Submitted:
16 October 2023
Posted:
18 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Experimental materials
2.2. Reagent preparation
2.3. MethylRAD experimental process
- (1)
- The enzyme digestion reaction system was as follows, a control group was set, and the reaction was performed at 37 ° C for 4 h.
- (2)
- 5 µl of each control group and enzyme digestion group were detected by 1% (wt/vol) agarose gel electrophoresis, 100 V electrophoresis for 10 - 15 min, The effect of digestion was observed under ultraviolet light.
- (3)
- The linking reaction system is as follows, and the reaction conditions are: 4 ℃ connection for 6-8 h.
- (4)
- The PCR reaction system and conditions is as follows.
- (5)
- 20 µl PCR product and 1 µl 100-bp DNA ladder were examined by electrophoresis with 8% polyacrylamide gel at 400V Swimming 35 mins
- (6)
- After electrophoresis, SYBR Safe DNA dye was used for 3 min to observe the brightness of the target band (100 bp).
- (7)
- Cut the desired strip and put it into a 1.5ml centrifuge tube, grind the glue with a grinding rod, add 30-40µl of pure water, and let it stand at 4°C 6-12 h.
- (8)
- PCR introduces the Barcode sequence and the PCR reaction system is as follows
- (9)
- Purify PCR products with QIAquick PCR Purification Kit, elute with 15 µl of pure water, and then determine with QubitQuantity. In general, the ideal concentration of purified products is 10 - 30 ng/µl.
- (10)
- If multiple libraries have been built, the libraries with different barcode numbers can be mixed according to the amount of sent measurement data, and finally mixed. The combined library concentration is more suitable at 5 - 10 ng/µl.
- (11)
- The mixed libraries were sequenced using the Illumina Novaseq PE150 sequencing platform.
| ingredient | Volume (μl/single sample; digestion group) | Volume (μl/single sample; control group) |
| DNA (1-200ng/μL) | 1 | 1 |
| 10×cut smart buffer | 1.5 | 1.5 |
| 30×Enzyme | 0.5 | 0.5 |
| activotor | ||
| FspEI (5U/μl) | 0.8 | 0 |
| Pure water | 11.2 | 12 |
| Total | 15 | 15 |
| ingredient | Volume (μl/single sample) |
| enzyme digestion product | 10 |
| 10× T4 ligase buffer | 1 |
| 10 m M ATP | 1 |
| Adaptor 1 (5µM) | 0.8 |
| Adaptor 2 (5µM)l | 0.8 |
| T4 DNA ligase (400 U/µl) | 2 |
| Pure water | 5.4 |
| Total | 20 |
| ingredient | Volume (μl/single sample) | Reaction conditions |
| Linked product | 7 | 98℃ ,5s; |
| 5×HF buffer | 4 | 60℃ ,20s |
| 10 Mm dNTP | 0.6 | 72℃ ,10s |
| Primer 1 (10 µM ) | 0.4 | 20 cycle |
| Primer 2 (10 µM ) | 0.4 | |
| Phusion high-fidelity DNA polymerase (2 U/µl) | 0.2 | |
| Pure water | 7.4 | |
| Total | 20 |
| ingredient | Volume (μl) | Reaction conditions |
| Linked product | 6 | 98℃ ,5s; |
| 5×HF buffer | 4 | 60℃ ,20s |
| 10 Mm dNTP | 0.6 | 72℃ ,10s |
| 10 μM Primer3 | 0.2 | 6 cycle |
| 10 μM Index Primer | 0.2 | |
| Phusion high-fidelity DNA polymerase (2 U/µl) | 0.2 | |
| Pure water | 8.8 | |
| Total | 20 |
| Adaptors and primers | Sequence (5’ to 3’) |
| Adap-1 sens | ACACTCTTTCCCTACACGACGCTCTTCCGATCT |
| Adap-1 antisense | NNNNAGATCGGAAGAGC(AminoC6) |
| Adap-2 sense | GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT |
| Adap-2 antisense | NNNNAGATCGGAAGAGC(AminoC6) |
| Primers | |
| Primer1 | ACACTCTTTCCCTACACGACGCT |
| Primer2 | GTGACTGGAGTTCAGACGTGTGCT |
| Primer3 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCT |
| Index primer | CAAGCAGAAGACGGCATACGAGATXXXXXXGTGACTGGAGTTCAGACGTGT |
2.4. Data analysis process
- (1)
- Check the raw data obtained from sequencing for quality. If there are more than 15% low-quality bases or sequences with too many N bases in the acquired reads, they must be removed.
- (2)
- Align Enzyme Reads to the reference genome using the bowtie2(version 2.3.4.1) software (parameter settings: -M = 4, –v = 2, –r = 0) to identify reliable methylation sites;
- (3)
- (4)
- Using the DESeq software, calculate the difference p value and difference multiple (Log2FC) of each site between the groups, combine the sequencing depth of each site in each sample, and compare the methylation levels between the two groups;
- (5)
- Screen the genes for which the difference between groups is p≦0.05 and |log2FC|>1 and organize their methylation level and annotation data;
- (6)
- GO and KEGG enrichment analysis of differential genes
3. Results
3.1. MethylRAD-Seq data and identification of A. japonicus DNA methylation sites
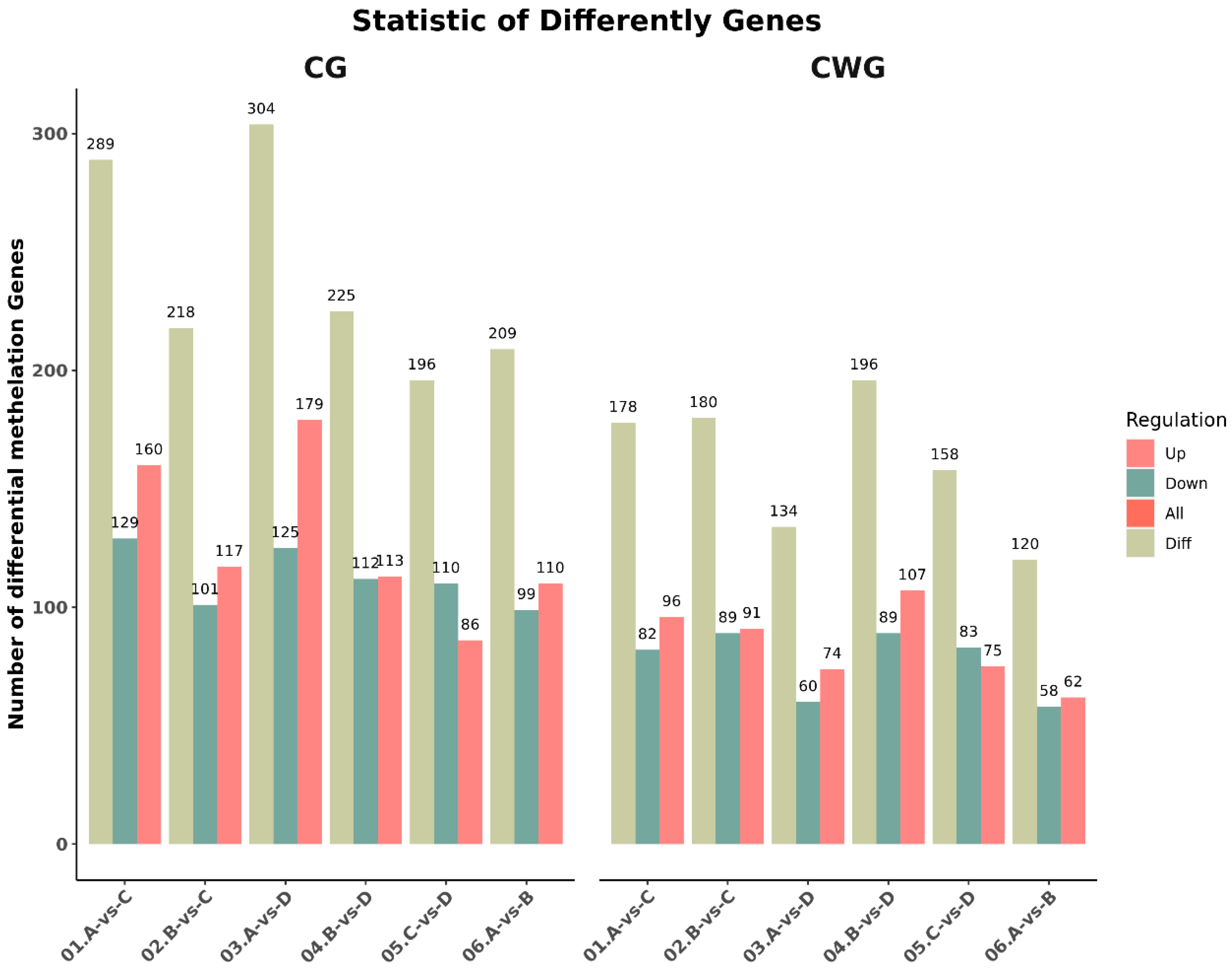
3.2. Identification of differentially methylated genes
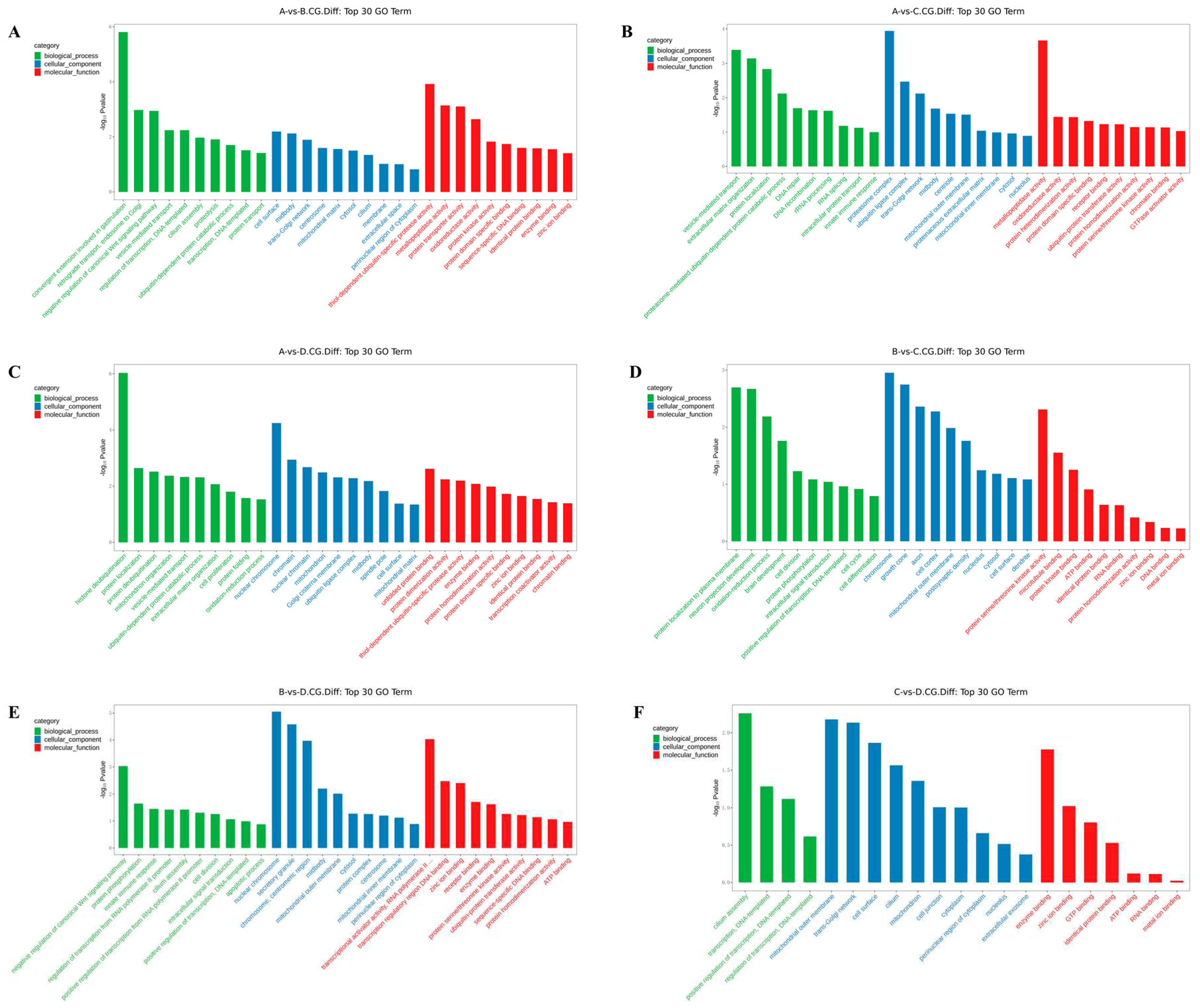
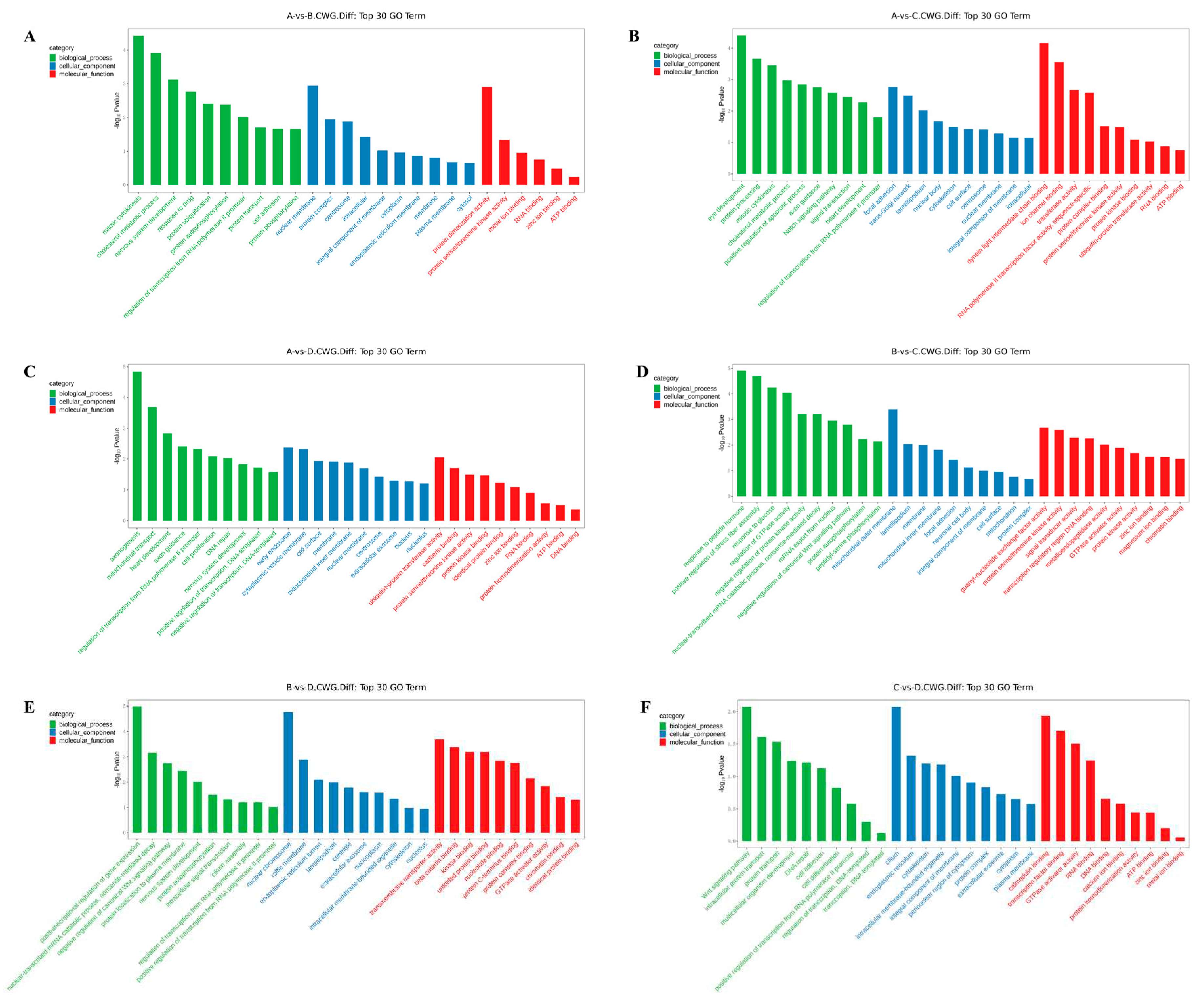
3.3. GO enrichment analysis of differentially methylated genes
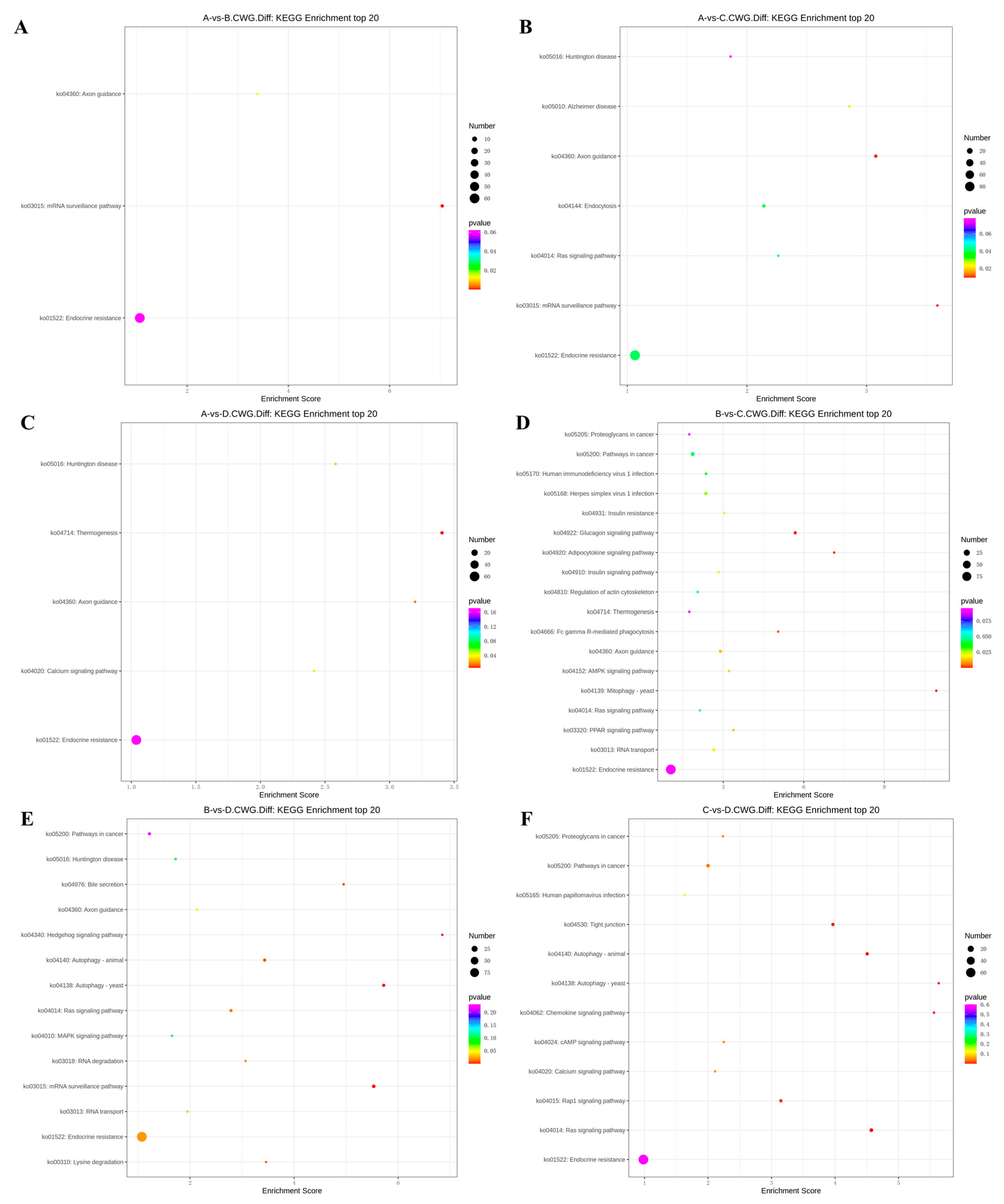
3.4. KEGG enrichment analysis of differentially methylated genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Liao, Y.L.; Zoology of China: Echinoderma: A. japonicuss.M. Science Press, 1997.
- Fisheries Administration of the Ministry of Agriculture and Rural Affairs; National Fisheries Technology Promotion Station; Chinese Fisheries Society. 2022 China Fisheries Statistical Yearbook.M. Beijing: China Agricultural Press, 2022:6.
- Zhang, L.H.; Ding, J.; Han, Z.H.; Chang, Y.Q.; Song, J.; Tian, Y.; Bai, X.Q.; Ding, W.J. Study on the species and morphology of imitation A. japonicus bone fragments. J. Marine Science.2015, 39 (4): 8-14. [CrossRef]
- Wang. J.J.; Liao, M.J.; Wang, Y.G.; Li, B.; Rong, X.J.; Ge, J.L.; Liu, Q.B.; Fan, R.Y. Study on the variation law of the species and structure of imitation A. japonicus bone fragments with age. J. Scientific Fish Farming.2023, (3): 73-75. [CrossRef]
- Montesanto, A. D; Aquila, P.; Lagani, V.; Paparazzo, E.; Passarino, G. A New Robust Epigenetic Model for Forensic Age Prediction. J. Journal of forensic sciences. 2020,65(5):1424-1431. [CrossRef]
- Anastasiadi, D; Piferrer, F.A clockwork fish: Age prediction using DNA methylation-based biomarkers in the European seabass. J.Molecular ecology resources.2020,20(2):387—397. [CrossRef]
- Zhang, L.; Jia, F.; Zhang, G.L.; Zeng, L.; Yi, Y.J.; Wang, J.S. Research progress of plant DNA methylation. J. Anhui Agricultural Sciences. 2012, (6): 3218-3221. [CrossRef]
- Lin, Z.K.; Xie, F.; Luo, J.Y.; Chen, T.; Xi, Q.Y.; Zhang, Y.L.; Sun, J.J. Methylation analysis of whole genome DNA of longissimus dorsi muscle in Lantang pigs and Everwhite pigs. J. Journal of Northwest A & F University (Natural Science Edition),.2023,51 (6): 1-10,17. [CrossRef]
- Wei, S.; Xie, L.L.; Zhu, H.; Zhang, Q.J.; Shen, Y.B.; Xu, X.Y.; Li, J.L. Differential methylation analysis of Asian grass carp populations.J. Chinese Journal of Fisheries. 2023, 47 (3): 99-111. [CrossRef]
- Wang, C.S.; Huang, X.D.; Cui, X.Y.; Ni, P.; Ye, S.G.; Wang, H.; Gao, D.X.; Lei, W. Effects of Vibrio harvei infection on DNA methylation of IL-6 gene of red-fin pufferfish.J. Journal of Dalian Ocean University.2022, 37 (2): 221-226. [CrossRef]
- Zhang, Y.L.; Zhou, C.J. DNA methylation and fish age.J. Henan Fisheries. 2021 (6): 20-23.
- Mcgaughey, D. M.; Abaan, H. O.; Miller, R. M.; Kropp, P. A.; Brody, L. C. Genomics of cpg methylation in developing and developed zebrafish. G3 (Bethesda, Md.) (5).2014. [CrossRef]
- Montesanto, A. D; Aquila, P.; Lagani, V.; Paparazzo, E.; Passarino, G. A New Robust Epigenetic Model for Forensic Age Prediction. J. Journal of forensic sciences. 2020,65(5):1424-1431. [CrossRef]
- De paoli-iseppi, R.; Deagle, B.E.; Polanowski, A.M.; McMahon, C.R.; Dickinson, J.L.; Hindell, Ma.A.; Jarman, S.N. Age estimation in a long-lived seabird (Ardenna tenuirostris) using DNA methylation-based biomarkers. J.Nature reviews Cancer. 2019,19(2):411-425. [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. J. Fly.2012,6(2):80-92. [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. J. Bioinformatics.2010,26(6):841-842. [CrossRef]
- Sharma, R.; Patnaik, S.K. Regulation of aspartate aminotransferase isoenzymes by hydrocortisone in the liver of aging rats. Archives of Gerontology & Geriatrics, (1).1987.27-32. [CrossRef]
- Ni, J.J.; Wu, Z.; Stoka, V.; Meng, J.; Hayashi, Y.; Peters, C.; Qing, H.; Turk, V.; Nakanishi, H. Increased expression and altered subcellular distribution of cathepsin B in microglia induce cognitive impairment through oxidative stress and inflammatory response in mice. J. Aging Cell.2019,18(1): n/a-n/a. [CrossRef]
- Meng J, Liu YC, Xie Z, Qing H, Lei P, Ni JJ. Nucleus distribution of cathepsin B in senescent microglia promotes brain aging through degradation of sirtuins. Neurobiol Aging, 2020, 96: 255–2.
- Sun, Q.Y.; Zhang, C.Z.; Mei, B.; Luo, X.; Zhu, Z.M.; Hua, T.M. Age-related retinal γ-aminobutyric acid and neurofilament protein expression in cats. Journal of Anatomy. 2005, 28(5), 4. [Google Scholar]
- Shu, J.B.; Jiang, S.Z.; Meng, Y.T. Research progress of succinic semialdehyde dehydrogenase deficiency. Continuing Medical Education. 2014, 28(10), 5. [Google Scholar]
- Al-Zghoul, M.B.; Ismail, Z.B.; Dalab, A.S.; Al-Ramadan, A.; Althnaian, T.A.; Al-ramadan, S.Y.; Ali, A.M.; Albokhadaim, I.F.; Al Busadah, K.A.; Eljarah, A.; Jawasreh, K.I.; Hannon, K.M. Hsp90, Hsp60 and HSF-1 genes expression in muscle, heart and brain of thermally manipulated broiler chicken. J. Research in Veterinary Science. 2015, 99105–111. [Google Scholar] [CrossRef]
- Bansal, G.S.; Norton, P.M.; Latchman, D.S. The 90-kDa heat shock protein protects mammalian cells from thermal stress but not from viral infection. Experimental Cell Research. 1991,195.2. [CrossRef]
- Boehm, A.K.; Seth, M.; Mayr, K.G.; Fortier, LA. Hsp90 mediates insulin-like growth factor 1 and interleukin-1beta signaling in an age-dependent manner in equine articular chondrocytes. Arthritis and rheumatism. 2007, 56.7.
- Johanna, S.S.; Bernard, R. Effects of Aging and Oxidative Stress on Spermatozoa of Superoxide-Dismutase 1- and Catalase-Null Mice1. Biology of Reproduction. 2016, 95.3.
- Miska, K.B.; Fetterer, R.H.; Wong, E.A. The mRNA expression of amino acid transporters, aminopeptidase N, and the di- and tri-peptide transporter PepT1 in the embryo of the domesticated chicken (Gallus gallus) shows developmental regulation. J. Poultry Science.2014,93(9):2262-2270. [CrossRef]
- Singh Yadav, R.N.; Singh, S.N. Regulation of NAD- and NADP-linked isocitrate dehydrogenase by hydrocortisone in the brain and liver of male rats of various ages." Biochimica et Biophysica Acta (BBA) - General Subjects. 1980, 633.3. [CrossRef]
- Markopoulos, G.S.; Roupakia, E.; Tokamani, M.; Vartholomatos, G.; Tzavaras, T.; Hatziapostolou, M.; Fackelmayer, F.O.; Sandaltzopoulos, R.; Polytarchou, C.; Kolettas, E. Senescence-associated microRNAs target cell cycle regulatory genes in normal human lung fibroblasts. J. Experimental Gerontology. 2017, 96110–122. [Google Scholar] [CrossRef] [PubMed]
- Pankiewicz, A.; Sledzinski, T.; Nogalska, A.; Swierczynski, J. Tissue specific, sex and age--related differences in the 6-phosphogluconate dehydrogenase gene expression. The international journal of biochemistry & cell biology. 2003,35(2), 235–245. [CrossRef]
- Kim, J.H.; Lee, H.K.; Takamiya, K.; Huganir, R.L. The role of synaptic GTPase-activating protein in neuronal development and synaptic plasticity. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003,23(4), 1119–1124. [CrossRef]
- Saito, S.; Kawamura, T.; Higuchi, M.; Kobayashi, T.; Yoshita-Takahashi, M.; Yamazaki, M.; Abe, M.; Sakimura, K.; Kanda, Y.; Kawamura, H.; Jiang, S.; Naito, M.; Yoshizaki, T.; Takahashi, M.; Fujii, M. RASAL3, a novel hematopoietic RasGAP protein, regulates the number and functions of NKT cells. European journal of immunology. 2015, 45(5), 1512–1523. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, K.; Tripathi, R.; Mishra, R. Age-dependent alterations in expression and co-localization of Pax6 and Ras-GAP in brain of aging mice. Journal of chemical neuroanatomy. 2018, 92, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Ethell, I. M.; Irie, F.; Kalo, M.S.; Couchman, J.R.; Pasquale, E.B.; Yamaguchi, Y. EphB/syndecan-2 signaling in dendritic spine morphogenesis. Neuron. 2001, 31(6), 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Morita, A.; Yamashita, N.; Sasaki, Y.; Uchida, Y.; Nakajima, O.; Nakamura, F.; Yagi, T.; Taniguchi, M.; Usui, H.; Katoh-Semba, R.; Takei, K.; Goshima, Y. Regulation of dendritic branching and spine maturation by semaphorin3A-Fyn signaling. The Journal of neuroscience: the official journal of the Society for Neuroscience.2006,26(11), 2971–2980. [CrossRef]
- Yoshida, J.; Kubo, T.; Yamashita, T. Inhibition of branching and spine maturation by repulsive guidance molecule in cultured cortical neurons. Biochemical and biophysical research communications. 2008, 72(4), 725–729. [Google Scholar] [CrossRef] [PubMed]
- El-Hoss, J.; Arabian, A.; Dedhar, S.; St-Arnaud, R. Inactivation of the integrin-linked kinase (ILK) in osteoblasts increases mineralization. Gene. 2014, 533(1), 246–252. [Google Scholar] [CrossRef] [PubMed]
- Weichhart, T. mTOR as Regulator of Lifespan, Aging, and Cellular Senescence: A Mini-Review. Gerontology. 2018, 64(2), 127–134. [Google Scholar] [CrossRef] [PubMed]
- Erdogan, C. S.; Hansen, B. W.; Vang, O. Are invertebrates relevant models in ageing research? Focus on the effects of rapamycin on TOR. Mechanisms of ageing and development. 2016, 153, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Zacharewicz, E.; Della Gatta, P.; Reynolds, J.; Garnham, A.; Crowley, T.; Russell, A.P.; Lamon, S. Identification of microRNAs linked to regulators of muscle protein synthesis and regeneration in young and old skeletal muscle. PloS one. 2014, 9(12), e114009. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Shi, R.; Ye, Y.; Zhang, Y.; Zhang, Y.; Wang, Z.; Gu, Y.; Yin, Y.; Chen, D.; Tang, J. Aging-Induced Down-Regulation of PKA/BKCa Pathway in Rat Cerebral Arteries. Physiological research. 2022, 71(6), 811–823. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Xu, W.; Zhu, H.; Dong, Z.; Dong, H.; Yin, S. Aging aggravates acetaminophen-induced acute liver injury and inflammation through inordinate C/EBPα-BMP9 crosstalk. Cell & bioscience. 2023,13(1), 61. [CrossRef]

| Sample | Raw_Reads | Clean_Reads | Percent |
| A | 75848311 | 36771351 | 48.48% |
| B | 62579986 | 32326130 | 51.66% |
| C | 65336130 | 30392328 | 46.52% |
| D | 76656509 | 30786727 | 40.16% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




