1. Introduction
Autophagy is an evolutionarily conserved catabolic process in all eukaryotes, from yeast to humans. In normal conditions, the autophagy is necessary to maintain cellular homeostasis through the degradation of dysfunctional organelles, protein aggregates or misfolded proteins, that are delivered to the lysosomes for recycling [
1]. One of the classes of autophagic pathway consists of macro-autophagy [
2], involving the sequestration of substrates within double-membrane cytosolic vesicles called “autophagosomes”. These organelles fuse with lysosomes, generating a single membrane organelle known as “autolysosome”, which represents the degradative compartment of the entire process [
3,
4]. Two main factors are involved in the regulation of the autophagic process, i.e., the
TOR complex-1 (TORC1) and the AMP-activated protein Kinase (AMPK), which play opposite roles. Indeed,
TORC1 negatively regulates autophagy, both in
S. cerevisiae and in mammals [
5]. Conversely, AMPK inhibits the TORC1 pathway through the phosphorylation of both its inhibitor protein TSC2 (tuberin) [
6] and Raptor, one further
TORC1 component [
7]. In addition, AMPK activates the Unc-5-like kinase 1 (ULK1), the serine/threonine protein kinase that triggers autophagy [
8].
Today, the role of autophagy in tumor remains a controversial issue. While numerous studies have shown that autophagy promote tumor suppression in the early stage of tumorigenesis and autophagy-associated death of cancer cells, other reports have highlighted the role of autophagy as a pro-tumor survival process under conditions of environmental stress [
9,
10].
Glioblastoma multiforme (GBM), the most lethal human brain tumor, has lower levels of protein markers of autophagy than low-grade astrocytic tumors [
11,
12]; in particular, ULK1 and ULK2 levels are significantly lower in GBM patients than in healthy controls [
13]. An analysis conducted according to the Karnofsky Performance Scale index, which allows to classify the patients based on their functional impairment, reported that a high expression of Microtubule-associated
protein 1A/1B-light chain 3 (LC3), a hallmark of autophagy, was correlated with an improved patient prognosis [
14].
In addition, hyper-phosphorylation of AKT and mTOR has been detected in patient with grade-III and grade-IV gliomas. These factors, when active, are associated with downregulation of autophagic process and increased cell proliferation and stemness of glioma stem cells [
15,
16]. Considering this evidence on the role of autophagy as a tumor suppressor, drugs with synergistic action with the chemotherapy agents already in use, notably temozolomide, are being developed [
10].
Since many years, our group has been studying the effects of M2 muscarinic acetylcholine receptor (M2 mAChR) activation in GBM. mAChRs are G protein coupled-receptors (GPCRs) involved in the regulation of several fundamental processes in both central and peripheral nervous systems [
17]. Moreover, the expression of mAChRs in different tumors as well as their strategic role in the modulation of cancer cell proliferation, survival and migration have been demonstrated [
18,
19,
20,
21,
22]. In this framework, we have demonstrated that the M2 mAChR activation by the orthosteric M2 agonist Arecaidine Propargyl Ester (APE) has negative effects on cell proliferation and survival, both in GBM cell lines and in GBM cancer stem cells (GSCs) [
23,
24,
25,
26]. Recently, we have also shown that treatment with an M2 agonist results in an increase of aberrant mitosis in U251 and U87 cell lines, caused by an altered mitotic spindle formation by reducing the ability of microtubules to bind chromosomes. GBM cell lines are unable to proliferate after APE treatment, due to the triggering of catastrophic mitotic events and apoptosis [
27].
To improve the therapeutic potential of mAChR ligands in different pathological conditions such as Alzheimer's disease [
28,
29], schizophrenia [
17,
30] and cancer [
18,
21,
23], novel hybrid molecules, named "dualsteric or bitopic agonists”, were designed, synthesized and tested. This kind of ligands, which have been reported also for various GPCR receptor families, recognize simultaneously the acetylcholine (orthosteric) binding site and an additional, allosteric site. Such a cooperative interaction may increase the degree of selectivity among the subtypes and also promote signaling bias, since binding to the allosteric site may favor a functionally relevant receptor structure that controls a specific signaling pathway [
31,
32,
33].
Iper-8-naphthalimide (N-8-Iper) is a synthetic dualsteric agonist for the M2 mAChR previously characterized by our research group [
33,
34,
35]. We demonstrated that N-8-Iper reduced GSCs proliferation in a comparable manner to APE in two GSC lines, GB7 cells and G166 cells. Quite interestingly, N-8-Iper is active at doses (3 μM for GB7 cells and 25 μM for G166 cells) that are significantly lower than that of APE (100 μM) [
34,
35].
Starting from our previous results, here we investigated the downstream molecular targets linked to the cytotoxic effects observed after M2 mAChR activation with APE and N-8-Iper [
23,
26,
35,
36]. Moreover, we analyzed the signal transduction pathways involved in these processes, that could be affected by the different recognition mode of the two investigated muscarinic agonists.
We detected an upregulation of the autophagy process both in the U251 cell line and in GB7 cells after N-8-Iper treatment at high and low doses, while APE triggered autophagy only in U251 cells. Moreover, we studied the downregulation of the phosphoinositide 3-kinase (PI3K)/AKT/TORC1 pathway and the upregulation of AMPKα expression, that have been identified as two of the main pathways involved in the modulation of the autophagy flux. Finally, the increased autophagy after M2 stimulation by N-8-Iper positively improved the apoptotic process, as shown by the increase in the cleaved form of Caspases 9 and 3.
2. Materials and Methods
2.1. Cell Cultures
The GSCs GB7 cells were obtained from human GBM biopsy [
37,
38,
39]. The cells were cultured on laminin-coated dishes (1 μg/mL; Sigma-Aldrich, St. Louis, MO, USA) and maintained in serum free medium consisting of DMEM/F12 (Sigma-Aldrich, St. Louis, MO, USA) and Neurobasal medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) (1:1; v:v) supplemented with 1% streptomycin, 50 IU/mL penicillin (Sigma-Aldrich, St. Louis, MO, USA), 1% glutamine (Sigma-Aldrich, St. Louis, MO, USA), 1% N2 supplement (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 2% B27 (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 20 ng/mL EGF (Recombinant Human Epidermal growth factor, Peprotec, London, UK), and 20 ng/mL FGF (Recombinant Human FGF-basic, ABM, Richmond, Canada). The GB7 cell culture was maintained at 37 °C in an atmosphere of 5% CO
2/95% air. Human glioblastoma U251MG cell line was cultured in DMEM (Sigma-Aldrich, St. Louis, MO, USA) containing 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA), 50 μg/mL streptomycin, 50 IU/mL penicillin, 2 mM glutamine (Sigma-Aldrich, St. Louis, MO, USA), 1% non-essential amino acids (Sigma-Aldrich, St. Louis, MO, USA), and maintained at 37 °C in a 10% CO
2 atmosphere.
2.2. Pharmacological Treatments
Arecaidine Propargyl Ester hydrochloride (APE, Sigma-Aldrich, Milan, Italy) is a synthetic compound obtained from modification of arecaidine, a natural alkaloid derived from areca nut. M2 agonist APE was used to selectively stimulate the M2 mAChR subtype. The ability of this agonist to bind the M2 mAChR subtype was previously demonstrated in GBM stable cell lines (U87 and U251 cell lines) and in GSCs (GB7 and GB8 cells) by pharmacological binding experiments and knockdown of the receptors by siRNA transfection pool [
23,
40]. Iper-8-naphthalimide (N-8-Iper) was synthesized according to a known literature procedure [
41]. Its ability to selectively bind M2 mAChR subtype has been showed by pharmacological binding and M2 mAChR knockdown experiments [
35]. To compare the activity of the two M2 receptor ligands, we first used both agonists at high concentration (100 μM), then adopted the lowest doses of N-8-Iper (25 μM and 3 μM) producing significant effects in U251 cell line and GB7 cells, as shown previously [
34].
2.3. U251 cell line transfection and immunofluorescence analysis
Transfection was performed on the U251 cell line using the following expression vectors: EGFP-LC3B and mRFP-EGFP-LC3B. Cells were seeded on 6-well plates at the density of 2 x 105/well, and transfected at 80% confluence using LipofectamineTM 3000 Reagent (Invitrogen, Waltham, MA, USA), according to the manufacturer's protocol. 48 h after transfection, the cells were maintained in complete medium supplemented with geneticine (800μg/ml; Sigma-Aldrich, St. Louis, MO, USA) to select only antibiotic-resistant clones containing EGFP-LC3B fusion protein or mRFP-EGFP-LC3B fusion protein.
Stable U251-EGFP-LC3 or U251-mRFP-EGFP-LC3 cells were plated on coverslips arranged in 24-well plates at the density of 2 x 104 cells. Cells were treated with 100 μM APE, 100 μM N-8-Iper, 25 μM N-8-Iper with or without 25 μM Chloroquine (CQ) for 72 h. Then, cells were washed 3 times with PBS and fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. After 3 washes in PBS, cells were incubated with Hoechst 33342 (1:1000, Sigma-Aldrich, St. Louis, MO, USA) for 10 min at room temperature (RT), for nuclei counterstaining, and then washed 3 times with PBS. At the end, coverslips were fixed on microscope slides with a PBS-glycerol (3:1; v/v) solution. The images were acquired with a Apotome Zeiss fluorescence microscope through Zeiss Zen lite software (Zeiss, Oberkochen, Germany).
2.4. Cell Viability Assay
U251 cells were seeded on 96-well plates at the density of 1.5 × 10
4 cells/well. After 24 h, cells were treated with N-8-Iper at different times (ranging from 24 to 96 h). Cell proliferation was evaluated by colorimetric assay based on 3-(4,5-dimethylthiazol-2-y1)-2,5-diphenyltetrazolium bromide (MTT, Sigma-Aldrich, St. Louis, MO, USA) metabolization. The MTT assay was performed according to the protocol optimized by Mosmann [
42]. MTT was dissolved in PBS at 5 mg/ml. The MTT stock solution (10×) was added and diluted (1×) in each well and then incubated at 37°C for 3 h. Isopropanol (+ 0.04 M HCl + 1% Triton X-100) to dissolve the dark blue crystals. For each well, the optical density (OD) at 570 nm was measured by Multiskan FC (Thermofisher Scientific, Waltham, MA, USA).
2.5. Protein Extraction and Western Blot Analysis
Cells were harvested in lysis buffer (Tris-EDTA 10mM, 0.5% NP40, NaCl 150 mM), containing a protease inhibitor, boiled for 5 min at 90°C. The protein extracts were run on SDS-polyacrilamide gel (SDS- PAGE) and transferred to Polyvinylidene Difluoride (PVDF) sheets (Merck Millipore, Darmstadt, Germany). Membranes were blocked for 40 min in 5% of non-fat milk powder (Sigma-Aldrich, St. Louis, MO, USA) in PBS containing 0.1% Tween-20 (PBS-Tween), and then incubated overnight at 4°C with one of the following primary antibodies: anti-LC3B (1:1500, Sigma-Aldrich, St. Louis, MO, USA), anti-PI3 Kinase p85 (dilution 1:800, Cell Signaling, Danvers, MA, USA), anti-Phospho AKTThr308 (1:800, Cell Signaling, Danvers, MA, USA), anti-AKT pan (1:800, Cell Signaling, Danvers, MA, USA), anti-Phospho AMPK𝛼1/2Thr172 (dilution 1:800, Cell Signaling, Danvers, MA, USA), anti-AMPK𝛼1 (dilution 1:1000, Immunological Science, Milan, Italy), anti-Phospho p70 S6 KinaseThr389 (dilution 1:600, Immunological Science, Milan, Italy), anti-p70S6 Kinase (dilution 1:1000, Immunological Science, Milan, Italy), anti-Caspase 9 (dilution 1:2000, Immunological Science, Milan, Italy), anti-Caspase 3 (dilution 1:2000, Immunological Science, Milan, Italy), anti-β-Actin (1:2000, Immunological Science, Milan, Italy). β-Actin was used as reference protein for loading control.
2.6. Statistical Analysis
Data are presented as the mean ± SEM. Statistical analysis was performed by One-way ANOVA followed by Dunnett multiple comparison post-test. Data were considered statistically significant at * p < 0.05, ** p < 0.01 and *** p < 0.001. Data analyses were performed with GraphPad Prism 8 (GraphPad Software, La Jolla, CA, USA).
4. Discussion
Over the past decade, various studies have shown that activation of mAChRs is critical in the regulation of cell proliferation and cancer progression [
47]
. In this context, we have been studying the role of the M2 mAChR
in cancer for many years, demonstrating that activation of this receptor subtype brings about anti-proliferative and cytotoxic effects on cells of different tumor types such as ovarian cancer [
19], breast cancer [
18], neuroblastoma [
21,
48], and GBM [
34]. As far as GBM we showed that, in the U251 cell line, activation of M2 mAChR by the orthosteric agonist APE caused cell cycle arrest as well as progressive accumulation of the cells in the G2/M phase [
25]. In GSCs cells, named GB7, APE and the dualsteric agonist N-8-Iper were capable to inhibit cell proliferation in a time- and dose-dependent manner [
23,
35]. Moreover, analysis of the U251 cell fraction with hypodiploid DNA content and higher granularity (SSC) obtained by flow cytometry, and the ELISA quantification of cytoplasmic nucleosomes, showed that treatment with 100 μM APE for 72 h caused an increase in apoptotic cells [
25]. Once the same analysis was performed in GB7 cells after 48 h and 72 h treatment with 100 μM N-8-Iper, this M2 mAChR activator produced an enhanced percentage of apoptotic cells [
35].
At present, the role of autophagy in cancer cells is receiving much attention since this process seems to have divergent effects depending on tumor type and/or grade. Indeed, it has been observed that autophagy plays a strategic role in the regulation of tumor progression, acting as a tumor suppressor in the early stages of tumor development and as a pro-survival mechanism in the later stages of tumorigenesis [
1]. A recent study has showed that combined treatment with temozolomide and rapamycin, an mTOR pathway inhibitor, causes overexpression of Beclin-1 and LC3-II, thus considerably increasing autophagy-induced cell death of U251 cells [
49]. Building on this evidence, by transfection of the constructs overexpressing LC3B-EGFP we demonstrated that the autophagic process is promoted after activation of M2 mAChRs in the U251 stable cell line by both M2 orthosteric (APE) and dualsteric (N-8-Iper) agonists (
Figure 2). With the pH-sensitive expression construct, mRFP-EGFP-LC3B, we demonstrated that M2 agonists upregulate autophagy without causing its arrest. In fact, the increase in LC3 aggregates was observed only after addition of CQ, an autophagy flux blocker, and no-summative effect between CQ and the applied M2 agonist was put in evidence (
Figure 3). The ability of M2 mAChR activation to promote autophagy was also confirmed by the analysis of LC3B protein expression by Western blotting (
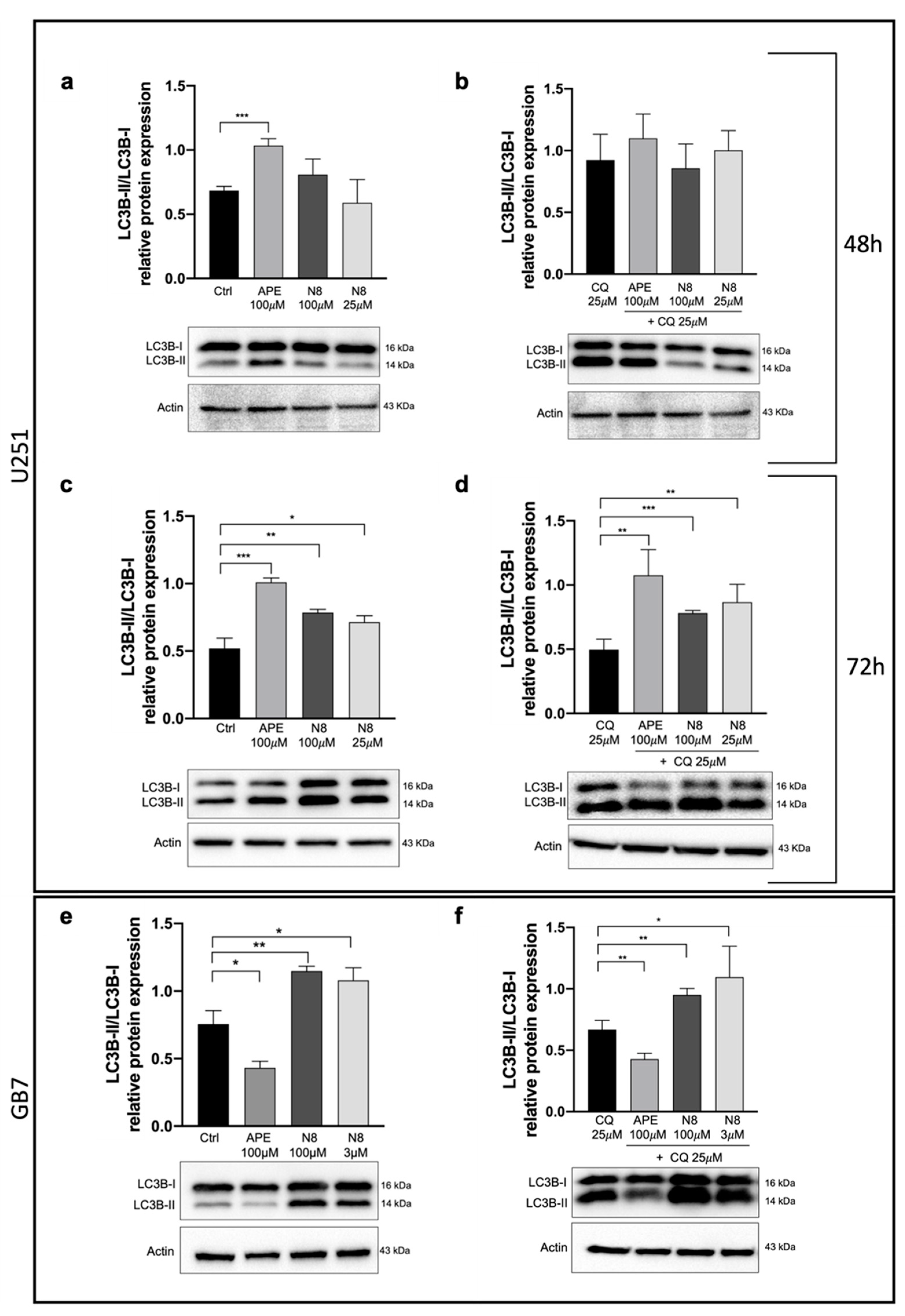
Figure 4). In this experiment, human GSCs were also assayed, to compare the effects of M2 agonists on a human established cell line (U251 cell line) with those on human GSCs (GB7 cells). Western blot analysis confirmed that N-8-Iper was able to induce autophagy both in U251 cell line and GB7 cells, while APE was able to increase LC3B-II compared with LC3B-I only in U251 cell line. The combined treatment with M2 agonists, that alone caused an increase in LC3B-II expression, and CQ significantly increased the LC3B-II/LC3B-I ratio when compared with the treatment with CQ alone (
Figure 4d,f). These results, together with those from the cells transfected with LC3-EGFP and mRFP-GFP-LC3B vectors, confirm the ability of M2 agonists to promote the autophagic flux and allow the fusion between autophagosomes and lysosomes; as a matter of fact, only CQ blocked the autophagy process at the autophagosome stage. However, a difference between the two studied M2 agonists clearly emerged since APE was found to induce autophagy in U251 cells only, whereas N-8-Iper was efficacious in both U251 and GB7 cells.
To further explore the different modulation pattern of autophagy by the two M2 agonists in both cell lines, we analyzed the expression of TORC1 and AMPK, two main factors involved in the regulation of this process. Treatment with N-8-Iper for 72 h, at high and low doses, downregulated in both cell lines the protein expression for PI3K p85 (
Figure 5a-c), the active form of AKT (phosphorylated at Thr308) (
Figure 5d-f), and the active form of the downstream effector of TORC1, p70 S6K phosphorylated at Thr389 (
Figure 5g-h). APE produced the same downstream effects but only in U251 cells, while in GB7 cells it negatively modulated the PI3K/AKT pathway but did not cause significant downregulation of the active form of p70 S6K, indicating a failure in TORC1 downregulation. Regarding the AMPKα protein level, an increase in the phosphorylated (active) form over the non-phosphorylated (inactive) one was observed after treatment with both APE and N-8-Iper, at high and low doses (
Figure 5i). In GB7 cells, only N-8-Iper was able to significantly raise the Phospho-AMPKα protein levels, further confirming the inability of APE to promote autophagy in GSCs (
Figure 5j). In
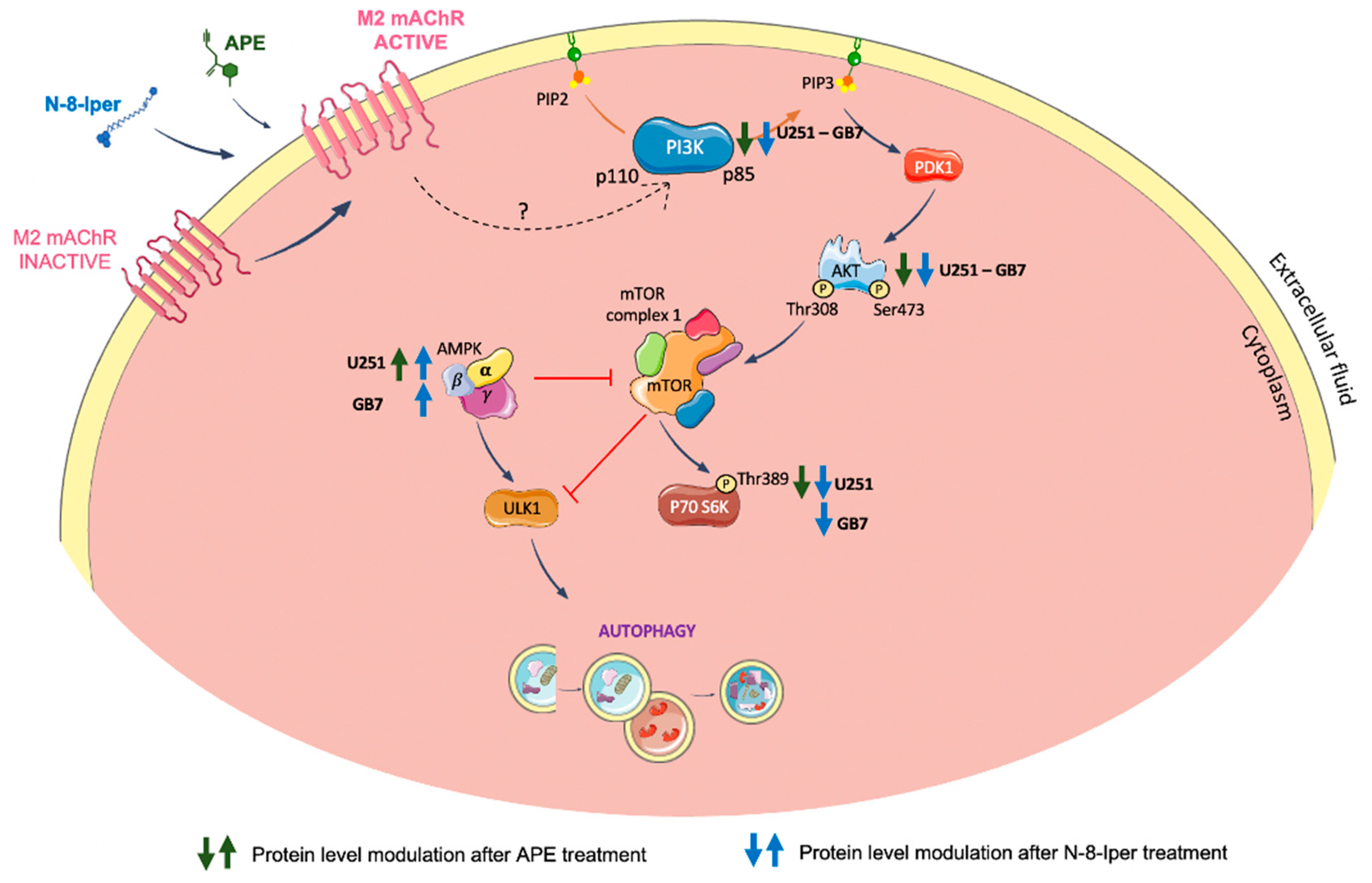
Figure 8, we have summarized the signaling pathways of the M2 receptor-induced autophagy process emerging from the above discussed data. Worth highlighting is the upregulation of AMPKα and downregulation of TORC1 activity following treatment with the dualsteric muscarinic agonist N-8-Iper in both cell lines. Conversely, the orthosteric agonist APE engendered a parallel modulation profile of the autophagy flux only in U251 cells and not in GB7 cells.
As already underlined, several literature data suggest that the autophagic flux may lead to cell death, including apoptosis [
46,
50]. In previous investigations, we demonstrated that the two M2 agonists induce cytotoxic effects [
26,
35] and by Florescence Activated Cell Sorting (FACS) analysis we found that they caused cell cycle arrest and apoptotic cell death [
25,
35,
36]. To further support these results and delve into the correlation between apoptotic cell death and M2 receptor-induced autophagy, we analyzed the expression of two of the proteins involved in the apoptotic process, i.e., Caspase-9 and Caspase-3 (
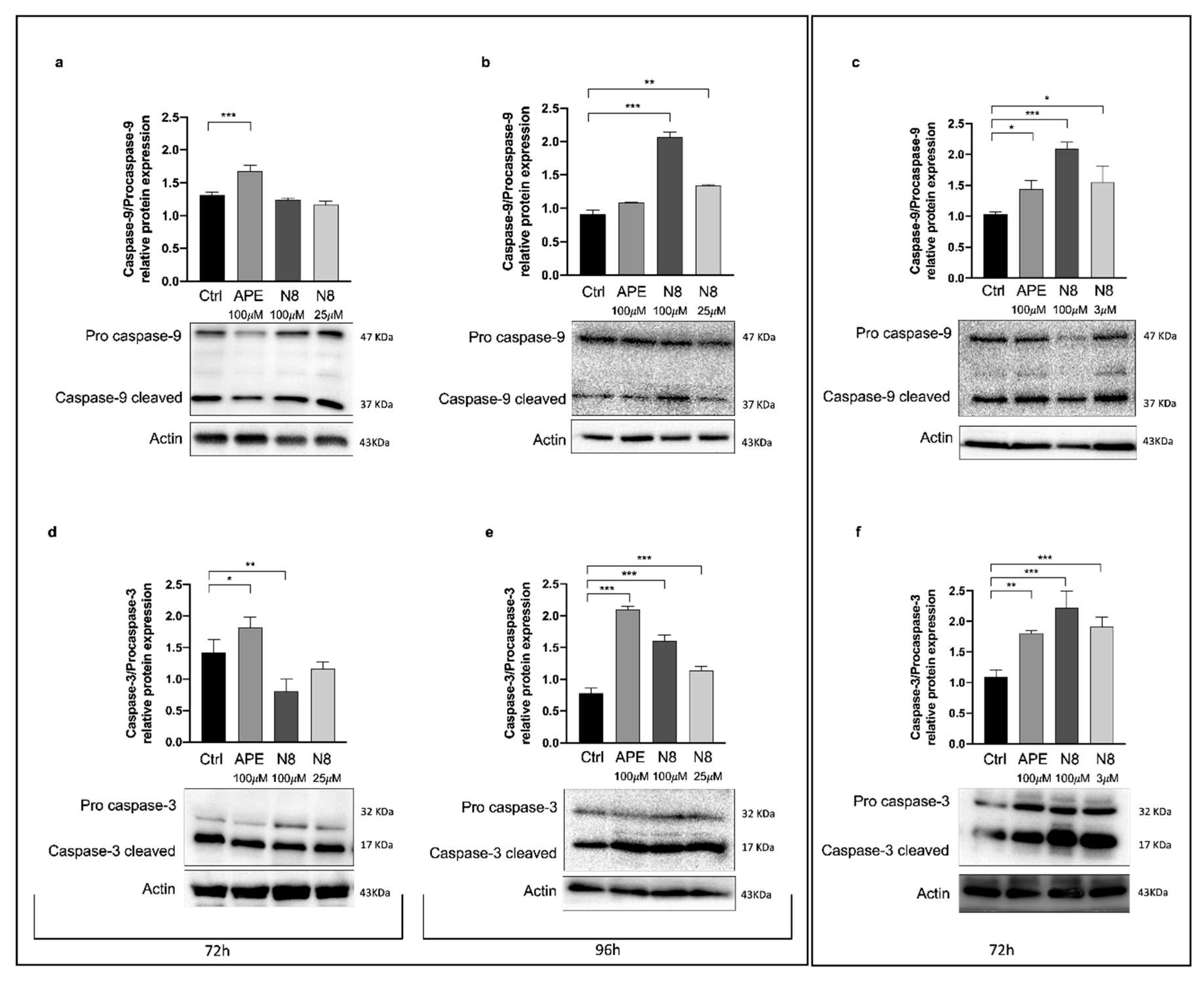
Figure 6). A treatment of 72 h with APE in U251 cells induced the increase of the cleaved forms of both Caspase-9 (
Figure 6a) and Caspase-3 (
Figure 6d), confirming our previous data. It was necessary to extend the treatment to 96 h to observe the increase of these proteins in N-8-Iper-treated U251 cells (
Figure 6b,e). In GB7 cells, the cleaved form of Caspases 9 (
Figure 6c) and 3 (
Figure 6f) were upregulated by N-8-Iper. The treatment duration is indicative of a direct sequence between autophagy and apoptotic events. Unlike N-8-Iper treatment, the upregulation of apoptotic proteins after APE treatment does not seem to be linked with autophagy in GB7 cells.
Treatment with the autophagy inhibitor CQ allowed to improve the correlation between autophagy and downstream apoptosis. Inhibition of the autophagic flux by CQ caused in fact a remarkable reduction or complete absence of caspases in their active form, differently from what we observed by treating cells with N-8-Iper or APE alone. These results suggest that prevention of the autophagy process, which is promoted by M2 agonists (N-8-Iper for the U251 cell line and GB7 cells, APE for U251 cells only), interferes with the activation of apoptosis.
According to the previous data both APE and N-8-Iper have antiproliferative effects, interfering with PI3K/AKT pathway, in both cell models [
23,
24,
35]. Downregulation of the PI3K/AKT/TORC1 pathway, involved in regulating the
proliferation, and activation of autophagy can lead to cell death by apoptosis in both GBM cell models after N-8-Iper treatments. Downregulation of the PI3K/AKT pathway after APE treatment in GB7 cells can explain the decreased cell proliferation previously described [
36] but it is not able to inhibit TORC1 action, as also suggested by the lack of increase AMPK expression. This evidence explains the lack of upregulation of the autophagic process in GB7 cells after APE treatment. Western blot analysis of caspases and previous results from APE-treated GB7 cells, in which an increased percentage of annexin V-positive cells was assessed [
36], suggest that 100 μM APE induces directly apoptosis without any correlation with autophagy [
36]. This different APE behavior in GB7 cells compared with U251 may in part dependent on genetic bacjground of two cells line. GB7 cells present p53 wild type differently by U251 cells that are p53 mutated. Considering the anti-prolifertive and the strong cytotoxic effects produced by APE in cells where p53 is active, according to its function of genome guardian and regulator of the cell cycle, it may drive directly the cells to apotptosis.
Differently N8-Iper and APE seems to trace the activation of the same pathway inducing autophagy followed by the apoptotic process in U251 cells but in GB7 cells the same effcts was evident only with N8-Iper.
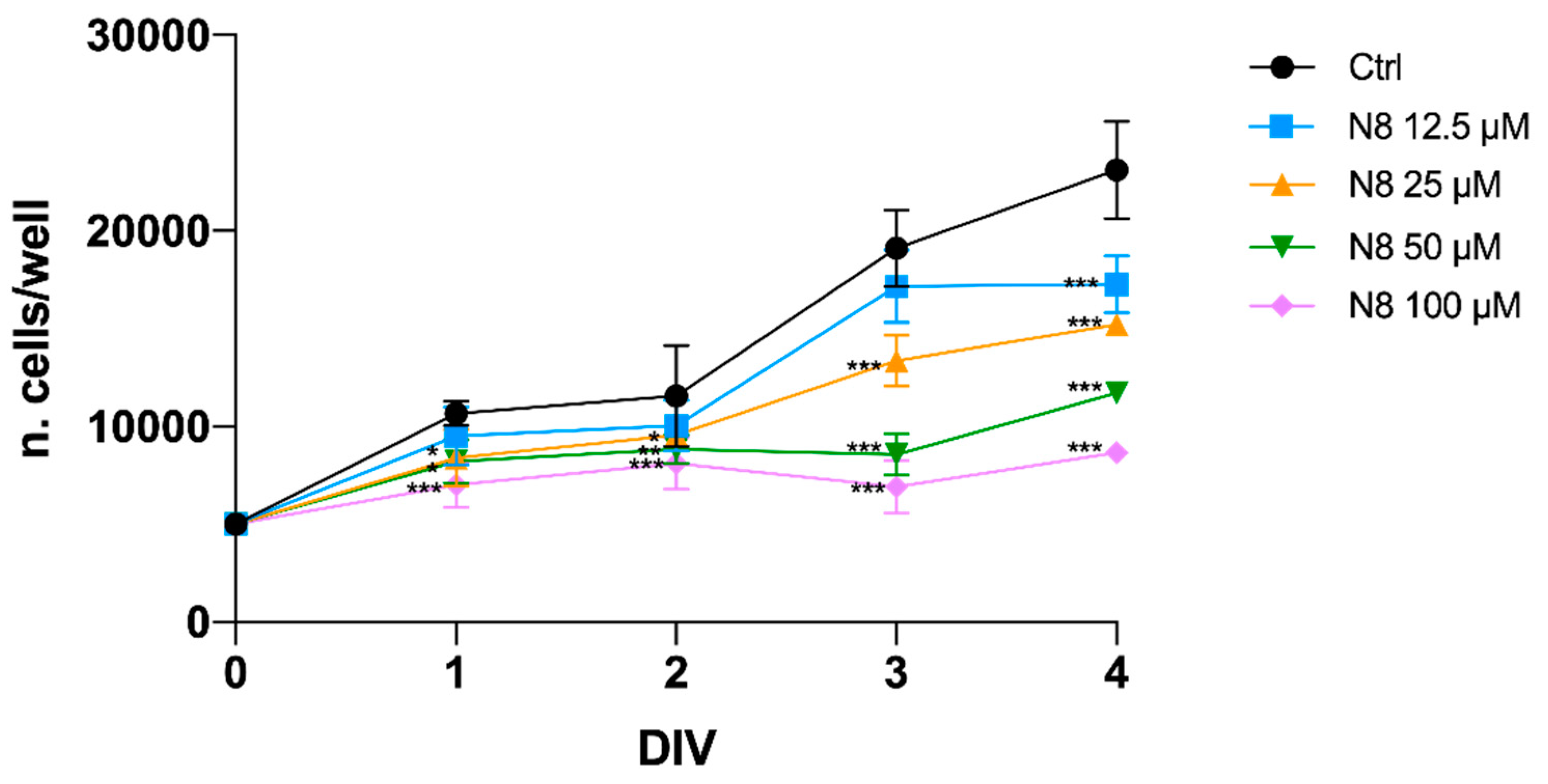
Figure 1.
Effect of N-8-Iper (12.5 μM, 25 μM, 50 μM and 100 μM) on U251 cell growth at different time points of treatment (ranging from 1 to 4 days in vitro, DIV). Data represent the mean (± SEM) of four different experiments performed in sextuplicate. ANOVA test was used followed by Dunnett’s post test (N-8-Iper treated cells vs. untreated cells, Ctrl. *** p < 0.001; ** p < 0.01; * p < 0.05).
Figure 1.
Effect of N-8-Iper (12.5 μM, 25 μM, 50 μM and 100 μM) on U251 cell growth at different time points of treatment (ranging from 1 to 4 days in vitro, DIV). Data represent the mean (± SEM) of four different experiments performed in sextuplicate. ANOVA test was used followed by Dunnett’s post test (N-8-Iper treated cells vs. untreated cells, Ctrl. *** p < 0.001; ** p < 0.01; * p < 0.05).
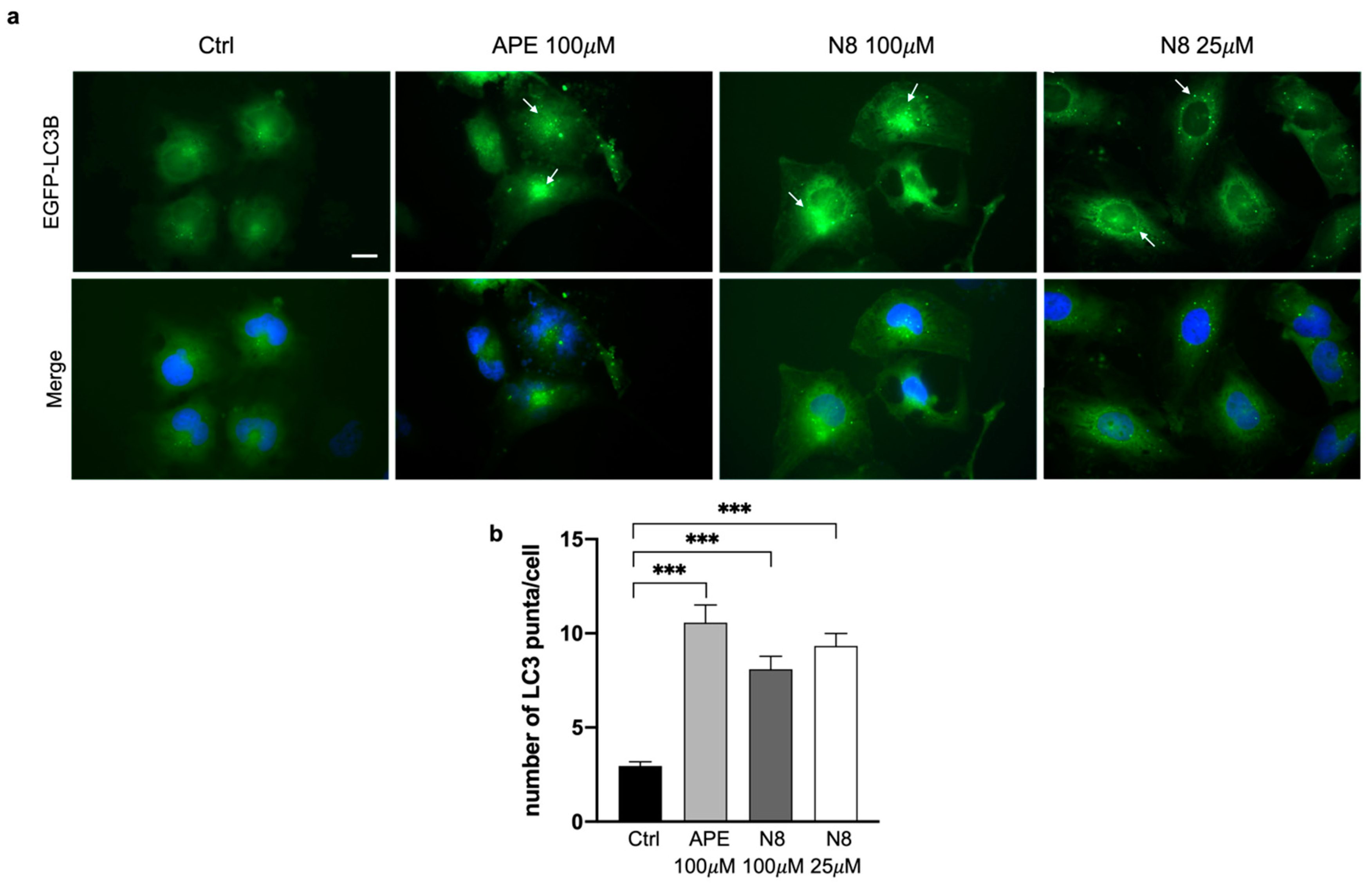
Figure 2.
(a) Representative images of fluorescence microscopy analysis of the U251 cell line transfected with the EGFP-LC3B expression vector, treated for 72 h with 100 μM APE, 100 μM N-8-Iper (N8) or 25 μM N-8-Iper (N8). Nuclei were stained with Hoechst 33342. White arrows indicate green spots. Scale bar = 10 μm. (b) The quantification has been performed counting the number of green spots within each cell in 13 photographic fields for each experimental condition. All the experiments were performed in triplicate. The values (mean ± SEM) are reported as number of green dots/cells. For each experimental condition, about 150 cells were analyzed. ANOVA test was used followed by Dunnett’s post test (untreated cells (Ctrl) vs M2 agonists treated cells; *** p < 0.001).
Figure 2.
(a) Representative images of fluorescence microscopy analysis of the U251 cell line transfected with the EGFP-LC3B expression vector, treated for 72 h with 100 μM APE, 100 μM N-8-Iper (N8) or 25 μM N-8-Iper (N8). Nuclei were stained with Hoechst 33342. White arrows indicate green spots. Scale bar = 10 μm. (b) The quantification has been performed counting the number of green spots within each cell in 13 photographic fields for each experimental condition. All the experiments were performed in triplicate. The values (mean ± SEM) are reported as number of green dots/cells. For each experimental condition, about 150 cells were analyzed. ANOVA test was used followed by Dunnett’s post test (untreated cells (Ctrl) vs M2 agonists treated cells; *** p < 0.001).
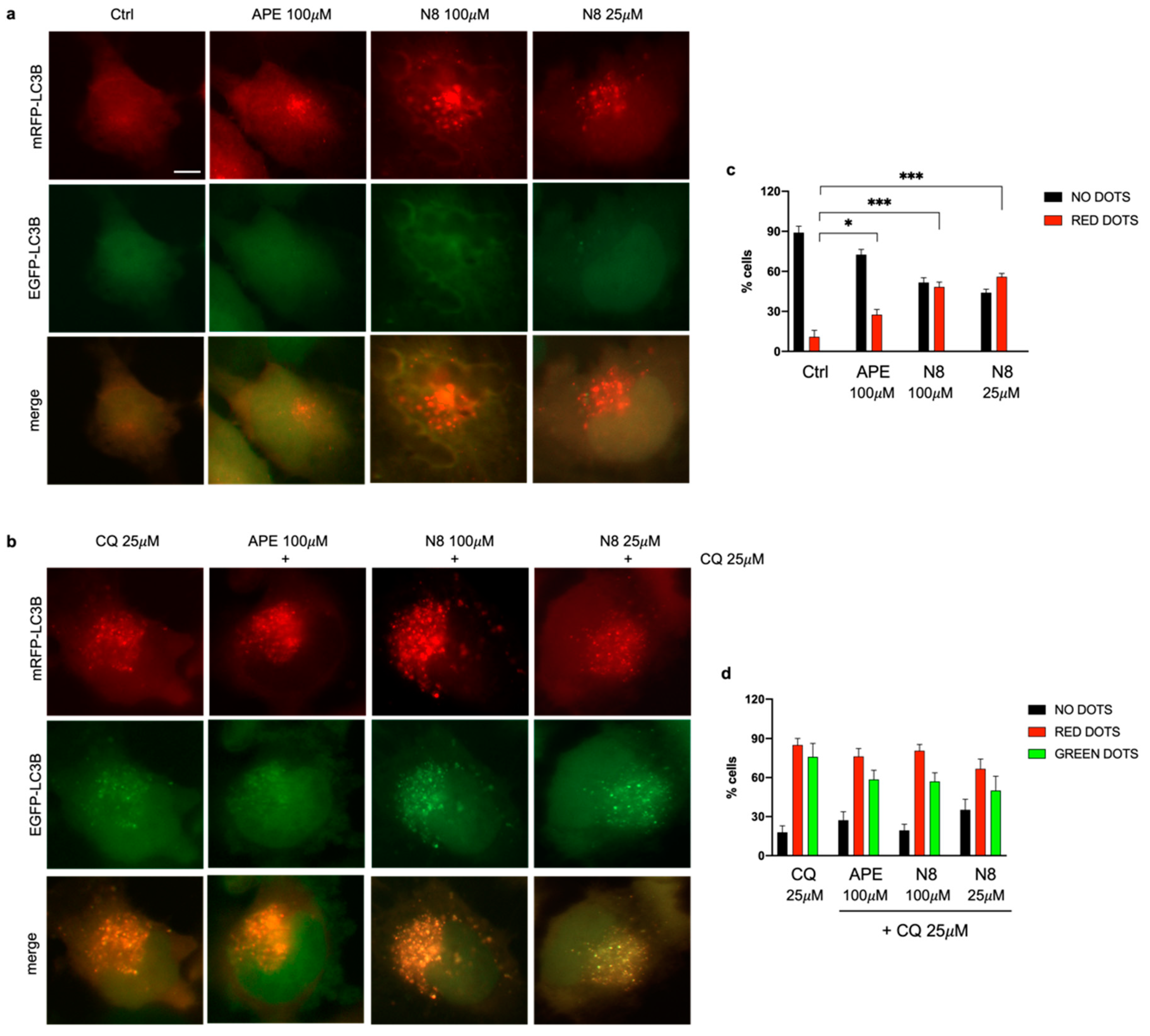
Figure 3.
Representative images of fluorescence microscopy analysis of the U251 cell line stably transfected with the mRFP-GFP-LC3B expression vector, treated for 72 h with 100 μM APE, 100 μM N-8-Iper (N8) or 25 μM N-8-Iper (N8) in (a) the absent or (b) presence of 25 μM CQ.. Scale bar=5 μm. (c, d) The quantification has been performed counting the number of cells without dots (black bars), with green dots or with red dots in 15 photographic fields for each experimental condition performed in triplicate. The values (mean ± SEM) are reported as percentage of red or green positive cells respect to total active cells. For each experimental condition, about 100 cells were analyzed. ANOVA test was used followed by Dunnett’s post test Ctrl (untreated cells) vs M2 agonists treated cells; *** p <0.001, * p <0.05. In d) any significant difference was observed between red and green dots for each experimental condition.
Figure 3.
Representative images of fluorescence microscopy analysis of the U251 cell line stably transfected with the mRFP-GFP-LC3B expression vector, treated for 72 h with 100 μM APE, 100 μM N-8-Iper (N8) or 25 μM N-8-Iper (N8) in (a) the absent or (b) presence of 25 μM CQ.. Scale bar=5 μm. (c, d) The quantification has been performed counting the number of cells without dots (black bars), with green dots or with red dots in 15 photographic fields for each experimental condition performed in triplicate. The values (mean ± SEM) are reported as percentage of red or green positive cells respect to total active cells. For each experimental condition, about 100 cells were analyzed. ANOVA test was used followed by Dunnett’s post test Ctrl (untreated cells) vs M2 agonists treated cells; *** p <0.001, * p <0.05. In d) any significant difference was observed between red and green dots for each experimental condition.
Figure 4.
Representative western blot relative to LC3B-II/I expression after 48 h (a) and 72h (c) of treatment with 100 μM APE, 100 μM N-8-Iper (N8), 25 μM N-8-Iper and after 48 h (b) and 72 h (d) of co-treatment with 25μM CQ and APE (100 μM) or N8 (100 μM and 25 μM) + CQ in the U251 cell line. (e) Representative western blot analysis of LC3-II/I expression after 48 h of treatment with 100 μM APE, 100 μM N-8-Iper, 3 μM N-8-Iper and (f) after 48 h of co-treatment with 25μM CQ and APE (100 μM) or N-8-Iper (100 μM and 3 μM) + CQ in the GB7 cells. Actin was used as internal reference protein. The graphs show the densitometric analysis of the bands of the western blot analysis for LC3B-II normalized with the bands of LC3B-I protein. The data are the average (mean ± SEM) of three independent experiments. ANOVA test was used followed by Dunnett’s post test (untreated cells (Ctrl) vs M2 agonists treated cells; *** p <0.001, ** p <0.01, * p <0.05).
Figure 4.
Representative western blot relative to LC3B-II/I expression after 48 h (a) and 72h (c) of treatment with 100 μM APE, 100 μM N-8-Iper (N8), 25 μM N-8-Iper and after 48 h (b) and 72 h (d) of co-treatment with 25μM CQ and APE (100 μM) or N8 (100 μM and 25 μM) + CQ in the U251 cell line. (e) Representative western blot analysis of LC3-II/I expression after 48 h of treatment with 100 μM APE, 100 μM N-8-Iper, 3 μM N-8-Iper and (f) after 48 h of co-treatment with 25μM CQ and APE (100 μM) or N-8-Iper (100 μM and 3 μM) + CQ in the GB7 cells. Actin was used as internal reference protein. The graphs show the densitometric analysis of the bands of the western blot analysis for LC3B-II normalized with the bands of LC3B-I protein. The data are the average (mean ± SEM) of three independent experiments. ANOVA test was used followed by Dunnett’s post test (untreated cells (Ctrl) vs M2 agonists treated cells; *** p <0.001, ** p <0.01, * p <0.05).
Figure 5.
(a) Representative western blot analysis for PI3K p85 expression after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper (N8), 25 μM N-8-Iper in U251 cell line and (b) after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper and (c) 3 μM N-8-Iper in GB7 cells. The graphs show the densitometric analysis of the bands of western blot analysis for PI3K p85 normalized with the bands of actin protein. (d) Representative western blot analysis of AKT phosphorylated in Thr 308 (Phospho-AKTThr308) expression after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper, 25 μM N-8-Iper in U251 cell line and (e) after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper and (f) 3 μM N-8-Iper in GB7 cells. The graphs show the densitometric analysis of the bands of western blot analysis for Phospho-AKTThr308 normalized with the bands of AKT pan. Actin was used as reference protein. (g) Representative western blot analysis of p70 S6K phosphorylated in Thr 389 (Phospho-p70 S6KThr389) expression after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper, 25 μM N-8-Iper in U251 cell line and (h) after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper and 3 μM N-8-Iper in GB7 cells. The graphs show the densitometric analysis of the bands of western blot analysis for Phospho-p70 S6KThr389 normalized with the bands of p70 S6K. Actin was used as reference protein. (i) Representative western blot analysis of AMPK𝛼 phosphorylated in Thr 172 (Phospho-AMPK𝛼 Thr172) expression after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper, 25 μM N-8-Iper in U251 cell line and (j) after 72 h of treatment in GB7 cells. The graphs show the densitometric analysis of the bands of western blot analysis for Phospho-AMPK𝛼Thr172 normalized with the bands of AMPK𝛼. Actin was used as reference protein. The data are the average (mean ± SEM) of three independent experiments. ANOVA test was used followed by Dunnett’s post test (untreated cells (Ctrl) vs M2 agonists treated cells; *** p <0.001, ** p <0.01, * p <0.05).
Figure 5.
(a) Representative western blot analysis for PI3K p85 expression after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper (N8), 25 μM N-8-Iper in U251 cell line and (b) after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper and (c) 3 μM N-8-Iper in GB7 cells. The graphs show the densitometric analysis of the bands of western blot analysis for PI3K p85 normalized with the bands of actin protein. (d) Representative western blot analysis of AKT phosphorylated in Thr 308 (Phospho-AKTThr308) expression after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper, 25 μM N-8-Iper in U251 cell line and (e) after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper and (f) 3 μM N-8-Iper in GB7 cells. The graphs show the densitometric analysis of the bands of western blot analysis for Phospho-AKTThr308 normalized with the bands of AKT pan. Actin was used as reference protein. (g) Representative western blot analysis of p70 S6K phosphorylated in Thr 389 (Phospho-p70 S6KThr389) expression after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper, 25 μM N-8-Iper in U251 cell line and (h) after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper and 3 μM N-8-Iper in GB7 cells. The graphs show the densitometric analysis of the bands of western blot analysis for Phospho-p70 S6KThr389 normalized with the bands of p70 S6K. Actin was used as reference protein. (i) Representative western blot analysis of AMPK𝛼 phosphorylated in Thr 172 (Phospho-AMPK𝛼 Thr172) expression after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper, 25 μM N-8-Iper in U251 cell line and (j) after 72 h of treatment in GB7 cells. The graphs show the densitometric analysis of the bands of western blot analysis for Phospho-AMPK𝛼Thr172 normalized with the bands of AMPK𝛼. Actin was used as reference protein. The data are the average (mean ± SEM) of three independent experiments. ANOVA test was used followed by Dunnett’s post test (untreated cells (Ctrl) vs M2 agonists treated cells; *** p <0.001, ** p <0.01, * p <0.05).
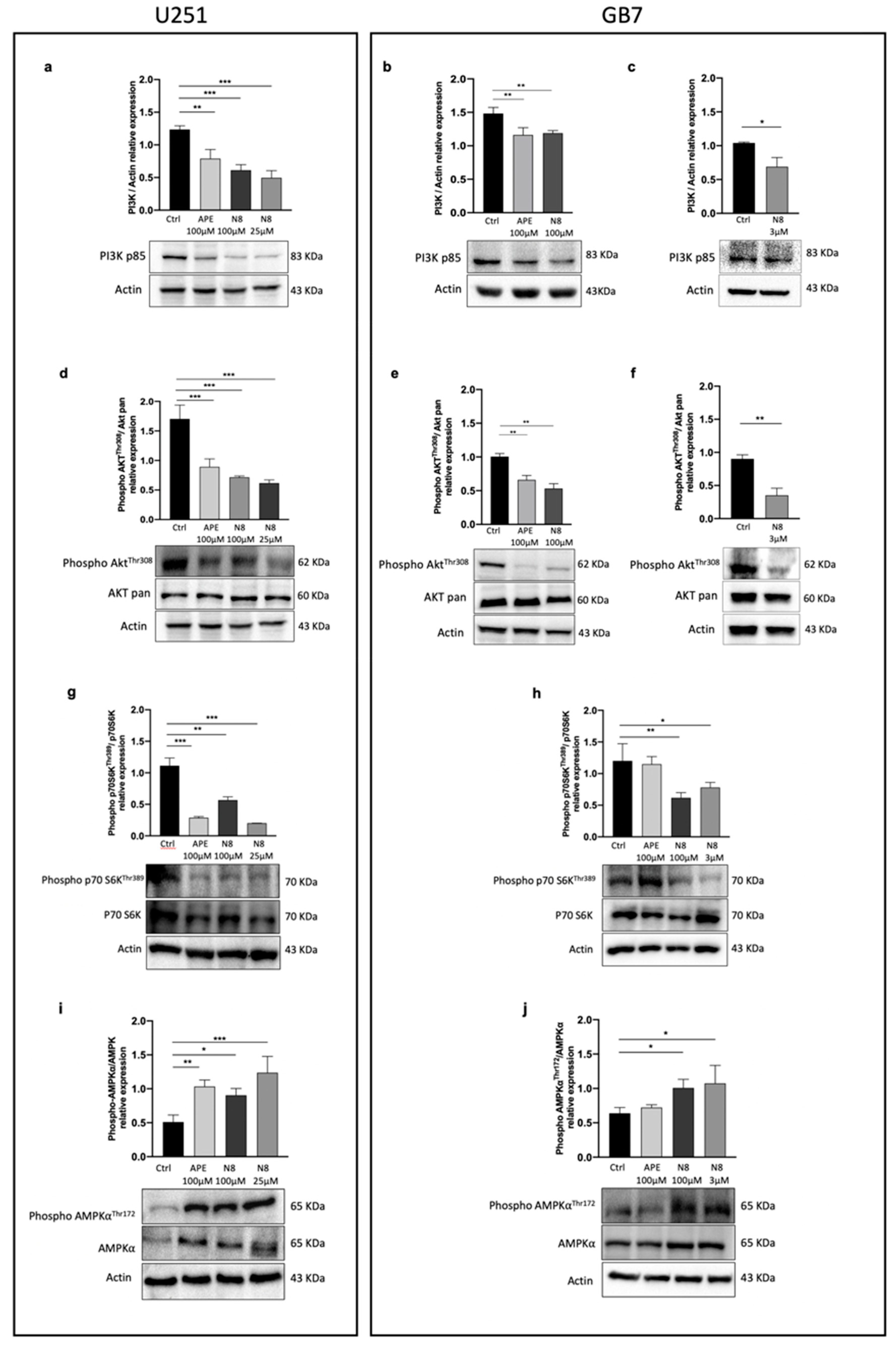
Figure 6.
Representative Western Blot analysis of Caspase-9 cleaved /Pro caspase-9 expression after 72 h of treatment (a) and after 96 h of treatment (b) with 100 μM APE, 100 μM N-8-Iper (N8), 25 μM N-8-Iper in the U251 cell line. (c) Representative western blot analysis of Caspase-9 cleaved /Pro caspase-9 expression after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper, 3 μM N-8-Iper in GB7 cells. The graphs show the densitometric analysis of the bands of western blot analysis for Caspase-9 cleaved normalized with the bands of Pro Caspase-9 protein. Actin was used as internal reference protein.
Figure 6.
Representative Western Blot analysis of Caspase-9 cleaved /Pro caspase-9 expression after 72 h of treatment (a) and after 96 h of treatment (b) with 100 μM APE, 100 μM N-8-Iper (N8), 25 μM N-8-Iper in the U251 cell line. (c) Representative western blot analysis of Caspase-9 cleaved /Pro caspase-9 expression after 72 h of treatment with 100 μM APE, 100 μM N-8-Iper, 3 μM N-8-Iper in GB7 cells. The graphs show the densitometric analysis of the bands of western blot analysis for Caspase-9 cleaved normalized with the bands of Pro Caspase-9 protein. Actin was used as internal reference protein.
Figure 7.
Representative Western blot analysis of Caspase-9 cleaved/Pro caspase-9 expression after 72 h of treatment (a) and after 96 h of treatment (b) with 25 μM CQ, 100 μM APE + 25 μM CQ, 100 μM N-8-Iper (N8) + 25 μM CQ, 25 μM N8 + 25 μM CQ in the U251 cell line. (c) Representative Western blot analysis of Caspase-9 cleaved/Pro caspase-9 expression after 72 h of treatment with 25 μM CQ, 100 μM N8 + 25 μM CQ, 3 μM N8 + 25 μM CQ in GB7 cells. Graphs show the densitometric analysis of the bands of Western blot analysis for Caspase-9 cleaved normalized with the bands of Pro Caspase-9 protein. Actin was used as the internal reference protein. (d) Representative Western blot analysis of Caspase-3 cleaved/Pro caspase-3 expression after 72 h of treatment. The graph shows the densitometric analysis of the bands of western blot analysis for Caspase-3 cleaved normalized with the bands of Pro Caspase-3 protein. Representative western blot analysis of Caspase-3 cleaved/Pro caspase-3 expression (e) after 96 h of treatment with 25 μM CQ, 100 μM APE + 25 μM CQ, 100 μM N-8-Iper (N8) + 25 μM CQ, 25 μM N8 + 25 μM CQ in the U251 cell line and (f) after 72 h of treatment with 25 μM CQ, 100 μM N8 + 25 μM CQ, 3 μM N8 + 25 μM CQ in GB7 cells. Graphs show the densitometric analysis of the bands of Western blot analysis for Pro Caspase-3 normalized with the bands of actin protein. Actin was used as the internal reference protein. e, f) No statical differences have been observed between all experimental conditions (n.s. p > 0.05). Data are the average (mean ± SEM) of three independent experiments. ANOVA test was used followed by Dunnett’s post-test (untreated cells (Ctrl) vs M2 agonists treated cells; ** p <0.01, * p <0.05).
Figure 7.
Representative Western blot analysis of Caspase-9 cleaved/Pro caspase-9 expression after 72 h of treatment (a) and after 96 h of treatment (b) with 25 μM CQ, 100 μM APE + 25 μM CQ, 100 μM N-8-Iper (N8) + 25 μM CQ, 25 μM N8 + 25 μM CQ in the U251 cell line. (c) Representative Western blot analysis of Caspase-9 cleaved/Pro caspase-9 expression after 72 h of treatment with 25 μM CQ, 100 μM N8 + 25 μM CQ, 3 μM N8 + 25 μM CQ in GB7 cells. Graphs show the densitometric analysis of the bands of Western blot analysis for Caspase-9 cleaved normalized with the bands of Pro Caspase-9 protein. Actin was used as the internal reference protein. (d) Representative Western blot analysis of Caspase-3 cleaved/Pro caspase-3 expression after 72 h of treatment. The graph shows the densitometric analysis of the bands of western blot analysis for Caspase-3 cleaved normalized with the bands of Pro Caspase-3 protein. Representative western blot analysis of Caspase-3 cleaved/Pro caspase-3 expression (e) after 96 h of treatment with 25 μM CQ, 100 μM APE + 25 μM CQ, 100 μM N-8-Iper (N8) + 25 μM CQ, 25 μM N8 + 25 μM CQ in the U251 cell line and (f) after 72 h of treatment with 25 μM CQ, 100 μM N8 + 25 μM CQ, 3 μM N8 + 25 μM CQ in GB7 cells. Graphs show the densitometric analysis of the bands of Western blot analysis for Pro Caspase-3 normalized with the bands of actin protein. Actin was used as the internal reference protein. e, f) No statical differences have been observed between all experimental conditions (n.s. p > 0.05). Data are the average (mean ± SEM) of three independent experiments. ANOVA test was used followed by Dunnett’s post-test (untreated cells (Ctrl) vs M2 agonists treated cells; ** p <0.01, * p <0.05).
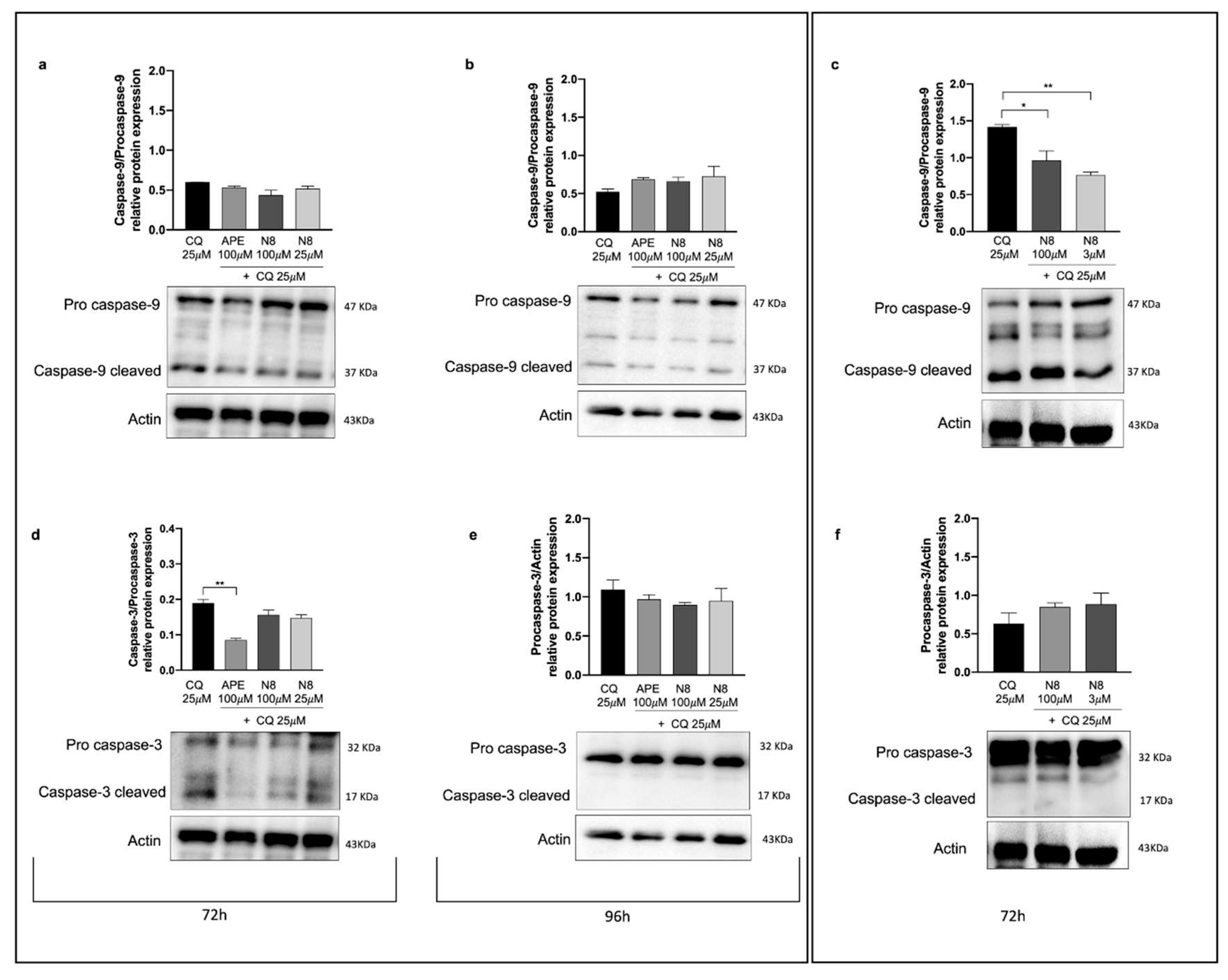
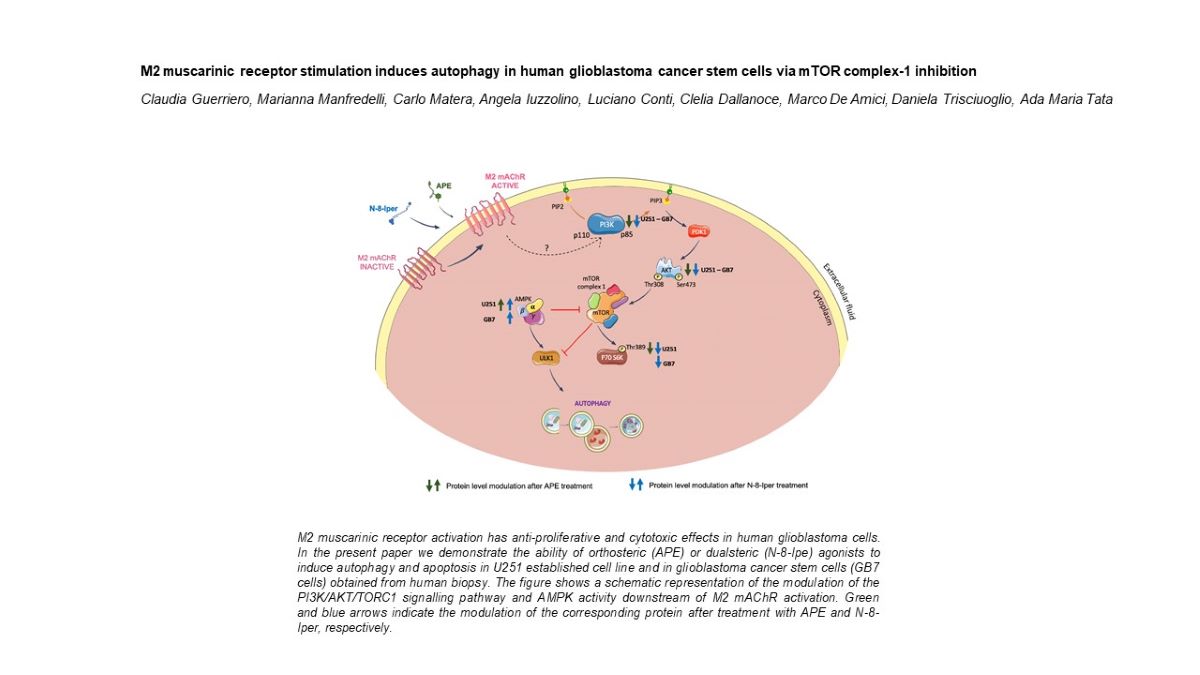
Figure 8.
Schematic representation of the modulation of PI3K/AKT/TORC1 signaling pathway and AMPK activity downstream of M2 mAChR activation by APE or N-8-Iper in U251 and GB7 cell lines. Green and blue arrows indicate the modulation of the corresponding protein after treatment with APE and N-8-Iper, respectively.
Figure 8.
Schematic representation of the modulation of PI3K/AKT/TORC1 signaling pathway and AMPK activity downstream of M2 mAChR activation by APE or N-8-Iper in U251 and GB7 cell lines. Green and blue arrows indicate the modulation of the corresponding protein after treatment with APE and N-8-Iper, respectively.