Z.; Zhu, Y.; Dong, J.; Tao, W.A.; Zhu, J.-K. MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Developmental Cell 2017, 43, 618–629.e5. [Google Scholar] [CrossRef] [PubMed]
- Hung, F.-Y.; Chen, F.-F.; Li, C.; Chen, C.; Chen, J.-H.; Cui, Y.; Wu, K. The LDL1/2-HDA6 histone modification complex interacts with TOC1 and regulates the core circadian clock components in Arabidopsis. Frontiers in Plant Science 2019, 10, 233. [Google Scholar] [CrossRef]
- Hong, W.-J.; Jiang, X.; Ahn, H.R.; Choi, J.; Kim, S.-R.; Jung, K.-H. Systematic analysis of cold Stress response and diurnal rhythm using transcriptome data in rice reveals the molecular networks related to various biological processes. International Journal of Molecular Sciences 2020, 21, 6872. [Google Scholar] [CrossRef]
- Li, Y.; Shi, Y.; Li, M.; Fu, D.; Wu, S.; Li, J.; Gong, Z.; Liu, H.; Yang, S. The CRY2–COP1–HY5–BBX7/8 module regulates blue light-dependent cold acclimation in Arabidopsis. The Plant Cell 2021, 33, 3555–3573. [Google Scholar] [CrossRef]
- Wu, H.; Wu, Z.; Wang, Y.; Ding, J.; Zheng, Y.; Tang, H.; Yang, L. Transcriptome and metabolome analysis revealed the freezing resistance mechanism in 60-year-old overwintering Camellia sinensis. Biology 2021, 10, 996. [Google Scholar] [CrossRef]
- Ren, C.; Fan, P.; Li, S.; Liang, Z. Advances in understanding cold tolerance in grapevine. Plant Physiology 2023, 192, 1733–1746. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kang, W.; Wu, F.; Miao, J.; Shi, S. Comparative transcriptome analysis reveals new insight of alfalfa (Medicago sativa L.) cultivars in response to abrupt freezing stress. Frontiers in Plant Science 2022, 13, 798118. [Google Scholar] [CrossRef]
- Manara, A.; DalCorso, G.; Furini, A. The role of the atypical kinases ABC1K7 and ABC1K8 in abscisic acid responses. Frontiers in Plant Science 2016, 7, 366. [Google Scholar] [CrossRef]
- Wu, Q.; Han, T.; Yang, L.; Wang, Q.; Zhao, Y.; Jiang, D.; Ruan, X. The essential roles of OsFtsH2 in developing the chloroplast of rice. BMC Plant Biology 2021, 21, 445. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Fan, Z.; Zhang, Q.; Wang, C.; Gao, Y.; Deng, Y.; Zhu, B.; Zhu, H.; Chen, J.; Shan, W.; Yin, X.; Zhong, S.; Grierson, D.; Jiang, C.Z.; Luo, Y.; Fu, D.Q. BEL1-LIKE HOMEODOMAIN 11 regulates chloroplast development and chlorophyll synthesis in tomato fruit. The Plant Journal 2018, 94, 1126–1140. [Google Scholar] [CrossRef] [PubMed]
- Ke, N.J.; Kumka, J.E.; Fang, M.X.; Weaver, B.; Burstyn, J.N.; Bauer, C.E. RedB, a member of the CRP/FNR family, functions as a transcriptional redox brake. Microbiology spectrum 2022, 10, e0235322. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Bürger, M.; Chory, J.; Wang, X. The role of ethylene in plant temperature stress response. Trends in Plant Science 2023, 28, 808–824. [Google Scholar] [CrossRef]
- Wang, X.; Yu, C.; Liu, Y.; Yang, L.; Li, Y.; Yao, W.; Cai, Y.; Yan, X.; Li, S.; Cai, Y.; Li, S.; Peng, X. GmFAD3A, a ω-3 fatty acid desaturase gene, enhances cold tolerance and seed germination rate under low temperature in rice. International Journal of Molecular Sciences 2019, 20, 3796. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, N.; Zhang, Z.; Liu, W.; Xie, W. Identification of flowering regulatory networks and Hub genes expressed in the leaves of Elymus sibiricus L. using comparative transcriptome analysis. Frontiers in Plant Science 2022, 13, 877908. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.Z.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; Pesseat, S.; Quinn, A.F.; Sangrador-Vegas, A.; Scheremetjew, M.; Yong, S.Y.; Lopez, R.; Hunter, S. InterProScan 5: genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–63. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome biology 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 2008, 9, 559. [Google Scholar] [CrossRef]
- Adnan, M.; Morton, G.; Hadi, S. Analysis of rpoS and bolA gene expression under various stress-induced environments in planktonic and biofilm phase using 2(-ΔΔCT) method. Molecular and cellular biochemistry 2011, 357, 275–282. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




