Submitted:
29 September 2023
Posted:
30 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Study area and dataset
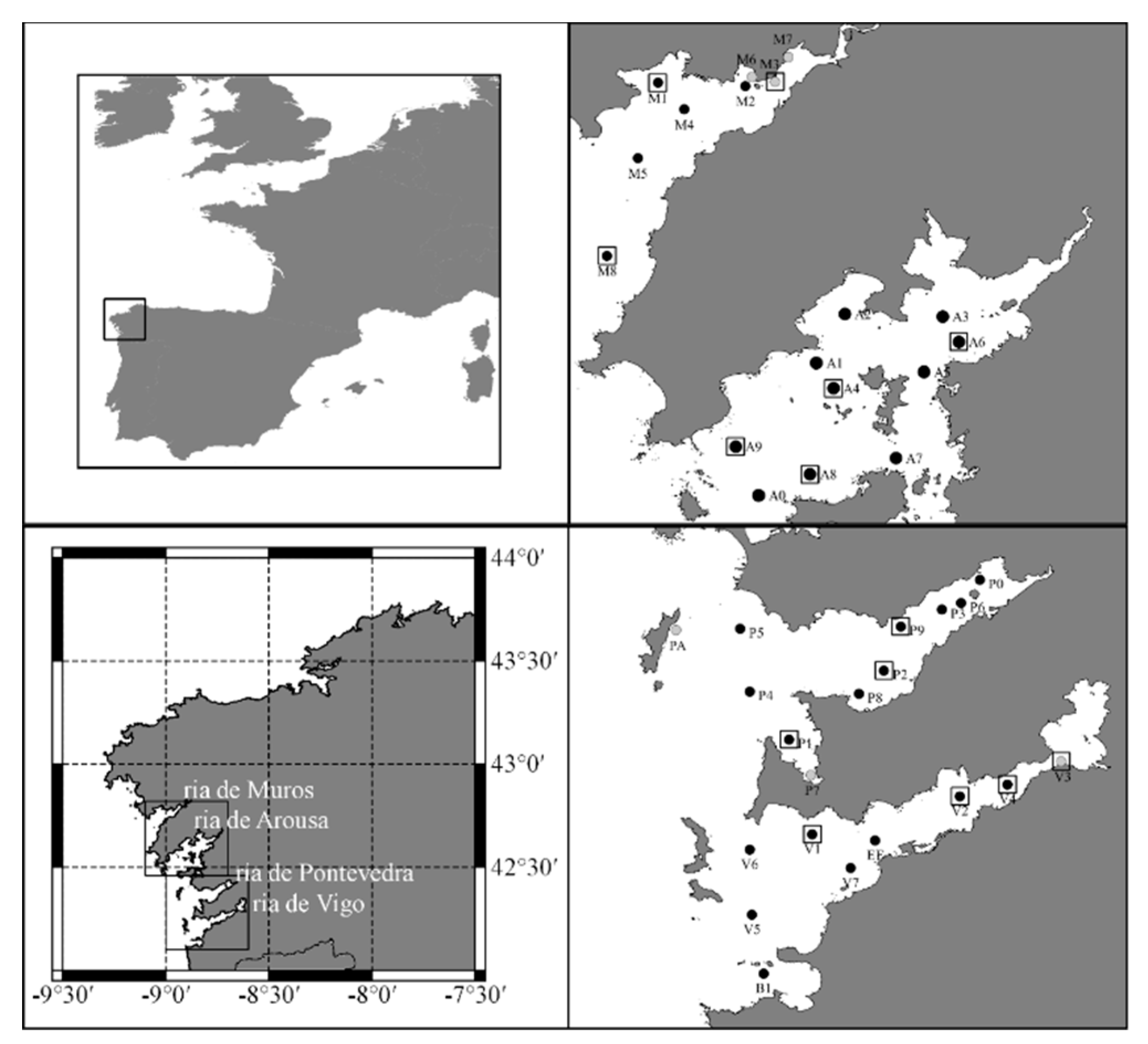
2.1. The Rias Baixas
2.2. Sentinel-3 imagery
2.3. INTECMAR data
3. Methods
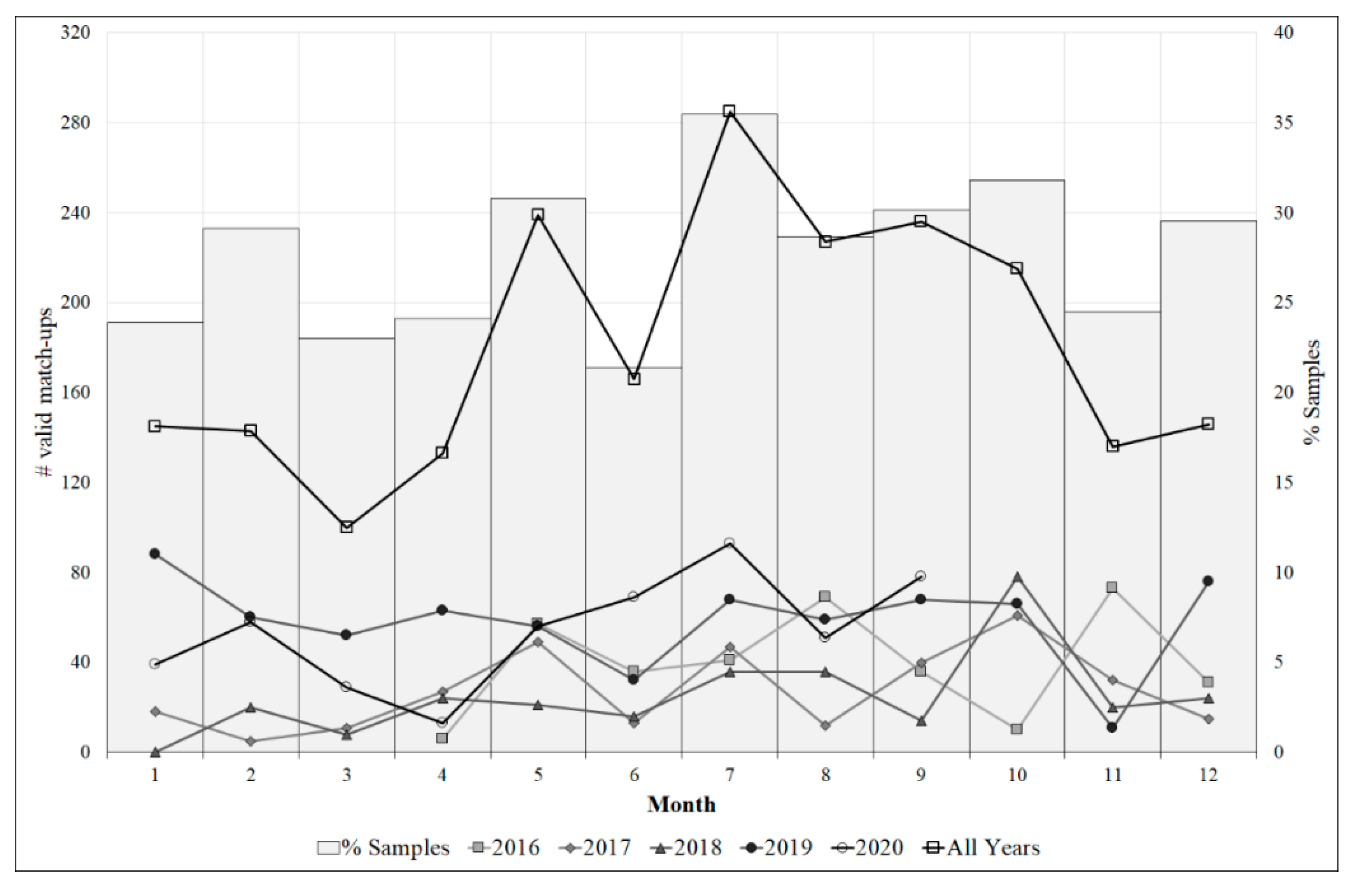
3.1. Image processing and dataset generation
3.2. Support Vector Machine models
3.2.1. Scaling and imbalance effect
3.2.2. Model selection
3.2.3. Model evaluation
4. Results
4.1. Spatial and temporal distribution of Pseudo-nitzschia spp. abundance
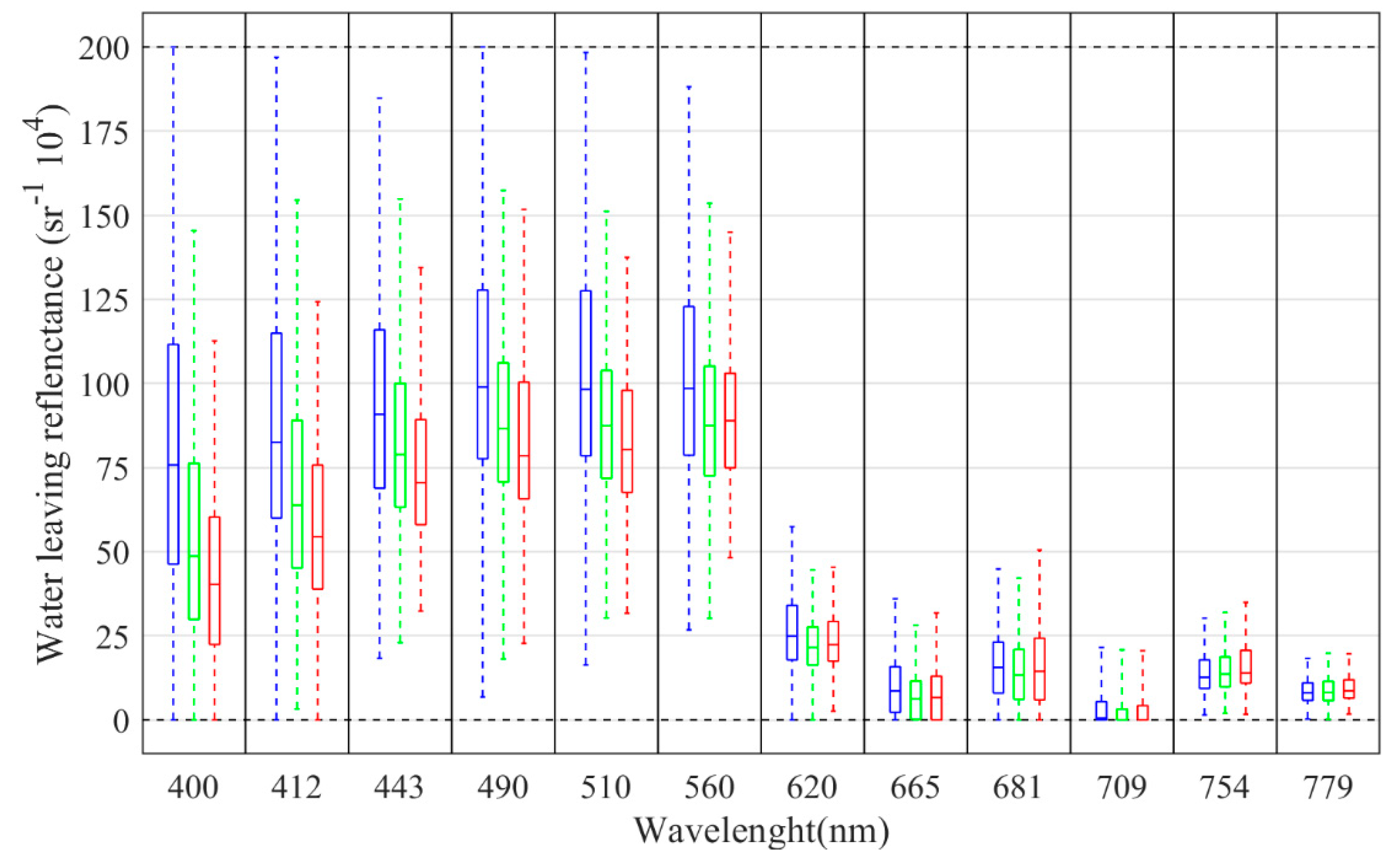
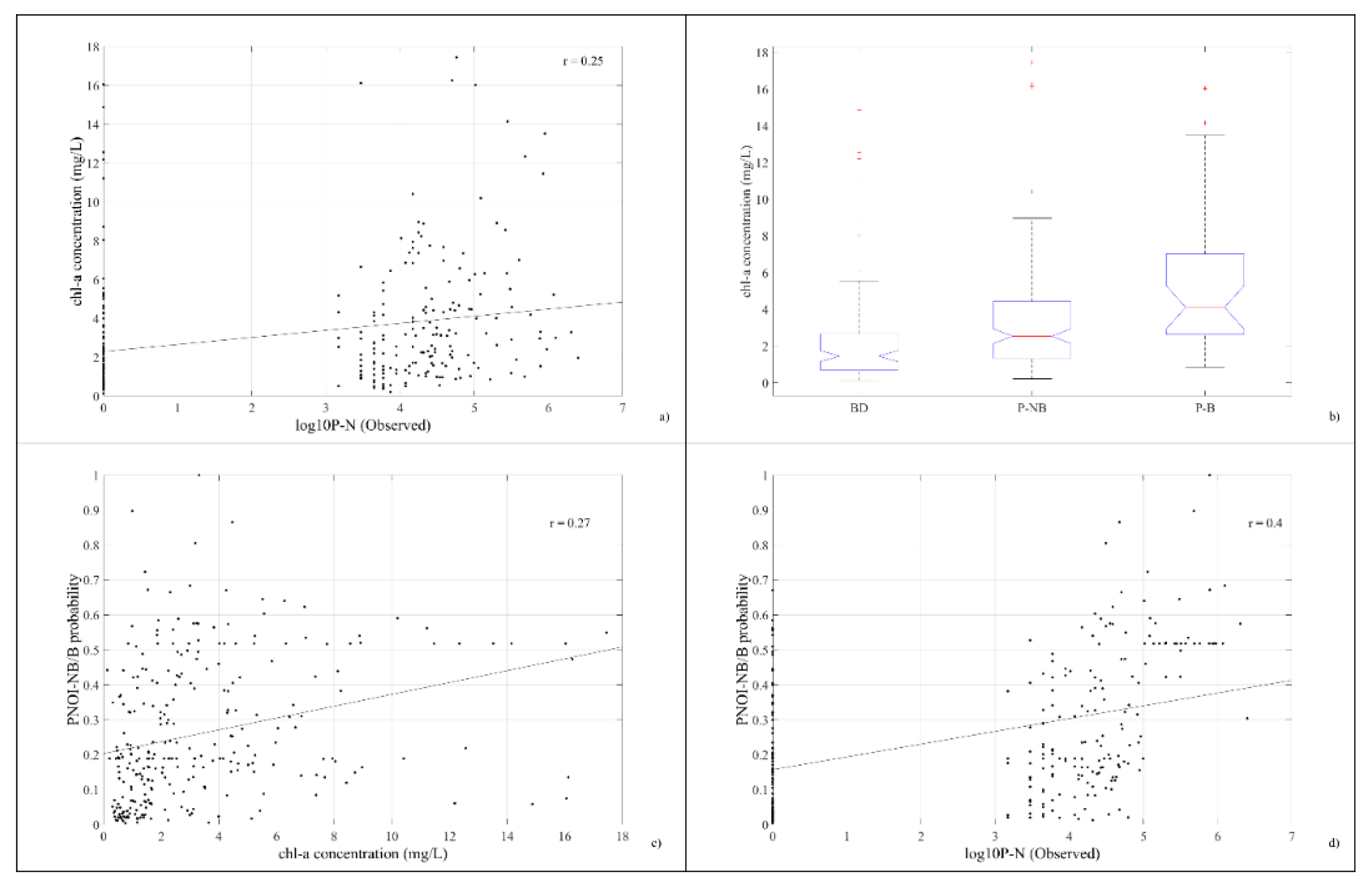
4.2. Relationship between Pseudo-nitzschia spp. abundance and Sentinel-3 data
4.3. SVM models
4.4. Comparison with the linear model and threshold analysis
5. Discussion
5.1. Relationships between Pseudo-nitzschia spp. abundance and Sentinel-3 data
5.2. Models performance
5.3. Model evaluation and filling observational gaps of Pseudo-nitzschia spp. evolution
5.4. Pseudo-nitzschia spp. maps
6. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Gobler, C.J., Doherty, O.M., Hattenrath-Lehmann, T.K., Griffith, A.W., Kang, Y., Litaker, R.W. Ocean warming since 1982 has expanded the niche of toxic algal blooms in the North Atlantic and North Pacific oceans. Proc. Natl. Acad. Sci. 2017, 114, 4975–4980. [CrossRef] [PubMed]
- Griffith, A.W., Gobler, C.J. Harmful algal blooms: A climate change co-stressor in marine and freshwater ecosystems. Harmful Algae 2020, 91, 101590. [CrossRef] [PubMed]
- Anderson, D. HABs in a changing world: a perspective on harmful algal blooms, their impacts, and research and management in a dynamic era of climactic and environmental change. In Proceedings of the 15th International Conference on Harmful Algae: CECO; Kim, H. G., Reguera, B, Hallegraeff, Lee, C.K., Eds.; 29 October - 2 November 2012; pp. 3–17.
- Gobler, C.J. Climate Change and Harmful Algal Blooms: Insights and perspective. Harmful Algae 2020, 91, 101731. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M. Approaches to monitoring, control and management of harmful algal blooms (HABs). Ocean Coast Manag. 2009, 52, 342. [Google Scholar] [CrossRef]
- Anderson, D., Cembella, A., Hallegraeff, G. Progress in understanding harmful algal blooms: paradigm shifts and new technologies for research, monitoring, and management. Ann. Rev. Mar. Sci. 2012, 4, 143–176. [CrossRef]
- Tyler, A.N., Hunter, P.D., Spyrakos, E., Groom, S., Constantinescu, A.M., Kitchen, J. Developments in Earth observation for the assessment and monitoring of inland, transitional, coastal and shelf-sea waters. Sci. Total Environ. 2016, 572, 1307–1321. [CrossRef]
- Spyrakos, E., Hunter, P., Simis, S., Neil, C., Riddick, C., Wang, S., et al. Moving towards global satellite based products for monitoring of inland and coastal waters. Regional examples from Europe and South America. In Proceedings of the 2020 IEEE Latin American GRSS & ISPRS Remote Sensing Conference (LAGIRS); Santiago, Chile, 2020, pp. 363–368.
- Spyrakos, E., González Vilas, L., Torres Palenzuela, J.M., Barton, E.D. Remote sensing chlorophyll a of optically complex waters (rias Baixas, NW Spain): Application of a regionally specific chlorophyll a algorithm for MERIS full resolution data during an upwelling cycle. Remote Sens. Environ. 2011, 115, 2471–2485.
- Amin, R., Zhou, J., Gilerson, A., Gross, B., Moshary, F., Ahmed, S. Novel optical techniques for detecting and classifying toxic dinoflagellate Karenia brevis blooms using satellite imagery. Opt. Express 2009, 17, 9126–9144. [CrossRef]
- Kurekin, A.A., Miller, P.I., Woerd, H.J.V.d. Satellite discrimination of Karenia mikimotoi and Phaeocystis harmful algal blooms in European coastal waters: Merged classification of ocean colour data. Harmful Algae 2014, 31, 163–176. [CrossRef]
- Moore, T.S., Dowell, M.D., Franz, B.A. Detection of coccolithophore blooms in ocean color satellite imagery: A generalized approach for use with multiple sensors. Remote Sens. Environ. 2012, 117, 249–263. [CrossRef]
- Dierssen, H.M., Kudela, R.M., Ryan, J.P., Zimmerman, R.C. Red and black tides: Quantitative analysis of water-leaving radiance and perceived color for phytoplankton, colored dissolved organic matter, and suspended sediments. Limnol. Oceanogr 2006, 51, 2646–2659. [CrossRef]
- Cannizzaro, J.P., Carder, K.L., Chen, F.R., Heil, C.A., Vargo, G.A. A novel technique for detection of the toxic dinoflagellate, Karenia brevis, in the Gulf of Mexico from remotely sensed ocean color data. Cont. Shelf Res. 2008, 28, 137–158. [CrossRef]
- Brown, C.W., Podestá, G.P. Remote sensing of coccolithophore blooms in the Western South atlantic ocean. Remote Sens. Environ. 1997, 60, 83–91. [CrossRef]
- Kopelevich, O., Burenkov, V., Sheberstov, S., Vazyulya, S., Kravchishina, M., Pautova, L., Silkinb, V., Artemieva, V., Grigorieva, A. Satellite monitoring of coccolithophore blooms in the Black Sea from ocean color data. Remote Sens. Environ. 2014, 146, 113–123. [CrossRef]
- Shutler, J.D., Grant, M.G., Miller, P.I., Rushton, E., Anderson, K. Coccolithophore bloom detection in the north east Atlantic using SeaWiFS: Algorithm description, application and sensitivity analysis. Remote Sens. Environ. 2010, 114, 1008–1016. [CrossRef]
- Dupouy, C., Benielli-Gary, D., Neveux, J., Dandonneau, Y., Westberry, T.K. An algorithm for detecting Trichodesmium surface blooms in the South Western Tropical Pacific. Biogeosciences 2011, 8, 3631–3647. [CrossRef]
- Gower, J., King, S., Young, E. Global remote sensing of Trichodesmium. Int. J. Remote Sens. 2014, 35, 5459–5466. [CrossRef]
- Hu, C., Cannizzaro, J., Carder, K.L., Muller-Karger, F., Hardy, R. Remote detection of Trichodesmium blooms in optically complex coastal waters: Examples with MODIS full-spectral data. Remote Sens. Environ. 2010, 114, 2048–2058. [CrossRef]
- Kudela, R.M., Stumpf, R.P., Petrov, P. Acquisition and analysis of remote sensing imagery of harmful algal blooms. In Harmful Algal Blooms (HABs) and Desalination: A Guide to Impacts, Monitoring and Management. Intergovernmental Oceanographic Commission of UNESCO; IOC Manuals and Guides No. 78; Anderson, D.M., Boerlage, S.F., Dixon, M.B., Eds.; 2017; pp. 119–132. [Google Scholar]
- Spyrakos, E., O'Donnell, R., Hunter, P.D., Miller, C., Scott, M., Simis, S.G.H., et al. Optical types of inland and coastal waters. Limnol. Oceanogr. 2018, 63, 846–870.
- Shen, L., Xu, H., Guo, X. Satellite remote sensing of harmful algal blooms (HABs) and a potential synthesized framework. Sensors 2012, 12, 7778–7803. [CrossRef]
- Blondeau-Patissier, D., Gower, J.F.R., Dekker, A.G., Phinn, S.R., Brando, V.E. A review of ocean color remote sensing methods and statistical techniques for the detection, mapping and analysis of phytoplankton blooms in coastal and open oceans. Prog. Oceanogr. 2014, 123, 123–144. [CrossRef]
- Mountrakis, G., Im, J., Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [CrossRef]
- Vapnik, V.N. An overview of statistical learning theory. IEEE Trans. Neural. Netw. 1999, 10, 988–999. [Google Scholar] [CrossRef]
- Zhu, G., Blumberg, D.G. Classification using ASTER data and SVM algorithms; The case study of Beer Sheva, Israel. Remote Sens. Environ. 2002, 80, 233–240. [CrossRef]
- Bates, S.S., Hubbard, K.A., Lundholm, N., Montresor, M., Leaw, C.P. Pseudo-nitzschia, Nitzschia, and domoic acid: New research since 2011. Harmful Algae, 2018, 79, 3–43. [CrossRef] [PubMed]
- Lelong, A., Hégaret, H., Soudant, P., Bates, S.S. Pseudo-nitzschia (Bacillariophyceae) species, domoic acid and amnesic shellfish poisoning: revisiting previous paradigms. Phycologia 2012, 51, 168–216. [CrossRef]
- Trainer, V.L., Bates, S.S., Lundholm, N., Thessen, A.E., Cochlan, W.P., Adams, N.G., Trick, G.C. Pseudo-nitzschia physiological ecology, phylogeny, toxicity, monitoring and impacts on ecosystem health. Harmful Algae 2012, 14, 271–300. [CrossRef]
- Ryan, J.P., Kudela, R.M., Birch, J.M., Blum, M., Bowers, H.A., Chavez, F.P., Doucette, G.J., Hayashi, K., Marin III, R., Mikulski, C.M., Pennington, J.T., Scholin, C.A., Smith, G.J., Woods, A., Zhang, Y. Causality of an extreme harmful algal bloom in Monterey Bay, California, during the 2014–2016 northeast Pacific warm anomaly. Geophys. Res. Lett. 2017, 44, 5571–5579.
- Anderson, C.R., Sapiano, M.R.P., Prasad, M.B.K., Long, W., Tango, P.J., Brown, C.W., Murtugudde, R. Predicting potentially toxigenic Pseudo-nitzschia blooms in the Chesapeake Bay. J. Mar. Syst. 2010, 83, 127–140. [CrossRef]
- Anderson, C.R., Siegel, D.A., Kudela, R.M., Brzezinski, M.A. Empirical models of toxigenic Pseudo-nitzschia blooms: Potential use as a remote detection tool in the Santa Barbara Channel. Harmful Algae 2009, 8, 478–492. [CrossRef]
- Margalef, R. Estructura y dinámica de la “purga de mar” en Ría de Vigo. Investigacion Pesquera 1956, 5, 113–134. [Google Scholar]
- Torres Palenzuela, J.M., González Vilas, L., Bellas, F.M., Garet, E., González-Fernández, Á., Spyrakos, E. Pseudo-nitzschia Blooms in a Coastal Upwelling System: Remote Sensing Detection, Toxicity and Environmental Variables. Water 2019, 11, 1954. [CrossRef]
- González Vilas, L., Spyrakos, E., Torres Palenzuela, J.M., Pazos, Y. Support Vector Machine-based method for predicting Pseudo-nitzschia spp. blooms in coastal waters (Galician rias, NW Spain). Prog. Oceanogr. 2014, 124, 66–77. [CrossRef]
- Pitcher, G.C., Figueiras, F. G., Hickey, B.M., Moita, M.T. The physical oceanography of upwelling systems and the development of harmful algal blooms. Prog. Oceanogr. 2010, 85, 5–32. [CrossRef]
- Labarta, U., Fernández-Reiriz, M.J. The Galician mussel industry: Innovation and changes in the last forty years. Ocean Coast Manag. 2019, 167, 208–218. [CrossRef]
- Fraga, S., Alvarez, M.J., Míguez, A., Fernández, M.L., Costas, E., Lopez-Rodas, V. Pseudo-nitzschia species isolated from Galician waters: toxicity, DNA content and lectin binding assay. In Harmful Algae. Xunta de Galicia and Intergovernmental Commission of UNESCO; Reguera, B., Blanco, B., Fernández, M.L., Wyatt, T., Eds.; Santiago de Compostela, Spain, 1998; pp. 270–273. [Google Scholar]
- Rodríguez, G.R., Villasante, S., Garcí a-Negro, M.C. Are red tides affecting economically the commercialization of the Galician (NW Spain) mussel farming? Mar. Policy 2011, 35, 252–257. [CrossRef]
- Donlon, C., Berruti, B., Buongiorno, A., Ferreira, M-H., Féménias, P., Frerick, J., Goryl, P., Klein, U., Laur, H., Mavrocordatos, C., Nieke, J., Rebhan, H., Seitz, B., Stroede, J., Sciarra, R. The Global Monitoring for Environment and Security (GMES) Sentinel-3 mission. Remote Sens. Environ. 2012, 120, 37–57.
- Utermöhl, H. Zur vervollkommnung der quantitativen phytoplankton-methodik. Mitteilung Internationale Vereinigung Theoretische unde Amgewandte Limnologie 1958, 9, 1–38. [Google Scholar] [CrossRef]
- Steinmetz, F., Deschamps, P.-Y., Ramon, D. Atmospheric correction on the presence of sun glint: application to MERIS. Opt. Express 2011, 19, 9783–9800. [CrossRef]
- Warren, M.A., Simis, S.G., Martinez-Vicente, V., Poser, K., Bresciani, M., Alikas, K., Spyrakos, E., Giardino, C., Ansper, A. Assessment of atmospheric correction algorithms for the Sentinel-2A MultiSpectral Imager over coastal and inland waters. Remote Sens. Environ. 2019, 225, 267–289. [CrossRef]
- González Vilas, L., Spyrakos, E., Torres Palenzuela, J.M. Neural network estimation of chlorophyll a from MERIS full resolution data for the coastal waters of Galician Rias (NW Spain). Remote Sens. Environ. 2011, 115, 524–535. [CrossRef]
- Chang, C.-C.L. and Lin C.-J. LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27:21–27:27.
- Camps-Valls, G., Bruzzone,. Kernel-based methods for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2005, 43, 1351–1362. [CrossRef]
- Cristianini, N., Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-based Learning Methods; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Sarle, W.S. Neural networks and statistical models. In SUGI19: Proceedings of the Nineteenth Annual SAS Users Group International Conference; SAS Institute: Cary, NC, USA, 1994; pp. 1538–1550. [Google Scholar]
- Liu, C., White, M., Newell, G. Measuring and comparing the accuracy of species distribution models with presence-absence data. Ecography 2011, 34, 232–243. [CrossRef]
- Allouche, O., Tsoar, A., Kadmon, R. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 2006, 43, 1223–1232. [CrossRef]
- Hastie, T., Tibshirani, R., Friedman, J. The elements of statistical learning. Data mining, inference and prediction, 2nd ed.; Springer: New York, 2017. [Google Scholar]
- Hosmer, D.W., Lemeshow, S. Applied Logistic Regression, 2nd ed.; John Wiley and Sons: New York, 2000. [Google Scholar]
- García-Roselló, E., Guisande, C, González-Vilas, L., González-Dacosta, J., Heine, J., Pérez-Costas, E., Lobo, J.M. A simple method to estimate the probable distribution of species. Ecography 2019, 42, 613–1622.
- Del Vecchio, R., Blough, N.V. Spatial and seasonal distribution of chromophoric dissolved organic matter and dissolved organic carbon in the Middle Atlantic Bight. Mar. Chem. 2004, 89, 169–187. [CrossRef]
- Kirk, J.T.O. Light and photosynthesis in aquatic ecosystems; Cambridge University Press: Cambridge, 1994. [Google Scholar]
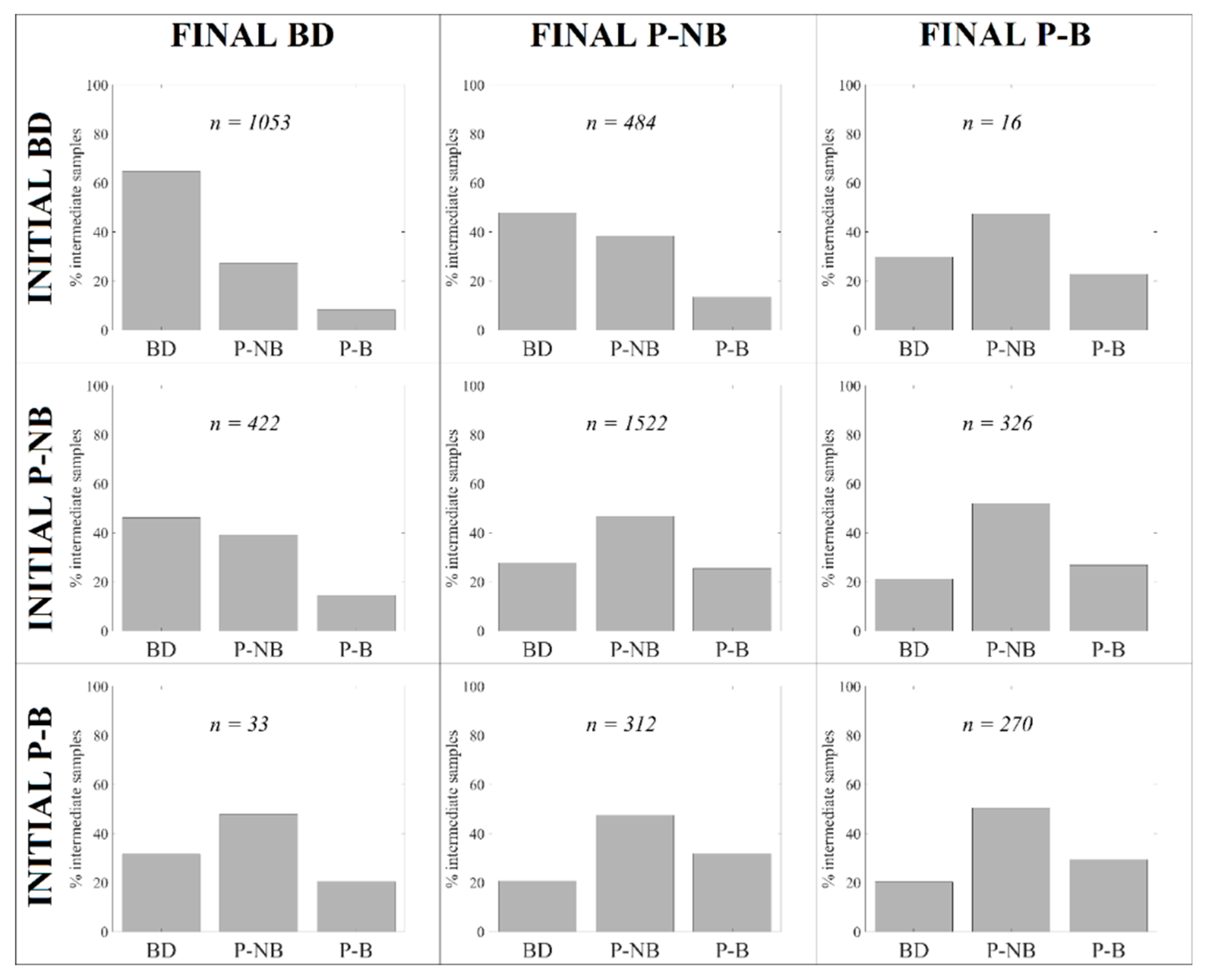
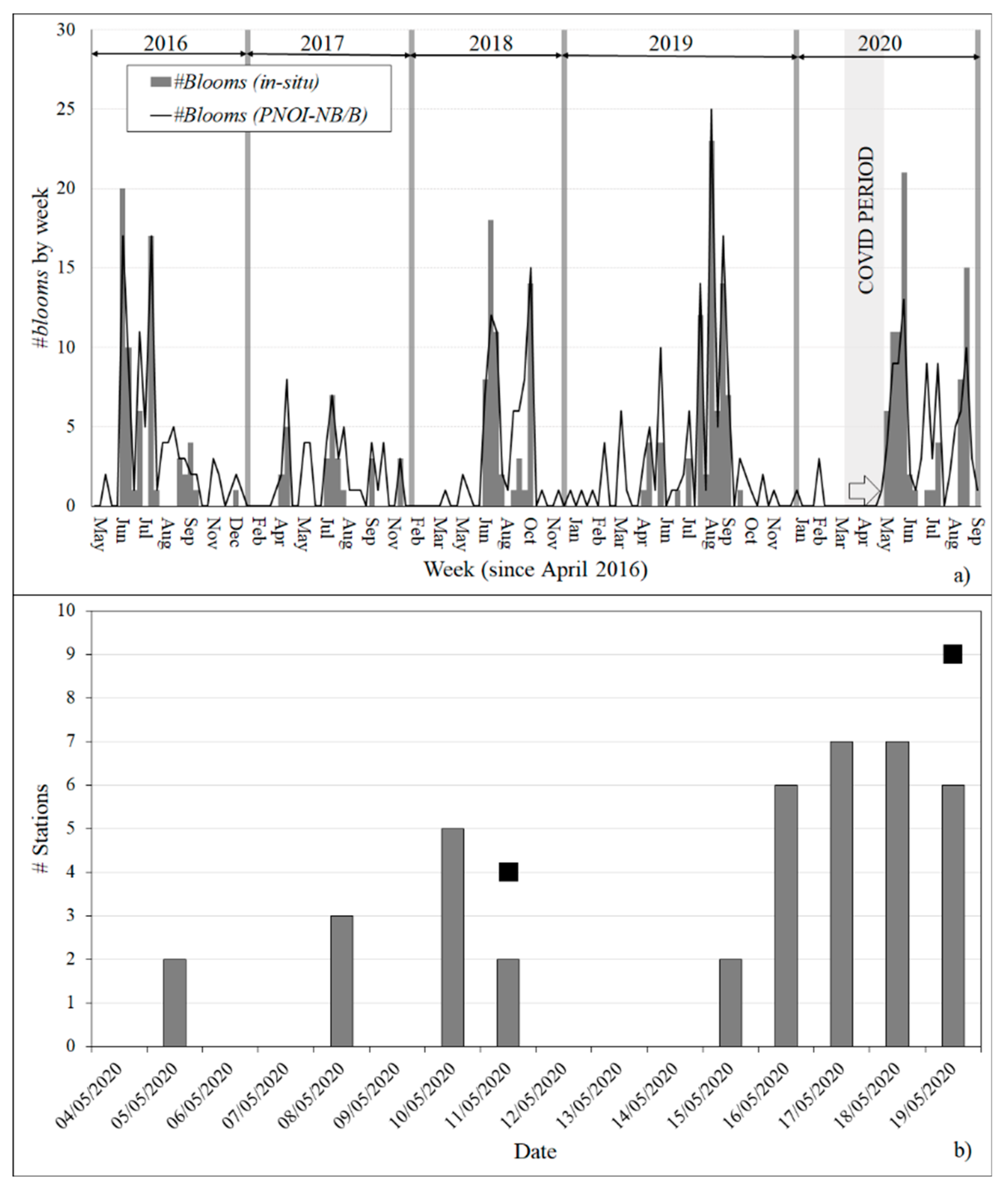
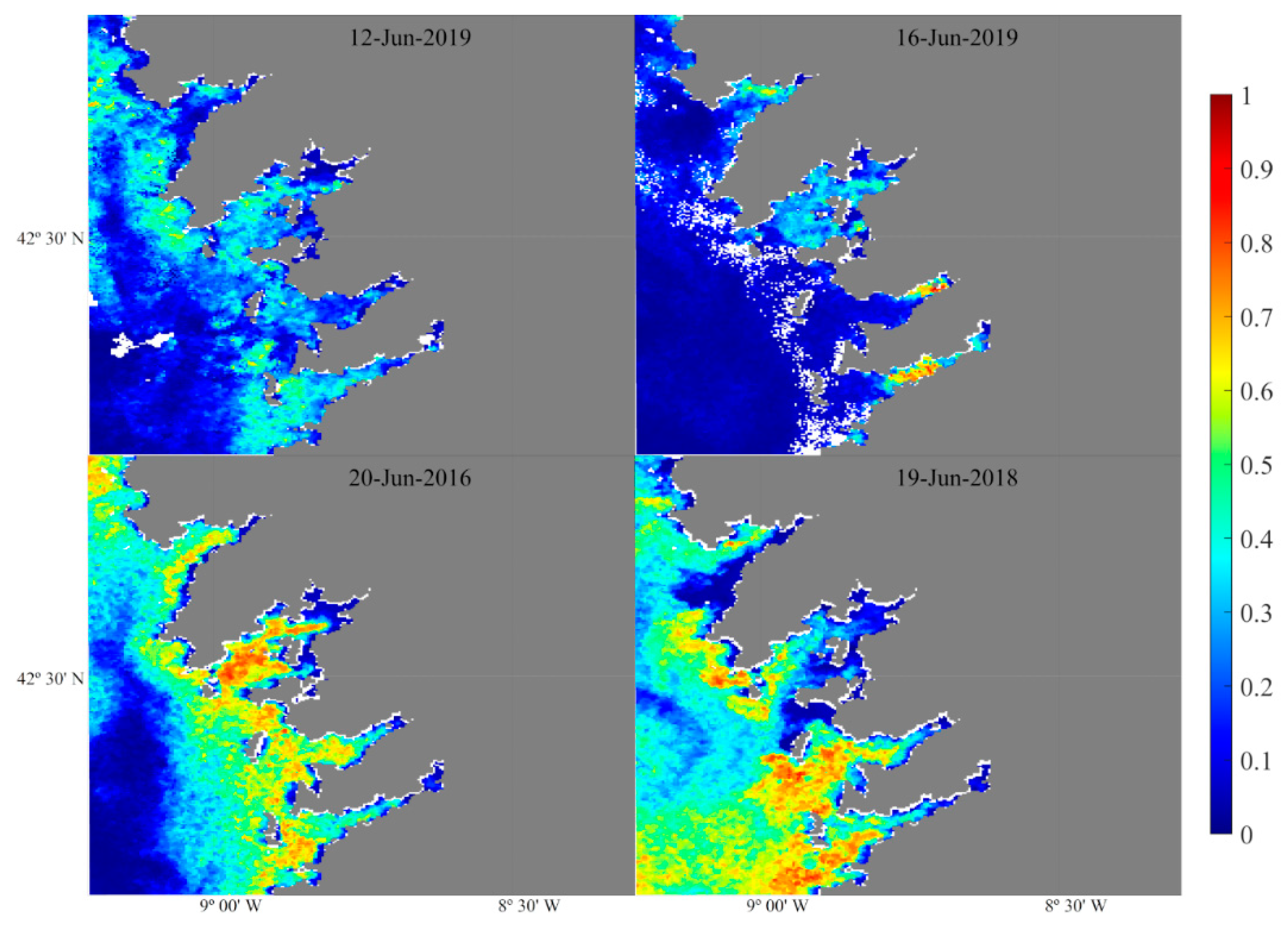
| Complete period |
2016 Apr-Dec |
2017 | 2018 | 2019 | 2020 Jan-Sep |
|
|---|---|---|---|---|---|---|
| # Images | 1446 | 159 | 220 | 241 | 471 | 355 |
| # Valid images | 989 | 118 | 158 | 142 | 317 | 254 |
| # Images with match-ups | 260 | 36 | 41 | 36 | 81 | 66 |
| # Sampling days | 465 | 77 | 103 | 104 | 103 | 78 |
| # Samples | 7740 | 1328 | 1679 | 1790 | 1771 | 1172 |
| # Match-ups | 4607 | 638 | 704 | 666 | 1482 | 1117 |
| # Valid match-ups | 3008 | 482 | 472 | 393 | 961 | 700 |
| # Valid HQ match -ups | 2171 | 359 | 330 | 297 | 699 | 486 |
| Study area |
Vigo (175 km2) |
Pontev. (145 km2) |
Arousa (230 km2) |
Muros (120 km2) |
||
| # Stations / HQ | 38 / 32 | 9 /8 | 11 /9 | 10 /10 | 8 /5 | |
| # Samples | 7740 | 1775 | 2361 | 2174 | 1430 | |
| # Match-ups | 4607 | 1006 | 1356 | 1306 | 939 | |
| # Valid match-ups | 3008 | 666 | 830 | 1020 | 492 | |
| # Valid HQ match-ups | 2171 | 485 | 456 | 890 | 340 | |
| #Samples #HQ match-ups |
log10P-n | BD | P-NB | P-B | |
|---|---|---|---|---|---|
| Vigo | 1775 | 2.59±2.27 | 747 (42.08%) | 783 (44.11%) | 245 (13.80%) |
| 485 | 2.76±2.25 | 186 (38.35%) | 227 (46.8%) | 72 (14.85%) | |
| Pontevedra | 2361 | 2.58±2.21 | 969 (41.04%) | 1138 (48.2%) | 254 (10.76%) |
| 456 | 2.82±2.17 | 163 (35.75%) | 224 (49.12%) | 69 (15.13%) | |
| Arousa | 2174 | 2.53±2.22 | 919 (42.27%) | 1047 (48.16%) | 208 (9.57%) |
| 890 | 2.64±2.24 | 361 (40.56%) | 421 (47.3%) | 108 (12.13%) | |
| Muros | 1430 | 2.98±2.23 | 489 (34.20%) | 689 (48.18%) | 252 (17.62%) |
| 340 | 2.94±2.28 | 122 (35.88%) | 154 (45.29%) | 64 (18.82%) | |
| Study area | 7740 | 2.59±2.27 | 3124 (40.36%) | 3657 (47.25%) | 959 (12.39%) |
| 2171 | 2.75±2.23 | 832 (38.32%) | 1026 (47.26%) | 313 (14.42%) |
| #Samples #HQ match-ups |
log10P-n | BD | P-NB | P-B | |
|---|---|---|---|---|---|
| 2016 | 1328 359 |
2.73±2.24 2.80±2.25 |
515 (38.78%) 133 (37.05%) |
609 (45.86%) 160 (44.57%) |
204 (15.36%) 66 (18.38%) |
| 2017 | 1679 330 |
3.12±2.00 3.35±1.79 |
468 (27.87%) 69 (20.91%) |
1049 (62.48%) 234 (70.91%) |
162 (9.65%) 27 (8.18%) |
| 2018 | 1790 297 |
2.67±2.31 2.95±2.33 |
737 (41.17%) 110 (37.04%) |
781 (43.63%) 129 (43.43%) |
272 (15.20%) 58 (19.53%) |
| 2019 | 1771 699 |
2.34±2.23 2.42±2.25 |
826 (46.64%) 315 (45.06%) |
773 (43.65%) 303 (43.35%) |
172 (9.71%) 81 (11.59%) |
| 2020 | 1172 486 |
2.29±2.31 2.66±2.32 |
578 (49.32%) 205 (42.18%) |
445(37.97%) 200 (41.15%) |
149 (12.71%) 81 (16.67%) |
| Below detection limit/presence (PNOI-BD/P) | ||||||||
|---|---|---|---|---|---|---|---|---|
| #BD | #P | Sens. | Spec. | Prec. | TSS | F1 | AUC | |
| LOU CV | 973 | 655 | 0.73 | 0.71 | 0.79 | 0.43 | 0.75 | 0.78 |
| Training set | 0.84 | 0.82 | 0.87 | 0.66 | 0.86 | 0.91 | ||
| Test set | 366 | 177 | 0.70 | 0.63 | 0.79 | 0.32 | 0.74 | 0.68 |
| No bloom / Bloom (PNOI-NB/B) | ||||||||
| #NB | #B | Sens. | Spec. | Prec. | TSS | F1 | AUC | |
| LOU CV | 1393 | 235 | 0.73 | 0.72 | 0.30 | 0.43 | 0.45 | 0.76 |
| Training set | 0.92 | 0.90 | 0.61 | 0.82 | 0.74 | 0.94 | ||
| Test set | 465 | 78 | 0.72 | 0.79 | 0.37 | 0.51 | 0.48 | 0.80 |
| Presence detection | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| #BD | #P | Model | Threshold | Sens. | Spec. | Prec. | TSS | F1 | AUC | |
| 832 | 1339 | BD/P | F1 | 0.504 | 0.91 | 0.64 | 0.80 | 0.55 | 0.86 | 0.86 |
| TSS | 0.640 | 0.80 | 0.78 | 0.85 | 0.58 | 0.83 | ||||
| Linear | 3 | 0.56 | 0.74 | 0.78 | 0.30 | 0.73 | ||||
| Bloom detection | ||||||||||
| #NB | #B | Model | Threshold | Sens. | Spec. | Prec. | TSS | F1 | AUC | |
| 1858 | 313 | NB/B | TSS | 0.430 | 0.87 | 0.87 | 0.54 | 0.75 | 0.67 | 0.90 |
| F1 | 0.512 | 0.79 | 0.93 | 0.65 | 0.72 | 0.72 | ||||
| Linear | 5 | 0.01 | 0.99 | 0.30 | 0.01 | 0.33 | ||||
|
BD/P + NB/B |
F1 + TSS | 0.86 | 0.88 | 0.54 | 0.74 | 0.66 | ||||
| F1 + F1 | 0.78 | 0.93 | 0.65 | 0.71 | 0.71 | |||||
| TSS + TSS | 0.81 | 0.88 | 0.54 | 0.70 | 0.65 | |||||
| TSS + F1 | 0.74 | 0.93 | 0.65 | 0.68 | 0.69 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





