Submitted:
24 October 2023
Posted:
25 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Patient and Sample Collection
2.2. Data Analysis
3. Results
3.1. Distinct Protein Expression and Site Phosphorylation Patterns in RELAPSE Patients for AML FAB Subtypes M1/M2 and M4/M5
3.2. High Mitochondrial Protein Expression Splits Relapsing from Non-Relapsing AML Patients with the FAB Subtypes M4/M5
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dohner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef] [PubMed]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.M.; Catovsky, D.; Daniel, M.T.; Flandrin, G.; Galton, D.A.; Gralnick, H.R.; Sultan, C. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br. J. Haematol. 1976, 33, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Dohner, H.; Estey, E.H.; Amadori, S.; Appelbaum, F.R.; Buchner, T.; Burnett, A.K.; Dombret, H.; Fenaux, P.; Grimwade, D.; Larson, R.A.; et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 2010, 115, 453–474. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Boissel, N.; Renneville, A.; Biggio, V.; Philippe, N.; Thomas, X.; Cayuela, J.M.; Terre, C.; Tigaud, I.; Castaigne, S.; Raffoux, E.; et al. Prevalence, clinical profile, and prognosis of NPM mutations in AML with normal karyotype. Blood 2005, 106, 3618–3620. [Google Scholar] [CrossRef] [PubMed]
- Rau, R.; Brown, P. Nucleophosmin (NPM1) mutations in adult and childhood acute myeloid leukaemia: Towards definition of a new leukaemia entity. Hematol. Oncol. 2009, 27, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Tsykunova, G.; Reikvam, H.; Hovland, R.; Bruserud, O. The surface molecule signature of primary human acute myeloid leukemia (AML) cells is highly associated with NPM1 mutation status. Leukemia 2012, 26, 557–559. [Google Scholar] [CrossRef]
- Reikvam, H.; Aasebo, E.; Brenner, A.K.; Bartaula-Brevik, S.; Gronningsaeter, I.S.; Forthun, R.B.; Hovland, R.; Bruserud, O. High Constitutive Cytokine Release by Primary Human Acute Myeloid Leukemia Cells Is Associated with a Specific Intercellular Communication Phenotype. J. Clin. Med. 2019, 8. [Google Scholar] [CrossRef]
- Brenner, A.K.; Tvedt, T.H.; Nepstad, I.; Rye, K.P.; Hagen, K.M.; Reikvam, H.; Bruserud, O. Patients with acute myeloid leukemia can be subclassified based on the constitutive cytokine release of the leukemic cells; the possible clinical relevance and the importance of cellular iron metabolism. Expert Opin. Ther. Targets 2017, 21, 357–369. [Google Scholar] [CrossRef]
- Bullinger, L.; Dohner, K.; Bair, E.; Frohling, S.; Schlenk, R.F.; Tibshirani, R.; Dohner, H.; Pollack, J.R. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N. Engl. J. Med. 2004, 350, 1605–1616. [Google Scholar] [CrossRef]
- Alcalay, M.; Tiacci, E.; Bergomas, R.; Bigerna, B.; Venturini, E.; Minardi, S.P.; Meani, N.; Diverio, D.; Bernard, L.; Tizzoni, L.; et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood 2005, 106, 899–902. [Google Scholar] [CrossRef]
- Handschuh, L.; Kazmierczak, M.; Milewski, M.C.; Goralski, M.; Luczak, M.; Wojtaszewska, M.; Uszczynska-Ratajczak, B.; Lewandowski, K.; Komarnicki, M.; Figlerowicz, M. Gene expression profiling of acute myeloid leukemia samples from adult patients with AML-M1 and -M2 through boutique microarrays, real-time PCR and droplet digital PCR. Int. J. Oncol. 2018, 52, 656–678. [Google Scholar] [CrossRef]
- Luczak, M.; Kazmierczak, M.; Handschuh, L.; Lewandowski, K.; Komarnicki, M.; Figlerowicz, M. Comparative proteome analysis of acute myeloid leukemia with and without maturation. J. Proteomics 2012, 75, 5734–5748. [Google Scholar] [CrossRef]
- Casado, P.; Cutillas, P.R. Proteomic Characterization of Acute Myeloid Leukemia for Precision Medicine. Mol. Cell. Proteom. 2023, 22, 100517. [Google Scholar] [CrossRef] [PubMed]
- Casado, P.; Wilkes, E.H.; Miraki-Moud, F.; Hadi, M.M.; Rio-Machin, A.; Rajeeve, V.; Pike, R.; Iqbal, S.; Marfa, S.; Lea, N.; et al. Proteomic and genomic integration identifies kinase and differentiation determinants of kinase inhibitor sensitivity in leukemia cells. Leukemia 2018, 32, 1818–1822. [Google Scholar] [CrossRef] [PubMed]
- Hijazi, M.; Smith, R.; Rajeeve, V.; Bessant, C.; Cutillas, P.R. Reconstructing kinase network topologies from phosphoproteomics data reveals cancer-associated rewiring. Nat. Biotechnol. 2020, 38, 493–502. [Google Scholar] [CrossRef]
- Hernandez-Valladares, M.; Wangen, R.; Aasebo, E.; Reikvam, H.; Berven, F.S.; Selheim, F.; Bruserud, O. Proteomic Studies of Primary Acute Myeloid Leukemia Cells Derived from Patients before and during Disease-Stabilizing Treatment Based on All-Trans Retinoic Acid and Valproic Acid. Cancers 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Aasebo, E.; Brenner, A.K.; Hernandez-Valladares, M.; Birkeland, E.; Berven, F.S.; Selheim, F.; Bruserud, O. Proteomic Comparison of Bone Marrow Derived Osteoblasts and Mesenchymal Stem Cells. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef]
- Aasebo, E.; Brenner, A.K.; Birkeland, E.; Tvedt, T.H.A.; Selheim, F.; Berven, F.S.; Bruserud, O. The Constitutive Extracellular Protein Release by Acute Myeloid Leukemia Cells-A Proteomic Study of Patient Heterogeneity and Its Modulation by Mesenchymal Stromal Cells. Cancers 2021, 13. [Google Scholar] [CrossRef]
- Jayavelu, A.K.; Wolf, S.; Buettner, F.; Alexe, G.; Haupl, B.; Comoglio, F.; Schneider, C.; Doebele, C.; Fuhrmann, D.C.; Wagner, S.; et al. The proteogenomic subtypes of acute myeloid leukemia. Cancer Cell 2022, 40, 301–317.e312. [Google Scholar] [CrossRef]
- Caplan, M.; Wittorf, K.J.; Weber, K.K.; Swenson, S.A.; Gilbreath, T.J.; Willow Hynes-Smith, R.; Amador, C.; Hyde, R.K.; Buckley, S.M. Multi-omics reveals mitochondrial metabolism proteins susceptible for drug discovery in AML. Leukemia 2022, 36, 1296–1305. [Google Scholar] [CrossRef]
- Kramer, M.H.; Zhang, Q.; Sprung, R.; Day, R.B.; Erdmann-Gilmore, P.; Li, Y.; Xu, Z.; Helton, N.M.; George, D.R.; Mi, Y.; et al. Proteomic and phosphoproteomic landscapes of acute myeloid leukemia. Blood 2022, 140, 1533–1548. [Google Scholar] [CrossRef]
- Stratmann, S.; Vesterlund, M.; Umer, H.M.; Eshtad, S.; Skaftason, A.; Herlin, M.K.; Sundstrom, C.; Eriksson, A.; Hoglund, M.; Palle, J.; et al. Proteogenomic analysis of acute myeloid leukemia associates relapsed disease with reprogrammed energy metabolism both in adults and children. Leukemia 2023, 37, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Emdal, K.B.; Palacio-Escat, N.; Wigerup, C.; Eguchi, A.; Nilsson, H.; Bekker-Jensen, D.B.; Ronnstrand, L.; Kazi, J.U.; Puissant, A.; Itzykson, R.; et al. Phosphoproteomics of primary AML patient samples reveals rationale for AKT combination therapy and p53 context to overcome selinexor resistance. Cell Rep. 2022, 40, 111177. [Google Scholar] [CrossRef]
- Aasebo, E.; Berven, F.S.; Bartaula-Brevik, S.; Stokowy, T.; Hovland, R.; Vaudel, M.; Doskeland, S.O.; McCormack, E.; Batth, T.S.; Olsen, J.V.; et al. Proteome and Phosphoproteome Changes Associated with Prognosis in Acute Myeloid Leukemia. Cancers 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Aasebo, E.; Vaudel, M.; Mjaavatten, O.; Gausdal, G.; Van der Burgh, A.; Gjertsen, B.T.; Doskeland, S.O.; Bruserud, O.; Berven, F.S.; Selheim, F. Performance of super-SILAC based quantitative proteomics for comparison of different acute myeloid leukemia (AML) cell lines. Proteomics 2014, 14, 1971–1976. [Google Scholar] [CrossRef]
- Hernandez-Valladares, M.; Aasebo, E.; Selheim, F.; Berven, F.S.; Bruserud, O. Selecting Sample Preparation Workflows for Mass Spectrometry-Based Proteomic and Phosphoproteomic Analysis of Patient Samples with Acute Myeloid Leukemia. Proteomes 2016, 4. [Google Scholar] [CrossRef]
- Vardiman, J.W.; Thiele, J.; Arber, D.A.; Brunning, R.D.; Borowitz, M.J.; Porwit, A.; Harris, N.L.; Le Beau, M.M.; Hellstrom-Lindberg, E.; Tefferi, A.; et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood 2009, 114, 937–951. [Google Scholar] [CrossRef]
- Kuusanmaki, H.; Leppa, A.M.; Polonen, P.; Kontro, M.; Dufva, O.; Deb, D.; Yadav, B.; Bruck, O.; Kumar, A.; Everaus, H.; et al. Phenotype-based drug screening reveals association between venetoclax response and differentiation stage in acute myeloid leukemia. Haematologica 2020, 105, 708–720. [Google Scholar] [CrossRef] [PubMed]
- Pei, S.; Pollyea, D.A.; Gustafson, A.; Stevens, B.M.; Minhajuddin, M.; Fu, R.; Riemondy, K.A.; Gillen, A.E.; Sheridan, R.M.; Kim, J.; et al. Monocytic Subclones Confer Resistance to Venetoclax-Based Therapy in Patients with Acute Myeloid Leukemia. Cancer Discov. 2020, 10, 536–551. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Nepusz, T.; Yu, H.; Paccanaro, A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat. Methods 2012, 9, 471–472. [Google Scholar] [CrossRef]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef]
- Hulsen, T.; de Vlieg, J.; Alkema, W. BioVenn—A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genom. 2008, 9, 488. [Google Scholar] [CrossRef] [PubMed]
- Nolfi-Donegan, D.; Braganza, A.; Shiva, S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. 2020, 37, 101674. [Google Scholar] [CrossRef]
- Ashton, T.M.; McKenna, W.G.; Kunz-Schughart, L.A.; Higgins, G.S. Oxidative Phosphorylation as an Emerging Target in Cancer Therapy. Clin. Cancer Res. 2018, 24, 2482–2490. [Google Scholar] [CrossRef]
- Papa, S.; Martino, P.L.; Capitanio, G.; Gaballo, A.; De Rasmo, D.; Signorile, A.; Petruzzella, V. The oxidative phosphorylation system in mammalian mitochondria. Adv. Exp. Med. Biol. 2012, 942, 3–37. [Google Scholar] [CrossRef]
- Walter, R.B.; Othus, M.; Burnett, A.K.; Lowenberg, B.; Kantarjian, H.M.; Ossenkoppele, G.J.; Hills, R.K.; van Montfort, K.G.; Ravandi, F.; Evans, A.; et al. Significance of FAB subclassification of “acute myeloid leukemia, NOS” in the 2008 WHO classification: Analysis of 5848 newly diagnosed patients. Blood 2013, 121, 2424–2431. [Google Scholar] [CrossRef]
- Canaani, J.; Beohou, E.; Labopin, M.; Socie, G.; Huynh, A.; Volin, L.; Cornelissen, J.; Milpied, N.; Gedde-Dahl, T.; Deconinck, E.; et al. Impact of FAB classification on predicting outcome in acute myeloid leukemia, not otherwise specified, patients undergoing allogeneic stem cell transplantation in CR1: An analysis of 1690 patients from the acute leukemia working party of EBMT. Am. J. Hematol. 2017, 92, 344–350. [Google Scholar] [CrossRef]
- Bostrom, B.; Brunning, R.D.; McGlave, P.; Ramsay, N.; Nesbit, M., Jr.; Woods, W.G.; Hurd, D.; Krivit, W.; Kim, T.; Goldman, A.; et al. Bone marrow transplantation for acute nonlymphocytic leukemia in first remission: Analysis of prognostic factors. Blood 1985, 65, 1191–1196. [Google Scholar] [CrossRef]
- Saultz, J.N.; Tyner, J.W. Chasing leukemia differentiation through induction therapy, relapse and transplantation. Blood Rev. 2023, 57, 101000. [Google Scholar] [CrossRef]
- Norgaard, J.M.; Olesen, L.H.; Olesen, G.; Meyer, K.; Kristensen, J.S.; Bendix, K.; Pedersen, B.; Kjeldsen, E.; Hokland, P. FAB M4 and high CD14 surface expression is associated with high cellular resistance to Ara-C and daunorubicin: Implications for clinical outcome in acute myeloid leukaemia. Eur. J. Haematol. 2001, 67, 221–229. [Google Scholar] [CrossRef]
- Wojtuszkiewicz, A.; van der Werf, I.; Hutter, S.; Walter, W.; Baer, C.; Kern, W.; Janssen, J.; Ossenkoppele, G.J.; Haferlach, C.; Cloos, J.; et al. Maturation State-Specific Alternative Splicing in FLT3-ITD and NPM1 Mutated AML. Cancers 2021, 13. [Google Scholar] [CrossRef]
- Alfayez, M.; Issa, G.C.; Patel, K.P.; Wang, F.; Wang, X.; Short, N.J.; Cortes, J.E.; Kadia, T.; Ravandi, F.; Pierce, S.; et al. The Clinical impact of PTPN11 mutations in adults with acute myeloid leukemia. Leukemia 2021, 35, 691–700. [Google Scholar] [CrossRef]
- Cheng, Z.; Hu, K.; Tian, L.; Dai, Y.; Pang, Y.; Cui, W.; Zhao, H.; Qin, T.; Han, Y.; Hu, N.; et al. Clinical and biological implications of mutational spectrum in acute myeloid leukemia of FAB subtypes M4 and M5. Cancer Gene Ther. 2018, 25, 77–83. [Google Scholar] [CrossRef]
- Miyajima, T.; Onozawa, M.; Yoshida, S.; Miyashita, N.; Kimura, H.; Takahashi, S.; Yokoyama, S.; Matsukawa, T.; Goto, H.; Sugita, J.; et al. Clinical implications of NUP98::NSD1 fusion at diagnosis in adult FLT3-ITD positive AML. Eur. J. Haematol. 2023, 111, 620–627. [Google Scholar] [CrossRef]
- Sano, H.; Shimada, A.; Taki, T.; Murata, C.; Park, M.J.; Sotomatsu, M.; Tabuchi, K.; Tawa, A.; Kobayashi, R.; Horibe, K.; et al. RAS mutations are frequent in FAB type M4 and M5 of acute myeloid leukemia, and related to late relapse: A study of the Japanese Childhood AML Cooperative Study Group. Int. J. Hematol. 2012, 95, 509–515. [Google Scholar] [CrossRef]
- Kornblau, S.M.; Tibes, R.; Qiu, Y.H.; Chen, W.; Kantarjian, H.M.; Andreeff, M.; Coombes, K.R.; Mills, G.B. Functional proteomic profiling of AML predicts response and survival. Blood 2009, 113, 154–164. [Google Scholar] [CrossRef]
- Aasebo, E.; Forthun, R.B.; Berven, F.; Selheim, F.; Hernandez-Valladares, M. Global Cell Proteome Profiling, Phospho-signaling and Quantitative Proteomics for Identification of New Biomarkers in Acute Myeloid Leukemia Patients. Curr. Pharm. Biotechnol. 2016, 17, 52–70. [Google Scholar] [CrossRef]
- Majumder, M.M.; Leppa, A.M.; Hellesoy, M.; Dowling, P.; Malyutina, A.; Kopperud, R.; Bazou, D.; Andersson, E.; Parsons, A.; Tang, J.; et al. Multi-parametric single cell evaluation defines distinct drug responses in healthy hematologic cells that are retained in corresponding malignant cell types. Haematologica 2020, 105, 1527–1538. [Google Scholar] [CrossRef]
- Attenhofer, C.; Vuilliomenet, A.; Richter, M.; Kaufmann, U.; Metzger, U.; Bertel, O. [Heart contusions: Pathological findings and clinical course]. Schweiz. Med. Wochenschr. 1992, 122, 1593–1599. [Google Scholar]
- Rundgren, I.M.; Ryningen, A.; Anderson Tvedt, T.H.; Bruserud, O.; Ersvaer, E. Immunomodulatory Drugs Alter the Metabolism and the Extracellular Release of Soluble Mediators by Normal Monocytes. Molecules 2020, 25. [Google Scholar] [CrossRef]
- Miari, K.E.; Guzman, M.L.; Wheadon, H.; Williams, M.T.S. Macrophages in Acute Myeloid Leukaemia: Significant Players in Therapy Resistance and Patient Outcomes. Front. Cell. Dev. Biol. 2021, 9, 692800. [Google Scholar] [CrossRef]
- Chew, A.; Buck, E.A.; Peretz, S.; Sirugo, G.; Rinaldo, P.; Isaya, G. Cloning, expression, and chromosomal assignment of the human mitochondrial intermediate peptidase gene (MIPEP). Genomics 1997, 40, 493–496. [Google Scholar] [CrossRef]
- Ogilvie, I.; Kennaway, N.G.; Shoubridge, E.A. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Investig. 2005, 115, 2784–2792. [Google Scholar] [CrossRef]
- Basak, N.P.; Banerjee, S. Mitochondrial dependency in progression of acute myeloid leukemia. Mitochondrion 2015, 21, 41–48. [Google Scholar] [CrossRef]
- Cherry, E.M.; Abbott, D.; Amaya, M.; McMahon, C.; Schwartz, M.; Rosser, J.; Sato, A.; Schowinsky, J.; Inguva, A.; Minhajuddin, M.; et al. Venetoclax and azacitidine compared with induction chemotherapy for newly diagnosed patients with acute myeloid leukemia. Blood Adv. 2021, 5, 5565–5573. [Google Scholar] [CrossRef]
- Pollyea, D.A.; Stevens, B.M.; Jones, C.L.; Winters, A.; Pei, S.; Minhajuddin, M.; D’Alessandro, A.; Culp-Hill, R.; Riemondy, K.A.; Gillen, A.E.; et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat. Med. 2018, 24, 1859–1866. [Google Scholar] [CrossRef]
- Tong, X.; Zhou, F. Integrated bioinformatic analysis of mitochondrial metabolism-related genes in acute myeloid leukemia. Front. Immunol. 2023, 14, 1120670. [Google Scholar] [CrossRef]
- Wu, S.; Akhtari, M.; Alachkar, H. Characterization of Mutations in the Mitochondrial Encoded Electron Transport Chain Complexes in Acute Myeloid Leukemia. Sci. Rep. 2018, 8, 13301. [Google Scholar] [CrossRef]
- Damm, F.; Bunke, T.; Thol, F.; Markus, B.; Wagner, K.; Gohring, G.; Schlegelberger, B.; Heil, G.; Reuter, C.W.; Pullmann, K.; et al. Prognostic implications and molecular associations of NADH dehydrogenase subunit 4 (ND4) mutations in acute myeloid leukemia. Leukemia 2012, 26, 289–295. [Google Scholar] [CrossRef]
- Romine, K.A.; Nechiporuk, T.; Bottomly, D.; Jeng, S.; McWeeney, S.K.; Kaempf, A.; Corces, M.R.; Majeti, R.; Tyner, J.W. Monocytic differentiation and AHR signaling as Primary Nodes of BET Inhibitor Response in Acute Myeloid Leukemia. Blood Cancer Discov. 2021, 2, 518–531. [Google Scholar] [CrossRef]
- Aasebo, E.; Berven, F.S.; Hovland, R.; Doskeland, S.O.; Bruserud, O.; Selheim, F.; Hernandez-Valladares, M. The Progression of Acute Myeloid Leukemia from First Diagnosis to Chemoresistant Relapse: A Comparison of Proteomic and Phosphoproteomic Profiles. Cancers 2020, 12. [Google Scholar] [CrossRef]
 ), REL_M1/2_all vs REL_M4/5_mut (
), REL_M1/2_all vs REL_M4/5_mut ( ) and REL_M1/2_all vs REL_M4/5_CN (
) and REL_M1/2_all vs REL_M4/5_CN ( ) comparisons. (b) Reactome pathways and (c) protein-protein interaction (PPI) network analyses of comparison-specific (61 and 25) and comparison-overlapping (78 and 22) regulated proteins.
) comparisons. (b) Reactome pathways and (c) protein-protein interaction (PPI) network analyses of comparison-specific (61 and 25) and comparison-overlapping (78 and 22) regulated proteins.
 ), REL_M1/2_all vs REL_M4/5_mut (
), REL_M1/2_all vs REL_M4/5_mut ( ) and REL_M1/2_all vs REL_M4/5_CN (
) and REL_M1/2_all vs REL_M4/5_CN ( ) comparisons. (b) Reactome pathways and (c) protein-protein interaction (PPI) network analyses of comparison-specific (61 and 25) and comparison-overlapping (78 and 22) regulated proteins.
) comparisons. (b) Reactome pathways and (c) protein-protein interaction (PPI) network analyses of comparison-specific (61 and 25) and comparison-overlapping (78 and 22) regulated proteins.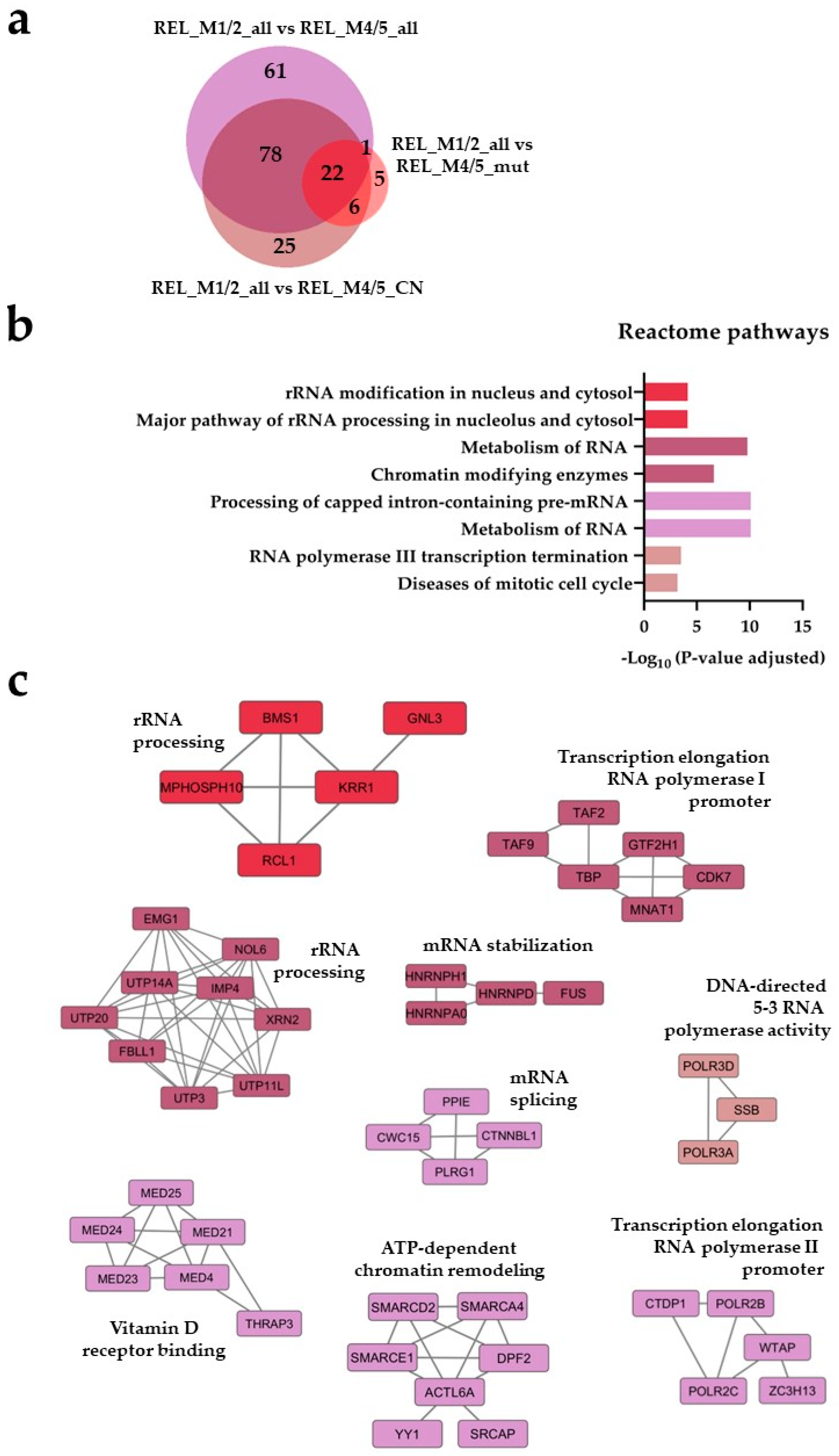
 ), REL_M4/5_mut vs REL_M1/2_all (
), REL_M4/5_mut vs REL_M1/2_all ( ) and REL_M4/5_CN vs REL_M1/2_all (
) and REL_M4/5_CN vs REL_M1/2_all ( ) comparisons. (b) Reactome pathways and (c) PPI network analyses of comparison-specific (30, 37 and 68) and comparison-overlapping (70, 166 and 78) regulated proteins. * It stands for Reactome pathways with unadjusted P-value < 0.05.
) comparisons. (b) Reactome pathways and (c) PPI network analyses of comparison-specific (30, 37 and 68) and comparison-overlapping (70, 166 and 78) regulated proteins. * It stands for Reactome pathways with unadjusted P-value < 0.05.
 ), REL_M4/5_mut vs REL_M1/2_all (
), REL_M4/5_mut vs REL_M1/2_all ( ) and REL_M4/5_CN vs REL_M1/2_all (
) and REL_M4/5_CN vs REL_M1/2_all ( ) comparisons. (b) Reactome pathways and (c) PPI network analyses of comparison-specific (30, 37 and 68) and comparison-overlapping (70, 166 and 78) regulated proteins. * It stands for Reactome pathways with unadjusted P-value < 0.05.
) comparisons. (b) Reactome pathways and (c) PPI network analyses of comparison-specific (30, 37 and 68) and comparison-overlapping (70, 166 and 78) regulated proteins. * It stands for Reactome pathways with unadjusted P-value < 0.05.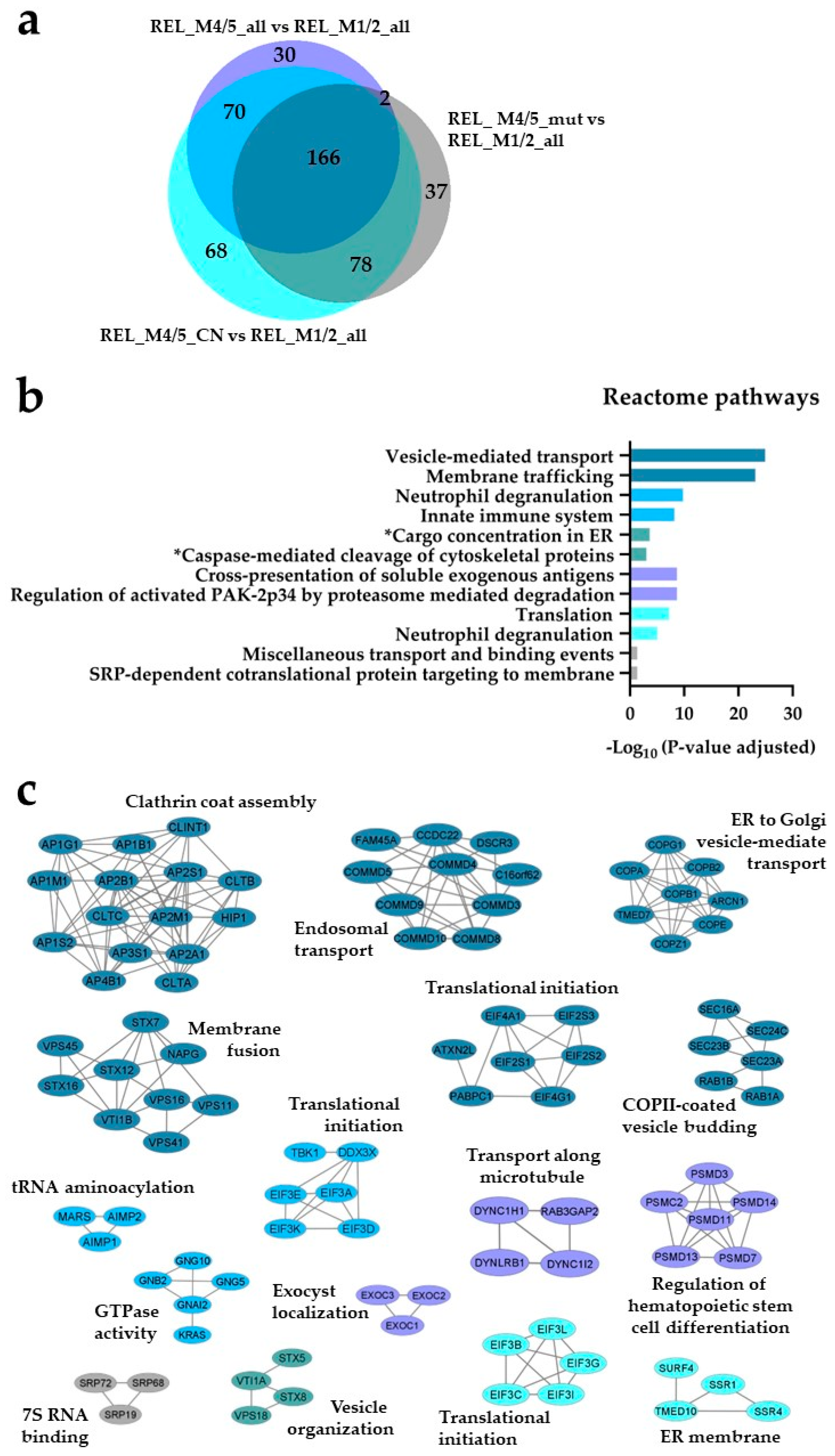
 ), REL_M1/2_all vs REL_M4/5_mut (
), REL_M1/2_all vs REL_M4/5_mut ( ) and REL_M1/2_all vs REL_M4/5_CN (
) and REL_M1/2_all vs REL_M4/5_CN ( ) comparisons. (b) Gene ontology (GO) with molecular function terms and KEGG pathway analyses of comparison-specific (16) and comparison-overlapping (11 and 13) differentially phosphorylated proteins. (c) Sequence motif analysis of the ±5 amino acids flanking the differentially regulated phosphorylation sites from the comparison-specific (16) and comparison-overlapping (11 and 13) datasets.
) comparisons. (b) Gene ontology (GO) with molecular function terms and KEGG pathway analyses of comparison-specific (16) and comparison-overlapping (11 and 13) differentially phosphorylated proteins. (c) Sequence motif analysis of the ±5 amino acids flanking the differentially regulated phosphorylation sites from the comparison-specific (16) and comparison-overlapping (11 and 13) datasets.
 ), REL_M1/2_all vs REL_M4/5_mut (
), REL_M1/2_all vs REL_M4/5_mut ( ) and REL_M1/2_all vs REL_M4/5_CN (
) and REL_M1/2_all vs REL_M4/5_CN ( ) comparisons. (b) Gene ontology (GO) with molecular function terms and KEGG pathway analyses of comparison-specific (16) and comparison-overlapping (11 and 13) differentially phosphorylated proteins. (c) Sequence motif analysis of the ±5 amino acids flanking the differentially regulated phosphorylation sites from the comparison-specific (16) and comparison-overlapping (11 and 13) datasets.
) comparisons. (b) Gene ontology (GO) with molecular function terms and KEGG pathway analyses of comparison-specific (16) and comparison-overlapping (11 and 13) differentially phosphorylated proteins. (c) Sequence motif analysis of the ±5 amino acids flanking the differentially regulated phosphorylation sites from the comparison-specific (16) and comparison-overlapping (11 and 13) datasets.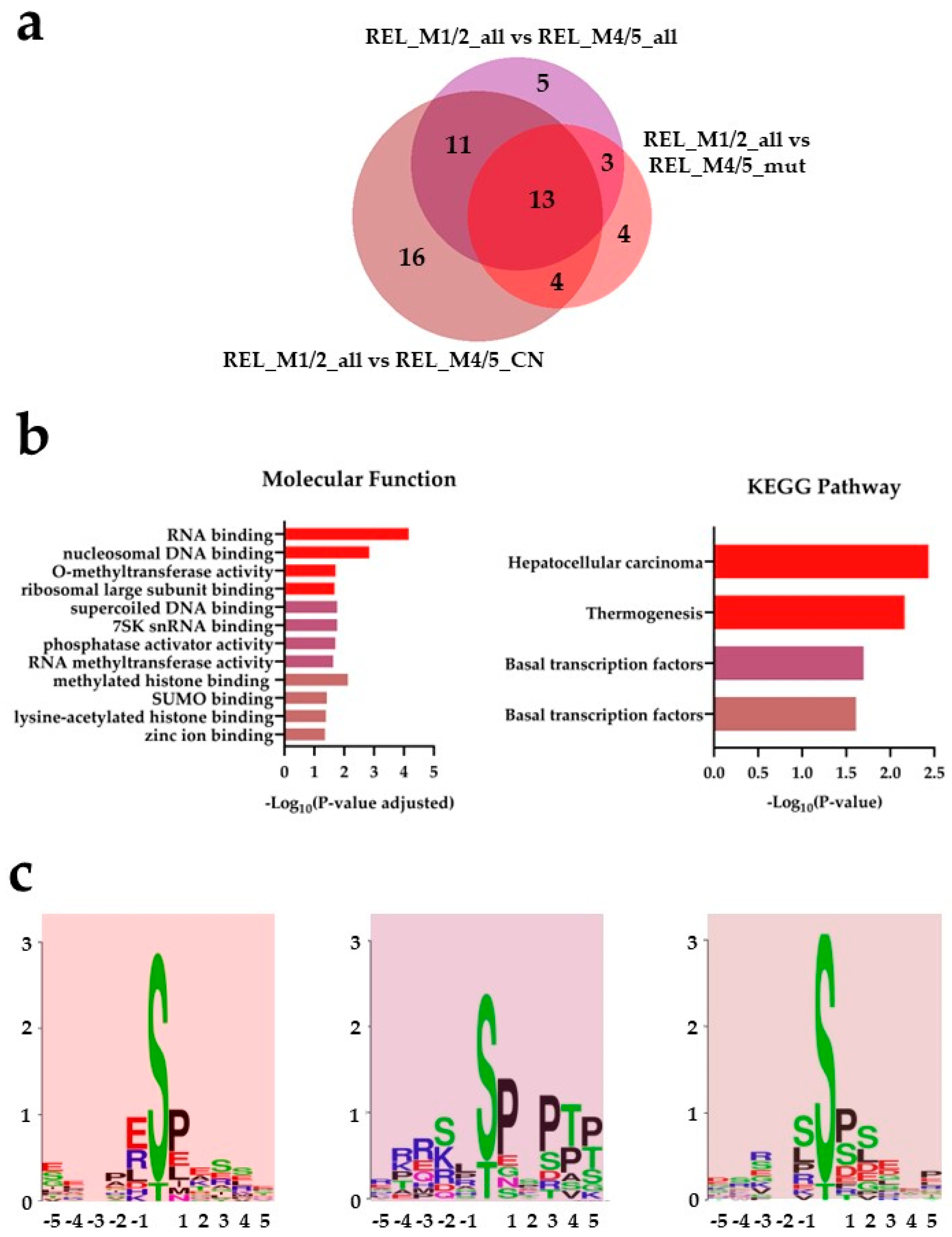
 ), REL_M4/5_mut vs REL_M1/2_all (
), REL_M4/5_mut vs REL_M1/2_all ( ) and REL_M4/5_CN vs REL_M1/2_all (
) and REL_M4/5_CN vs REL_M1/2_all ( ) comparisons. (b) GO with molecular function terms and KEGG pathway analyses of comparison-specific (22, 12 and 27) and comparison-overlapping (42 and 24) differentially phosphorylated proteins. (c) PPI network analyses of overlapping (42 and 24) differentially phosphorylated proteins. (d) Sequence motif analysis of the ± 5 amino acids flanking the differentially regulated phosphorylation sites from the comparison-specific (22, 12 and 27) and comparison-overlapping (42 and 24) datasets. * A shorter name for “Regulation of lipolysis in adipocytes” KEGG pathway is added for space purposes.
) comparisons. (b) GO with molecular function terms and KEGG pathway analyses of comparison-specific (22, 12 and 27) and comparison-overlapping (42 and 24) differentially phosphorylated proteins. (c) PPI network analyses of overlapping (42 and 24) differentially phosphorylated proteins. (d) Sequence motif analysis of the ± 5 amino acids flanking the differentially regulated phosphorylation sites from the comparison-specific (22, 12 and 27) and comparison-overlapping (42 and 24) datasets. * A shorter name for “Regulation of lipolysis in adipocytes” KEGG pathway is added for space purposes.
 ), REL_M4/5_mut vs REL_M1/2_all (
), REL_M4/5_mut vs REL_M1/2_all ( ) and REL_M4/5_CN vs REL_M1/2_all (
) and REL_M4/5_CN vs REL_M1/2_all ( ) comparisons. (b) GO with molecular function terms and KEGG pathway analyses of comparison-specific (22, 12 and 27) and comparison-overlapping (42 and 24) differentially phosphorylated proteins. (c) PPI network analyses of overlapping (42 and 24) differentially phosphorylated proteins. (d) Sequence motif analysis of the ± 5 amino acids flanking the differentially regulated phosphorylation sites from the comparison-specific (22, 12 and 27) and comparison-overlapping (42 and 24) datasets. * A shorter name for “Regulation of lipolysis in adipocytes” KEGG pathway is added for space purposes.
) comparisons. (b) GO with molecular function terms and KEGG pathway analyses of comparison-specific (22, 12 and 27) and comparison-overlapping (42 and 24) differentially phosphorylated proteins. (c) PPI network analyses of overlapping (42 and 24) differentially phosphorylated proteins. (d) Sequence motif analysis of the ± 5 amino acids flanking the differentially regulated phosphorylation sites from the comparison-specific (22, 12 and 27) and comparison-overlapping (42 and 24) datasets. * A shorter name for “Regulation of lipolysis in adipocytes” KEGG pathway is added for space purposes.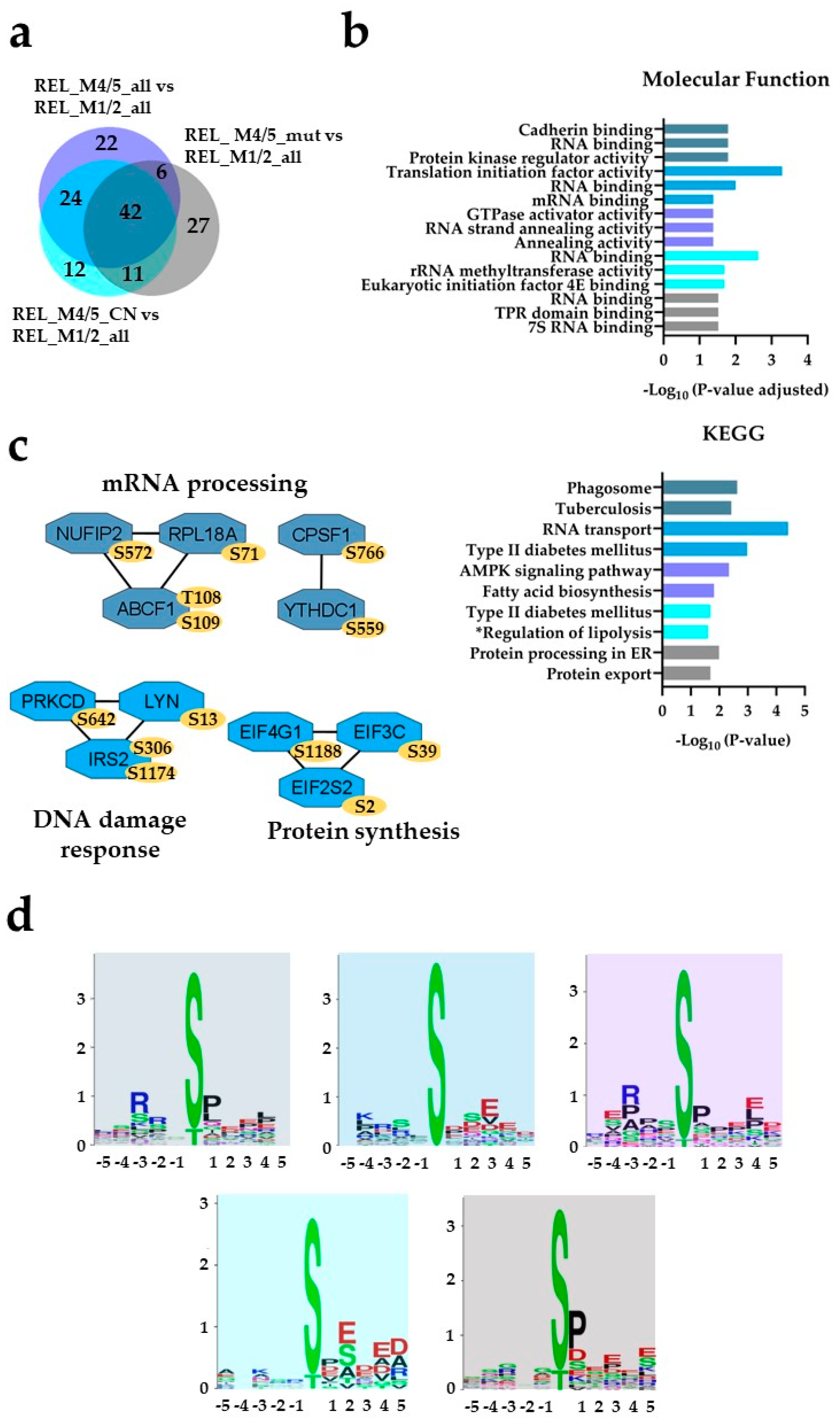
 ), REL_M4/5_mut vs REL_F_M4/5_mut (
), REL_M4/5_mut vs REL_F_M4/5_mut ( ) and REL_M4/5_CN vs REL_F_M4/5_CN (
) and REL_M4/5_CN vs REL_F_M4/5_CN ( ) comparisons. (b) Reactome pathway and (c) PPI network analyses of comparison-specific (119) and comparison-overlapping (15, 7 and 4) regulated proteins.
) comparisons. (b) Reactome pathway and (c) PPI network analyses of comparison-specific (119) and comparison-overlapping (15, 7 and 4) regulated proteins.
 ), REL_M4/5_mut vs REL_F_M4/5_mut (
), REL_M4/5_mut vs REL_F_M4/5_mut ( ) and REL_M4/5_CN vs REL_F_M4/5_CN (
) and REL_M4/5_CN vs REL_F_M4/5_CN ( ) comparisons. (b) Reactome pathway and (c) PPI network analyses of comparison-specific (119) and comparison-overlapping (15, 7 and 4) regulated proteins.
) comparisons. (b) Reactome pathway and (c) PPI network analyses of comparison-specific (119) and comparison-overlapping (15, 7 and 4) regulated proteins.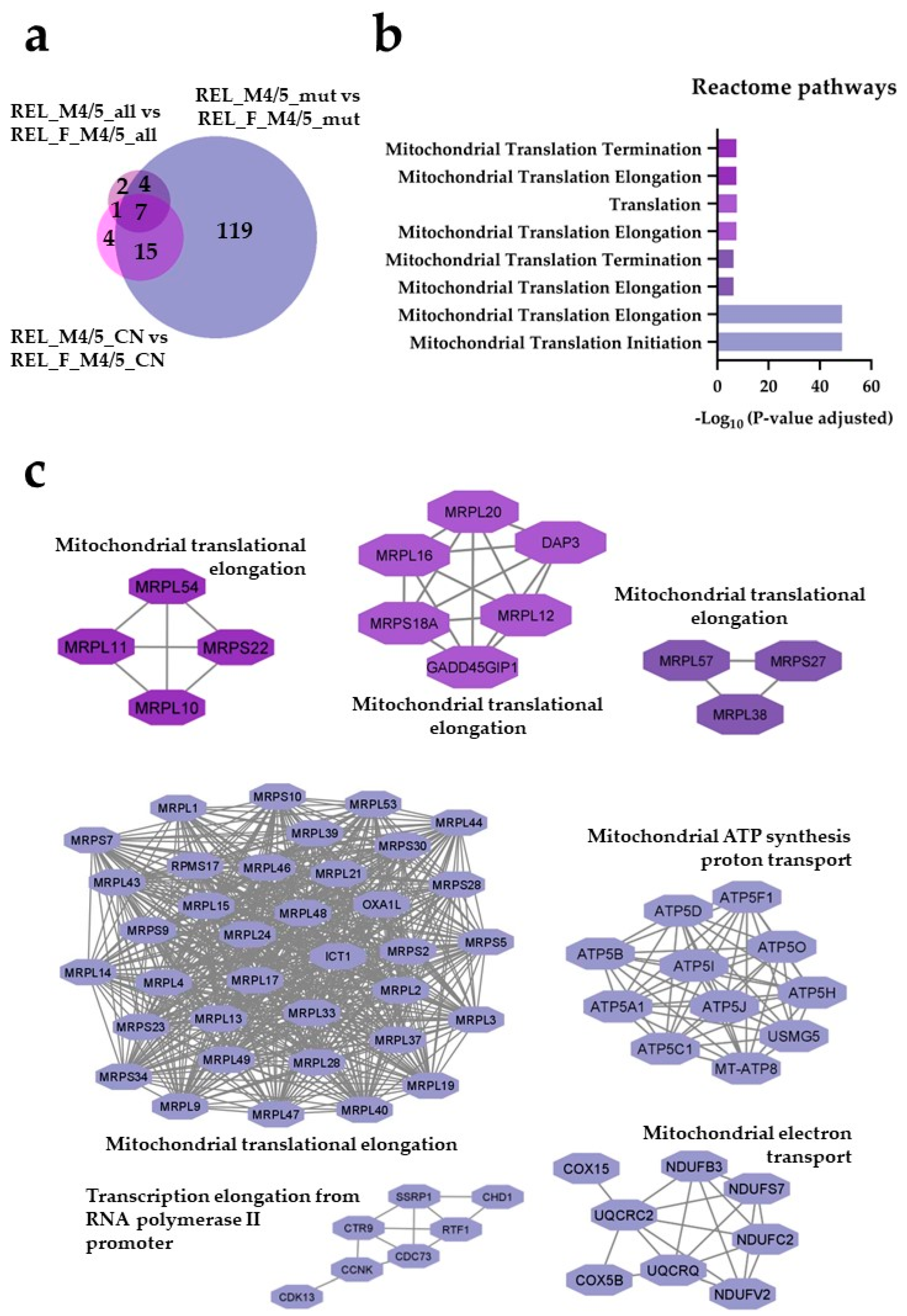
 ) and REL_F_M4/5_CN vs REL_M4/5_CN (
) and REL_F_M4/5_CN vs REL_M4/5_CN ( ) comparisons. (b) Reactome pathway and (c) PPI network analyses of comparison-specific (38 and 11) and comparison-overlapping (10) regulated proteins. * It stands for Reactome pathways with unadjusted P-value <0.05.
) comparisons. (b) Reactome pathway and (c) PPI network analyses of comparison-specific (38 and 11) and comparison-overlapping (10) regulated proteins. * It stands for Reactome pathways with unadjusted P-value <0.05.
 ) and REL_F_M4/5_CN vs REL_M4/5_CN (
) and REL_F_M4/5_CN vs REL_M4/5_CN ( ) comparisons. (b) Reactome pathway and (c) PPI network analyses of comparison-specific (38 and 11) and comparison-overlapping (10) regulated proteins. * It stands for Reactome pathways with unadjusted P-value <0.05.
) comparisons. (b) Reactome pathway and (c) PPI network analyses of comparison-specific (38 and 11) and comparison-overlapping (10) regulated proteins. * It stands for Reactome pathways with unadjusted P-value <0.05.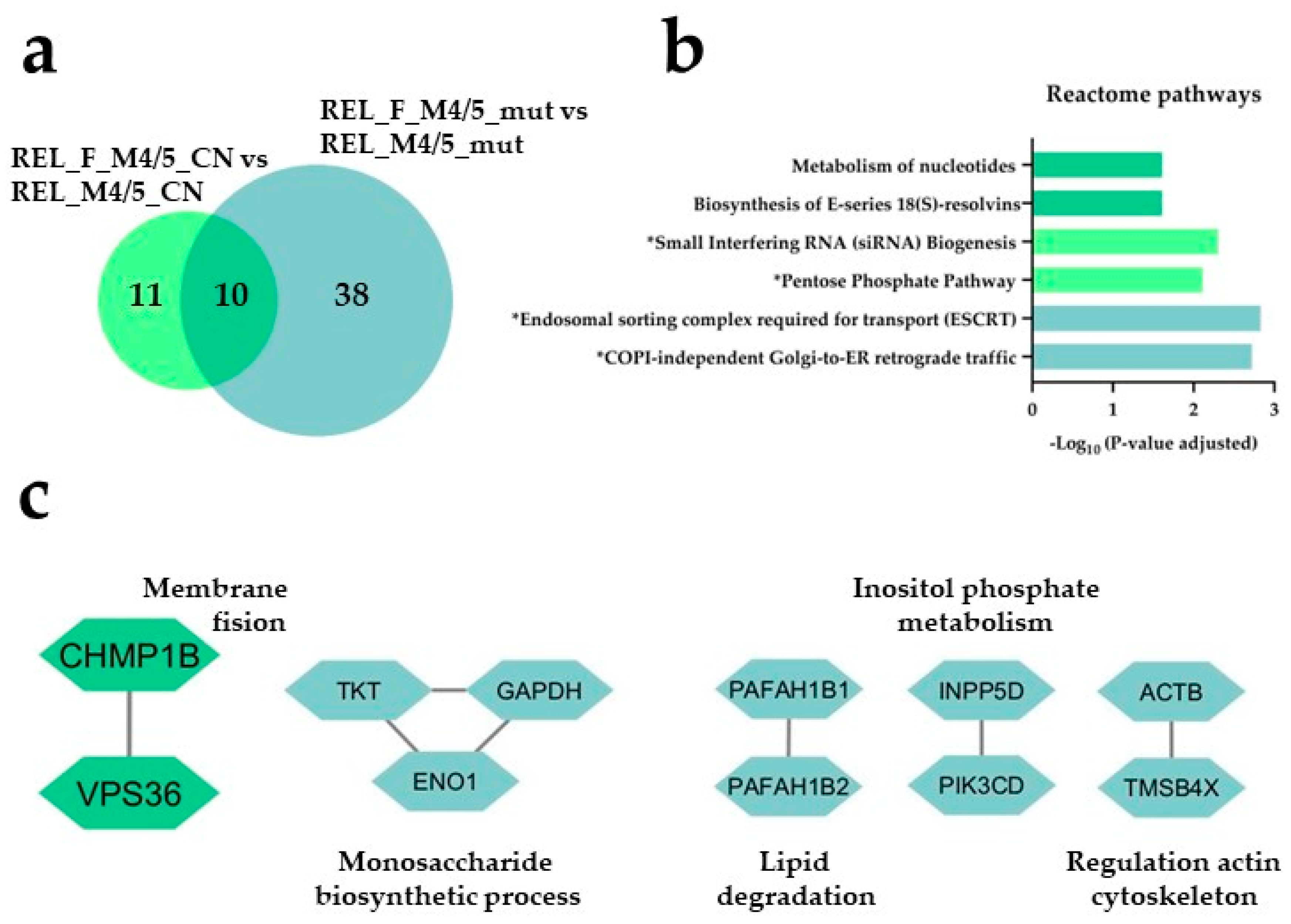
 ), REL_M4/5_mut vs REL_F_M4/5_mut (
), REL_M4/5_mut vs REL_F_M4/5_mut ( ) and REL_M4/5_CN vs REL_F_M4/5_CN (
) and REL_M4/5_CN vs REL_F_M4/5_CN ( ) comparisons. (b) GO with biological process (BP), molecular function (MF) and cellular compartment (CC) terms, KEGG and Reactome pathway analyses of 17 differentially higher phosphorylated proteins in the REL_M4/5_mut vs REL_F_M4/5_mut comparison. (c) Sequence motif analysis of the ± 5 amino acids flanking the differentially regulated phosphorylation sites in the REL_M4/5_mut vs REL_F_M4/5_mut comparison. * It stands for KEGG and Reactome pathways with unadjusted P-value < 0.05.
) comparisons. (b) GO with biological process (BP), molecular function (MF) and cellular compartment (CC) terms, KEGG and Reactome pathway analyses of 17 differentially higher phosphorylated proteins in the REL_M4/5_mut vs REL_F_M4/5_mut comparison. (c) Sequence motif analysis of the ± 5 amino acids flanking the differentially regulated phosphorylation sites in the REL_M4/5_mut vs REL_F_M4/5_mut comparison. * It stands for KEGG and Reactome pathways with unadjusted P-value < 0.05.
 ), REL_M4/5_mut vs REL_F_M4/5_mut (
), REL_M4/5_mut vs REL_F_M4/5_mut ( ) and REL_M4/5_CN vs REL_F_M4/5_CN (
) and REL_M4/5_CN vs REL_F_M4/5_CN ( ) comparisons. (b) GO with biological process (BP), molecular function (MF) and cellular compartment (CC) terms, KEGG and Reactome pathway analyses of 17 differentially higher phosphorylated proteins in the REL_M4/5_mut vs REL_F_M4/5_mut comparison. (c) Sequence motif analysis of the ± 5 amino acids flanking the differentially regulated phosphorylation sites in the REL_M4/5_mut vs REL_F_M4/5_mut comparison. * It stands for KEGG and Reactome pathways with unadjusted P-value < 0.05.
) comparisons. (b) GO with biological process (BP), molecular function (MF) and cellular compartment (CC) terms, KEGG and Reactome pathway analyses of 17 differentially higher phosphorylated proteins in the REL_M4/5_mut vs REL_F_M4/5_mut comparison. (c) Sequence motif analysis of the ± 5 amino acids flanking the differentially regulated phosphorylation sites in the REL_M4/5_mut vs REL_F_M4/5_mut comparison. * It stands for KEGG and Reactome pathways with unadjusted P-value < 0.05.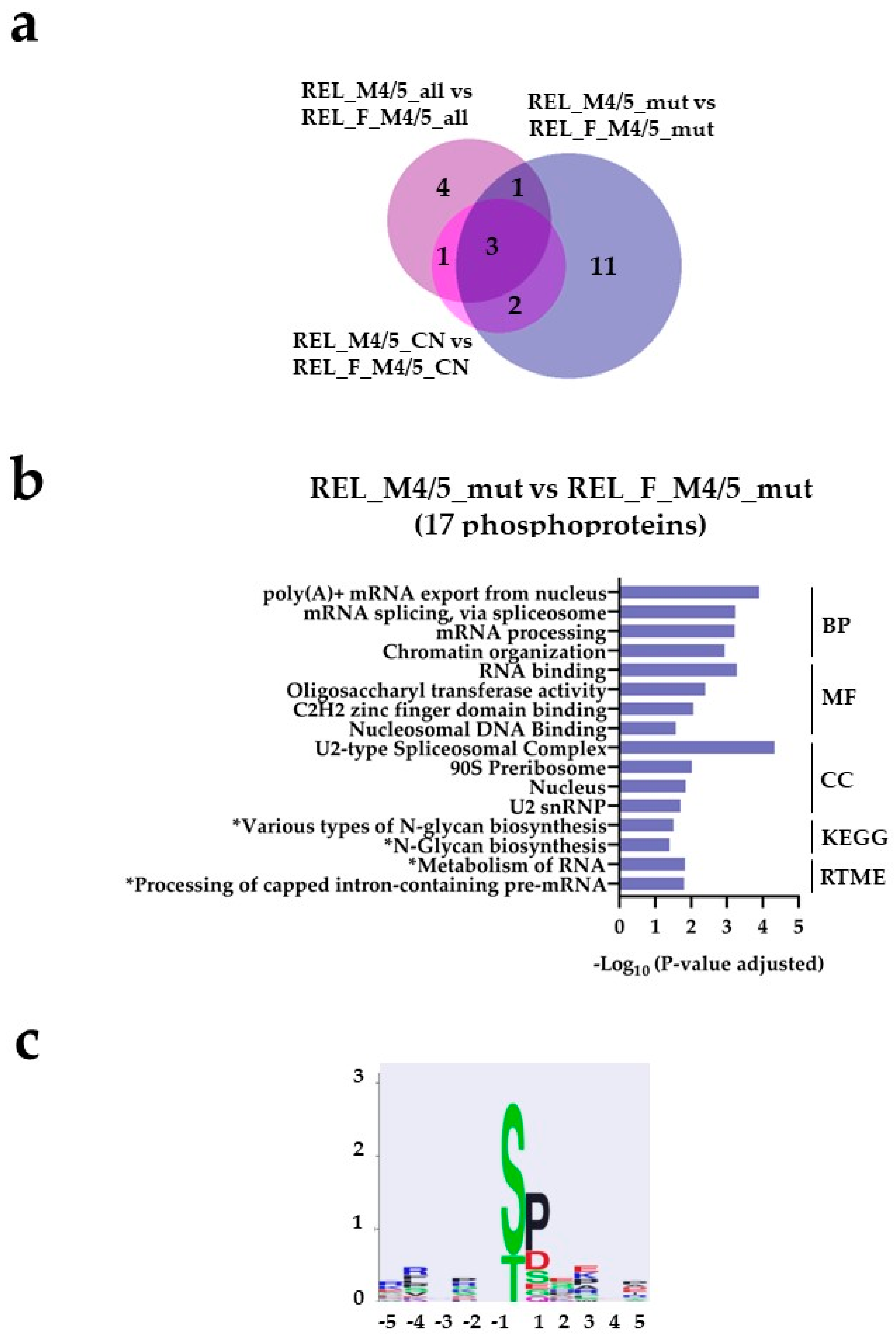
 ), REL_F_M4/5_mut vs REL_M4/5_mut (
), REL_F_M4/5_mut vs REL_M4/5_mut ( ) and REL_F_M4/5_CN vs REL_M4/5_CN (
) and REL_F_M4/5_CN vs REL_M4/5_CN ( ) comparisons. (b) GO with BP, MF and CC terms, KEGG and Reactome pathway analyses of 17 differentially higher phosphorylated proteins in the REL_F_M4/5_mut vs REL_M4/5_mut comparison. (c) Sequence motif analysis of the ± 5 amino acids flanking the differentially regulated phosphorylation sites in the REL_F_M4/5_mut subgroup when compared to REL_M4/5_mut patients.
) comparisons. (b) GO with BP, MF and CC terms, KEGG and Reactome pathway analyses of 17 differentially higher phosphorylated proteins in the REL_F_M4/5_mut vs REL_M4/5_mut comparison. (c) Sequence motif analysis of the ± 5 amino acids flanking the differentially regulated phosphorylation sites in the REL_F_M4/5_mut subgroup when compared to REL_M4/5_mut patients.
 ), REL_F_M4/5_mut vs REL_M4/5_mut (
), REL_F_M4/5_mut vs REL_M4/5_mut ( ) and REL_F_M4/5_CN vs REL_M4/5_CN (
) and REL_F_M4/5_CN vs REL_M4/5_CN ( ) comparisons. (b) GO with BP, MF and CC terms, KEGG and Reactome pathway analyses of 17 differentially higher phosphorylated proteins in the REL_F_M4/5_mut vs REL_M4/5_mut comparison. (c) Sequence motif analysis of the ± 5 amino acids flanking the differentially regulated phosphorylation sites in the REL_F_M4/5_mut subgroup when compared to REL_M4/5_mut patients.
) comparisons. (b) GO with BP, MF and CC terms, KEGG and Reactome pathway analyses of 17 differentially higher phosphorylated proteins in the REL_F_M4/5_mut vs REL_M4/5_mut comparison. (c) Sequence motif analysis of the ± 5 amino acids flanking the differentially regulated phosphorylation sites in the REL_F_M4/5_mut subgroup when compared to REL_M4/5_mut patients.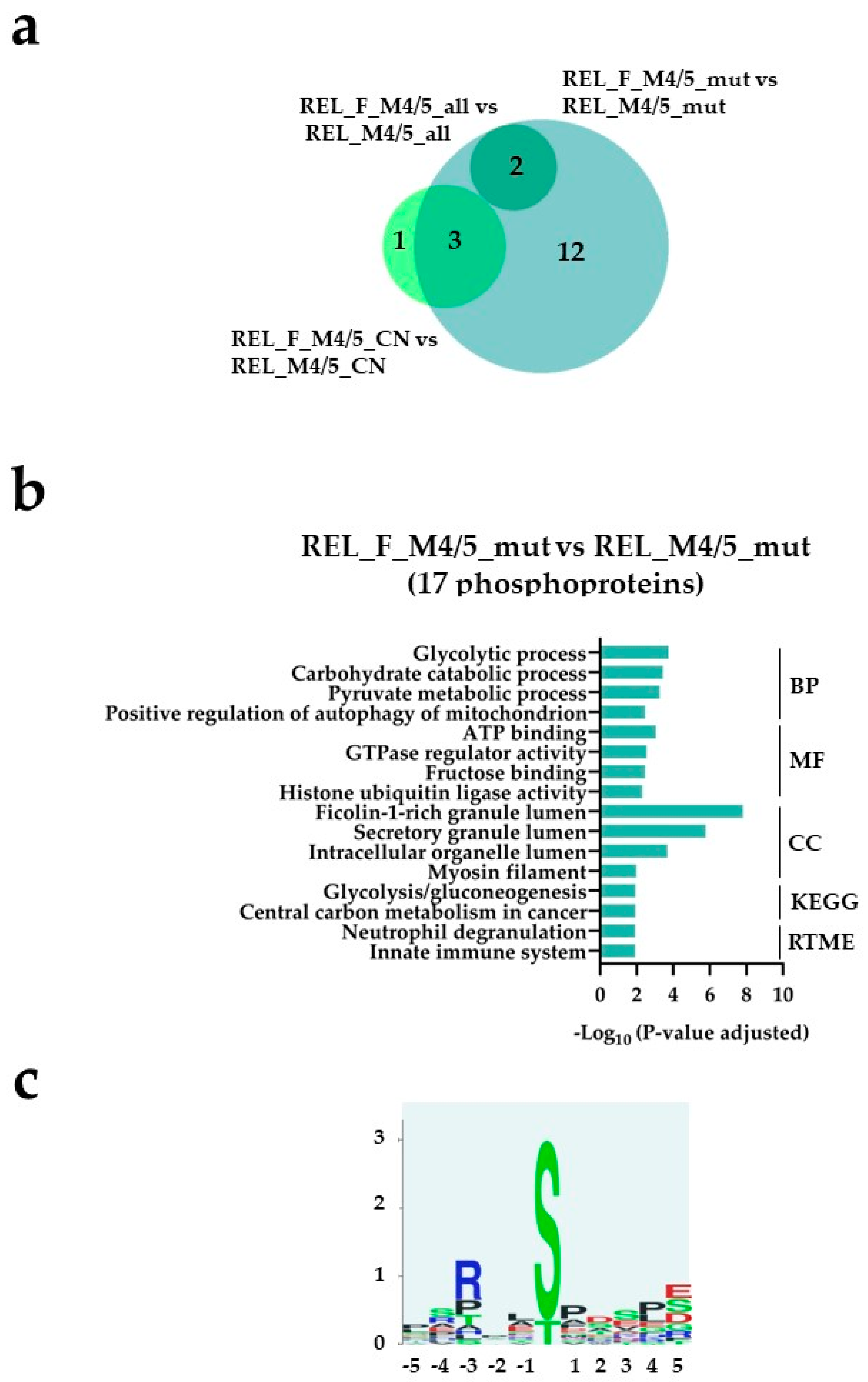
| Characteristic | RELAPSE | REL_FREE | |
|---|---|---|---|
| FAB classification | M1/M2 | 8 | 1 |
| M4/M5 | 12 | 14 | |
| NPM1 | WT | 6 | 6 |
| Ins | 5 | 8 | |
| CN | 46, XY or XX | 7 | 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





