Submitted:
21 September 2023
Posted:
22 September 2023
You are already at the latest version
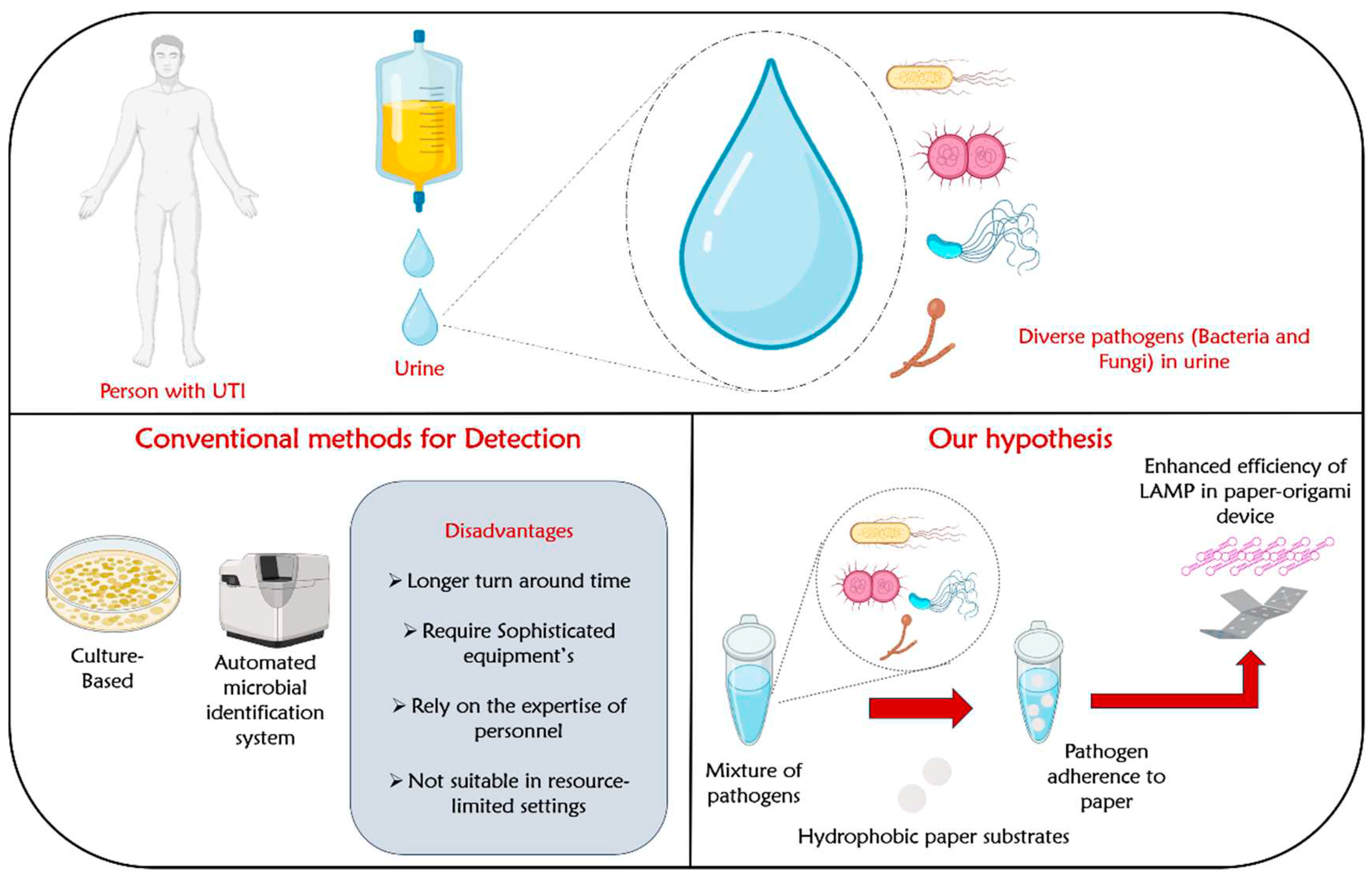
Abstract
Keywords:
Introduction
The Hypotheses
Evaluation of the Hypotheses
| Sl. No | Hydrophobic Material | Pathogen | Detection Method | Ref |
|---|---|---|---|---|
| 1 | Polymeric substrate film | S. aureus and E. coli | Fluorescence assay with Green Fluorescent Protein (GFP) and Bright Field Microscopy | [35] |
| 2 | Knitted polypropylene (PP) and poly-4-hydroxybutyrate (P4HB) | S. aureus and E. coli | Scanning Electron Microscopy (SEM) | [47] |
| 3 | Titanium dioxide (TiO2) surface | S. epidermidis | Fluorescence | [48] |
| 4 | Plastic surface | C. albicans | Hemacytometer measurement | [49] |
| 5 | Silane surface | Two strainsof E. coli, JM109 and D21 and two strains of B. cepacia, G4and Env435 | Column adhesion tests | [50] |
| 6 | Hydrophobic Steel Surface | E. coli | Scanning Electron Microscopy (SEM) | [51] |
Limitations
- (1)
- In the initial phase of adhesion, there is a possibility for capturing non-target microbes or metabolites. This is because when two different pathogens share similar hydrophobicity and diffusion coefficients, they typically exhibit similar adherence patterns to hydrophobic paper substrates.
- (2)
- Small variations in the fabrication of hydrophobic paper surfaces, or the surface properties of fabricated hydrophobic paper may lead to inconsistencies in microbial adhesion and pre-concentration efficiency. This would affect the reproducibility of results between different batches or experiments.
- (3)
- There can be various other factors apart from cell surface hydrophobicity (CSH) which could impact the adhesion of the cells to a particular surface. The impact of these confounding factors also needs to be considered. The rate of adhesion can differ depending on the culture conditions of the sample, and hence reproducibility of the results might become an issue.
- (4)
- Due to limited research in the field of microbial adhesion onto hydrophobic paper adsorbents, there may be significant aspects of the topic that have not been sufficiently addressed in this manuscript. Consequently, it is essential to conduct experimental verification of the hypothesis in future studies to ensure its validity.
- (5)
- Consequence of the Hypotheses and Discussion
Consent statement/Ethical approval
Data statement
Declaration of Competing Interest
Funding
References
- Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Hou K, Wu ZX, Chen XY, Wang JQ, Zhang D, Xiao C, et al. Microbiota in health and diseases. Signal Transduct Target Ther 2022, 7. [Google Scholar] [CrossRef]
- McLellan LK, Hunstad DA. Urinary Tract Infection: Pathogenesis and Outlook. Trends Mol Med 2016, 22, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Hasandka A, Singh AR, Prabhu A, Singhal HR, Nandagopal MSG, Mani NK. Paper and thread as media for the frugal detection of urinary tract infections (UTIs). Anal Bioanal Chem 2022, 414, 847–865. [Google Scholar] [CrossRef]
- Adrover-Jaume C, Rojo-Molinero E, Clemente A, Russell SM, Arranz J, Oliver A, et al. Mobile origami immunosensors for the rapid detection of urinary tract infections. Analyst 2020, 145, 7916–7921. [Google Scholar] [CrossRef]
- Simmering JE, Tang F, Cavanaugh JE, Polgreen LA, Polgreen PM. The Increase in Hospitalizations for Urinary Tract Infections and the Associated Costs in the United States, 1998-2011. Open Forum Infect Dis 2017, 4, ofw281. [CrossRef]
- Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Urinary tract infection caused by bacteria other than Escherichia coli. Arch Dis Child 2006, 91, 168.
- Sabih A, Leslie SW. Complicated Urinary Tract Infections., Treasure Island (FL): 2023.
- Torres-Sangiao E, Lamas Rodriguez B, Cea Pájaro M, Carracedo Montero R, Parajó Pazos N, García-Riestra C. Direct Urine Resistance Detection Using VITEK 2. Antibiot (Basel, Switzerland) 2022, 11. [CrossRef]
- Huang B, Zhang L, Zhang W, Liao K, Zhang S, Zhang Z, et al. Direct Detection and Identification of Bacterial Pathogens from Urine with Optimized Specimen Processing and Enhanced Testing Algorithm. J Clin Microbiol 2017, 55, 1488–1495. [Google Scholar] [CrossRef]
- Sudarsan S, Prabhu A, Prasad D, Mani NK. DNA Compaction Enhances Sensitivity of Fluorescence-Based Nucleic Acid Assays: Game changer in Point of Care Sensors? Analyst 2023.
- Kelkar N, Prabhu A, Prabhu A, Giri Nandagopal MS, Mani NK. Sensing of body fluid hormones using paper-based analytical devices. Microchem J 2022, 174, 107069. [Google Scholar] [CrossRef]
- Hasandka A, Prabhu A, Prabhu A, Singhal HR, Nandagopal M. S. G, Shenoy R, et al. “Scratch it out”: carbon copy based paper devices for microbial assays and liver disease diagnosis. Anal Methods 2021, 13, 3172–3180. [Google Scholar] [CrossRef]
- Prabhu A, Singhal H, Giri Nandagopal MS, Kulal R, Peralam Yegneswaran P, Mani NK. Knitting Thread Devices: Detecting Candida albicans Using Napkins and Tampons. ACS Omega 2021, 6, 12667–12675. [Google Scholar] [CrossRef]
- Singhal HR, Prabhu A, Giri Nandagopal MS, Dheivasigamani T, Mani NK. One-dollar microfluidic paper-based analytical devices: Do-It-Yourself approaches. Microchem J 2021, 165, 106126. [Google Scholar] [CrossRef]
- Prabhu A, Nandagopal M. S. G, Peralam Yegneswaran P, Prabhu V, Verma U, Mani NK. Thread integrated smart-phone imaging facilitates early turning point colorimetric assay for microbes. RSC Adv 2020, 10, 26853–26861. [Google Scholar] [CrossRef] [PubMed]
- Prabhu A, Giri Nandagopal MS, Peralam Yegneswaran P, Singhal HR, Mani NK. Inkjet printing of paraffin on paper allows low-cost point-of-care diagnostics for pathogenic fungi. Cellulose 2020, 27, 7691–7701. [Google Scholar] [CrossRef]
- Mani NK, Das SS, Dawn S, Chakraborty S. Electro-kinetically driven route for highly sensitive blood pathology on a paper-based device. Electrophoresis 2020, 41, 615–620. [Google Scholar] [CrossRef]
- Mani NK, Prabhu A, Biswas SK, Chakraborty S. Fabricating Paper Based Devices Using Correction Pens. Sci Rep 2019, 9, 1752. [Google Scholar] [CrossRef]
- Ray R, Goyal A, Prabhu A, Parekkh S, Maddasani S, Mani NK. Paper-based dots and smartphone for detecting counterfeit country eggs. Food Chem 2023, 403, 134484. [Google Scholar] [CrossRef]
- Ray R, Noronha C, Prabhu A, Mani NK. Latex-Based Paper Devices with Super Solvent Resistance for On-the-Spot Detection of Metanil Yellow in Food Samples. Food Anal Methods 2022, 15, 2664–2674. [Google Scholar] [CrossRef]
- Ray R, Prabhu A, Prasad D, Garlapati V kumar, Aminabhavi TM, Mani NK, et al. Paper-based microfluidic devices for food adulterants: Cost-effective technological monitoring systems. Food Chem 2022, 390, 133173. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi-Aghdam A, Majidi MR, Omidi Y. Microfluidic paper-based analytical devices (µPADs) for fast and ultrasensitive sensing of biomarkers and monitoring of diseases. Bioimpacts 2018, 8, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai RK, Pudasaini S, Sah M, Neupane BB, Giri B. Handmade Paper as a Paper Analytical Device for Determining the Quality of an Antidiabetic Drug. ACS Omega 2022, 7, 14074–14081. [Google Scholar] [CrossRef] [PubMed]
- Campbell JM, Balhoff JB, Landwehr GM, Rahman SM, Vaithiyanathan M, Melvin AT. Microfluidic and Paper-Based Devices for Disease Detection and Diagnostic Research. Int J Mol Sci 2018, 19, 2731. [Google Scholar] [CrossRef]
- A. , Shafiq CWNSSJRB. Paper-based analytical devices for clinical diagnosis. Physiol Behav 2017, 176, 139–148. [Google Scholar] [CrossRef]
- Sher M, Zhuang R, Demirci U, Asghar W. Paper-based analytical devices for clinical diagnosis: recent advances in the fabrication techniques and sensing mechanisms. Expert Rev Mol Diagn 2017, 17, 351–366. [Google Scholar] [CrossRef]
- Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Diagnostics for the Developing World: Microfluidic Paper-Based Analytical Devices. Anal Chem 2010, 82, 3–10. [Google Scholar] [CrossRef]
- St John A, Price CP. Existing and Emerging Technologies for Point-of-Care Testing. Clin Biochem Rev 2014, 35, 155–167. [Google Scholar]
- Notomi T, Mori Y, Tomita N, Kanda H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol 2015, 53, 1–5. [Google Scholar] [CrossRef]
- Geddes-McAlister J, Shapiro RS. New pathogens, new tricks: emerging, drug-resistant fungal pathogens and future prospects for antifungal therapeutics. Ann N Y Acad Sci 2019, 1435, 57–78. [Google Scholar] [CrossRef]
- Özay B, McCalla SE. A review of reaction enhancement strategies for isothermal nucleic acid amplification reactions. Sensors and Actuators Reports 2021, 3. [Google Scholar] [CrossRef]
- Zheng S, Bawazir M, Dhall A, Kim HE, He L, Heo J, et al. Implication of Surface Properties, Bacterial Motility, and Hydrodynamic Conditions on Bacterial Surface Sensing and Their Initial Adhesion. Front Bioeng Biotechnol 2021, 9, 1–22. [Google Scholar] [CrossRef]
- Yuan Y, Hays MP, Hardwidge PR, Kim J. Surface characteristics influencing bacterial adhesion to polymeric substrates. RSC Adv 2017, 7, 14254–14261. [Google Scholar] [CrossRef]
- Oh JK, Yegin Y, Yang F, Zhang M, Li J, Huang S, et al. The influence of surface chemistry on the kinetics and thermodynamics of bacterial adhesion. Sci Rep 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Tegoulia VA, Cooper SL. Staphylococcus aureus adhesion to self-assembled monolayers: effect of surface chemistry and fibrinogen presence. Colloids Surfaces B Biointerfaces 2002, 24, 217–228. [Google Scholar] [CrossRef]
- Krasowska A, Sigler K. How microorganisms use hydrophobicity and what does this mean for human needs? Front Cell Infect Microbiol 2014, 4, 1–7. [Google Scholar] [CrossRef]
- Heipieper HJ, Pepi M. Handbook of Hydrocarbon and Lipid Microbiology. Handb Hydrocarb Lipid Microbiol 2010. [CrossRef]
- Bayry J, Aimanianda V, Guijarro JI, Sunde M, Latgé J-P. Hydrophobins--unique fungal proteins. PLoS Pathog 2012, 8, e1002700. [Google Scholar] [CrossRef]
- Farniya F, Jamalli A, Dadgar T. Physicochemical surface characteristics in different pathogenic bacteria. Cogent Biol 2019, 5, 1638572. [Google Scholar] [CrossRef]
- 42. Nogueira BA, Olivella JGB, Sued-Karam BR, Ribeiro PMAP, Neves FPG, Fracalanzza SEL, et al. Biofilm formation, interaction and survival within A549 pneumocytes of Klebsiella pneumoniae clinical strains: identification of pulsotypes, multidrug-resistance and genes coding for adhesins. Brazilian J Dev 2022, 55259–55287. [CrossRef]
- Czerwonka G, Guzy A, Kałuża K, Grosicka M, Dańczuk M, Lechowicz Ł, et al. The role of Proteus mirabilis cell wall features in biofilm formation. Arch Microbiol 2016, 198, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Maikranz E, Spengler C, Thewes N, Thewes A, Nolle F, Jung P, et al. Different binding mechanisms of: Staphylococcus aureus to hydrophobic and hydrophilic surfaces. Nanoscale 2020, 12, 19267–19275. [Google Scholar] [CrossRef] [PubMed]
- Reifsteck F, Wee S, Wilkinson BJ. Hydrophobicity-hydrophilicity of staphylococci. J Med Microbiol 1987, 24, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Ellepola ANB, Samaranayake LP. Investigative methods for studying the adhesion and cell surface hydrophobicity of candida species: An overview. Microb Ecol Health Dis 2001, 13, 46–54. [Google Scholar] [CrossRef]
- Verhorstert KWJ, Guler Z, De Boer L, Riool M, Roovers JPWR, Zaat SAJ. In Vitro Bacterial Adhesion and Biofilm Formation on Fully Absorbable Poly-4-hydroxybutyrate and Nonabsorbable Polypropylene Pelvic Floor Implants. ACS Appl Mater Interfaces 2020, 12, 53646–53653. [Google Scholar] [CrossRef]
- Wassmann T, Kreis S, Behr M, Buergers R. The influence of surface texture and wettability on initial bacterial adhesion on titanium and zirconium oxide dental implants. Int J Implant Dent 2017, 3. [Google Scholar] [CrossRef]
- Klotz SA, Drutz DJ, Zajic JE. Factors governing adherence of Candida species to plastic surfaces. Infect Immun 1985, 50, 97–101. [Google Scholar] [CrossRef]
- Salerno MB, Logan BE, Velegol D. Importance of molecular details in predicting bacterial adhesion to hydrophobie surfaces. Langmuir 2004, 20, 10625–10629. [Google Scholar] [CrossRef]
- Arkan-Ozdemir S, Cansever N, Ilhan-Sungur E. Biofilm Formation of Escherichia coli on Hydrophobic Steel Surface Provided by Laser-Texturing. Johnson Matthey Technol Rev 2022, 186–196. [CrossRef]
- Kim HT, Jung SK, Kim D-E, Park CY, Lee S-Y. Wettability control of paper through substitution between the hydroxyl group and carbon elements using argon-carbon plasma treatment. Vacuum 2022, 205, 111398. [Google Scholar] [CrossRef]
- Gómez N, Quintana E, Villar JC. Effect of Paper Surface Properties on Coated Paper Wettability with Different Fountain Solutions. BioResources 2014, 9, 4226–4241. [Google Scholar] [CrossRef]
- Modaressi H, Garnier G. Mechanism of Wetting and Absorption of Water Droplets on Sized Paper: Effects of Chemical and Physical Heterogeneity. Langmuir 2002, 18, 642–649. [Google Scholar] [CrossRef]
- Wen Q, Guo F, Yang F, Guo Z. Green fabrication of coloured superhydrophobic paper from native cotton cellulose. J Colloid Interface Sci 2017, 497, 284–289. [Google Scholar] [CrossRef]
- Baidya A, Ganayee MA, Jakka Ravindran S, Tam KC, Das SK, Ras RHA, et al. Organic solvent-free fabrication of durable and multifunctional superhydrophobic paper from waterborne fluorinated cellulose nanofiber building blocks. ACS Nano 2017, 11, 11091–11099. [Google Scholar] [CrossRef] [PubMed]
- Balu B, Kim JS, Breedveld V, Hess DW. Tunability of the adhesion of water drops on a superhydrophobic paper surface via selective plasma etching. J Adhes Sci Technol 2009, 23, 361–380. [Google Scholar] [CrossRef]
- Hu Z, Zen X, Gong J, Deng Y. Water resistance improvement of paper by superhydrophobic modification with microsized CaCO3 and fatty acid coating. Colloids Surfaces A Physicochem Eng Asp 2009, 351, 65–70. [Google Scholar] [CrossRef]
- Werner O, Quan C, Turner C, Pettersson B, Wågberg L. Properties of superhydrophobic paper treated with rapid expansion of supercritical CO 2 containing a crystallizing wax. Cellulose 2010, 17, 187–198. [Google Scholar] [CrossRef]
- Yang H, Deng Y. Preparation and physical properties of superhydrophobic papers. J Colloid Interface Sci 2008, 325, 588–593. [Google Scholar] [CrossRef]
- Arbatan T, Zhang L, Fang X-Y, Shen W. Cellulose nanofibers as binder for fabrication of superhydrophobic paper. Chem Eng J 2012, 210, 74–79. [Google Scholar] [CrossRef]
- Carlmark A, Malmström EE. ATRP grafting from cellulose fibers to create block-copolymer grafts. Biomacromolecules 2003, 4, 1740–1745. [Google Scholar] [CrossRef]
- David S, Munteanu RE, Tițoiu AM, Petcu IC, Cernat IC, Leancu C, et al. Direct, Rapid Detection of Pathogens from Urine Samples. Materials (Basel) 2022, 15, 7640. [Google Scholar] [CrossRef] [PubMed]
- Wallis C, Melnick JL, Longoria CJ. Colorimetric method for rapid determination of bacteriuria. J Clin Microbiol 1981, 14, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Luka GS, Nowak E, Kawchuk J, Hoorfar M, Najjaran H. Portable device for the detection of colorimetric assays. R Soc Open Sci 2017, 4, 171025. [Google Scholar] [CrossRef] [PubMed]
- Qian S, Cui Y, Cai Z, Li L. Applications of smartphone-based colorimetric biosensors. Biosens Bioelectron X 2022, 11, 100173. [Google Scholar] [CrossRef]
| Sl. No | Type of pathogen | Name of pathogen | Favourable Surface | Ref |
|---|---|---|---|---|
| 1 | Gram negative | E. coli | Hydrophobic | [41] |
| 2 | Gram negative | Klebsiella pneumoniae | Hydrophobic | [42] |
| 3 | Gram negative | Pseudomonas aeruginosa | Hydrophobic | [41] |
| 4 | Gram negative | Proteus mirabilis | Hydrophobic | [43] |
| 5 | Gram positive | Staphylococcus aureus | Hydrophobic and Hydrophilic | [37,44,45] |
| 6 | Gram positive | Listeria monocytogenes | Hydrophobic | [41] |
| 7 | Yeast | Candida albicans | Hydrophobic | [46] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





