Submitted:
01 September 2023
Posted:
05 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
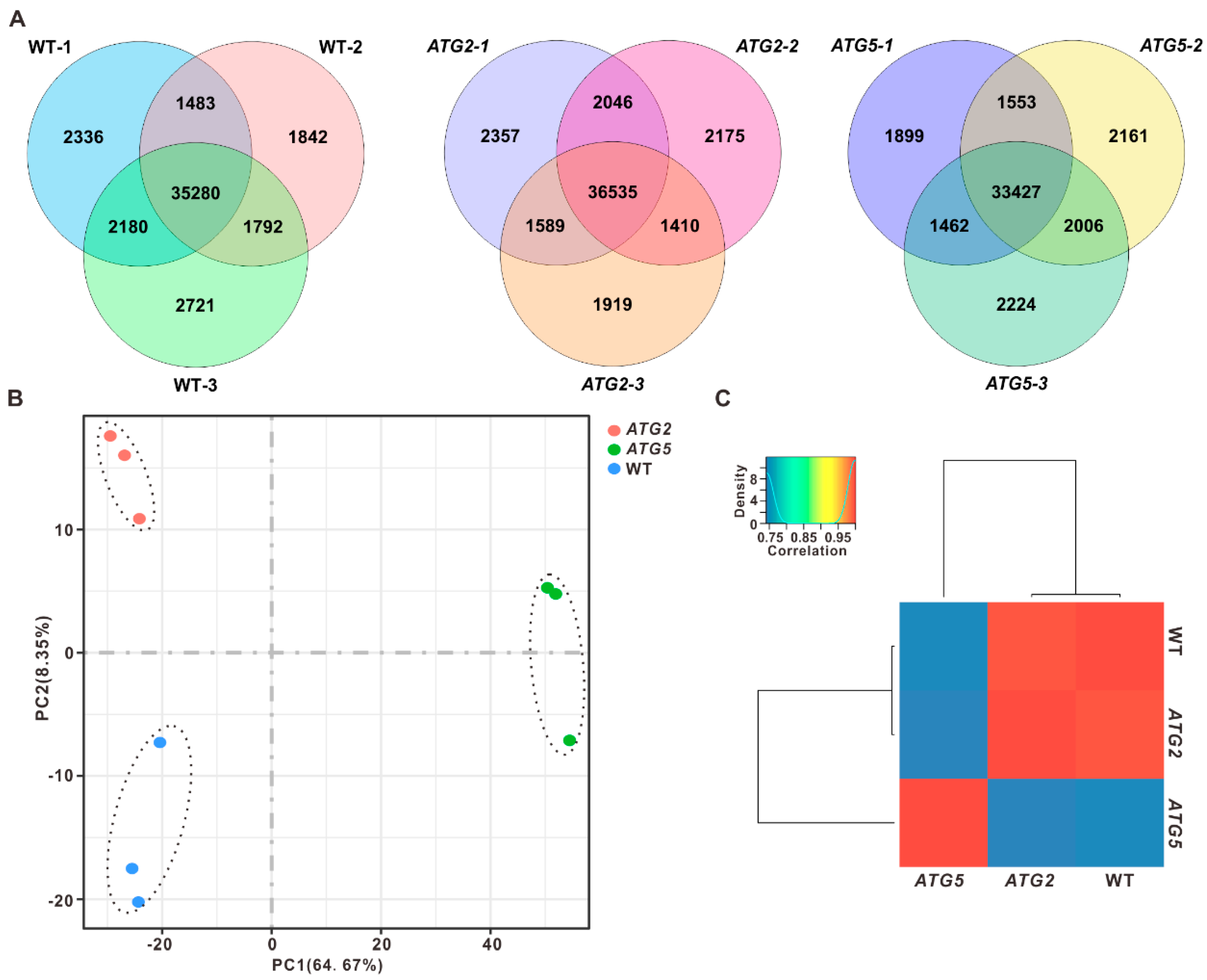
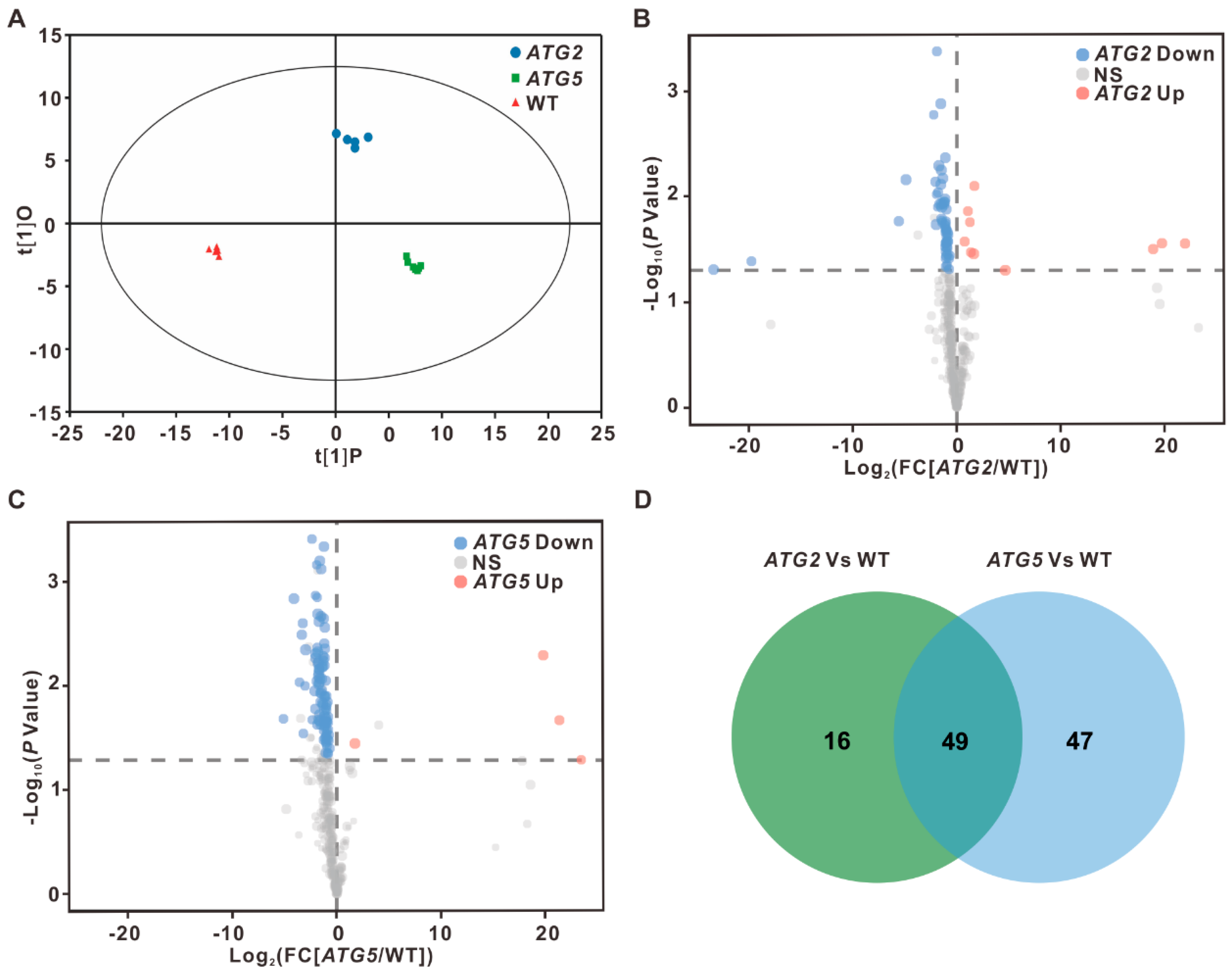
2.1. Global impact of silencing of ATG2 and ATG5 on pollen transcriptome
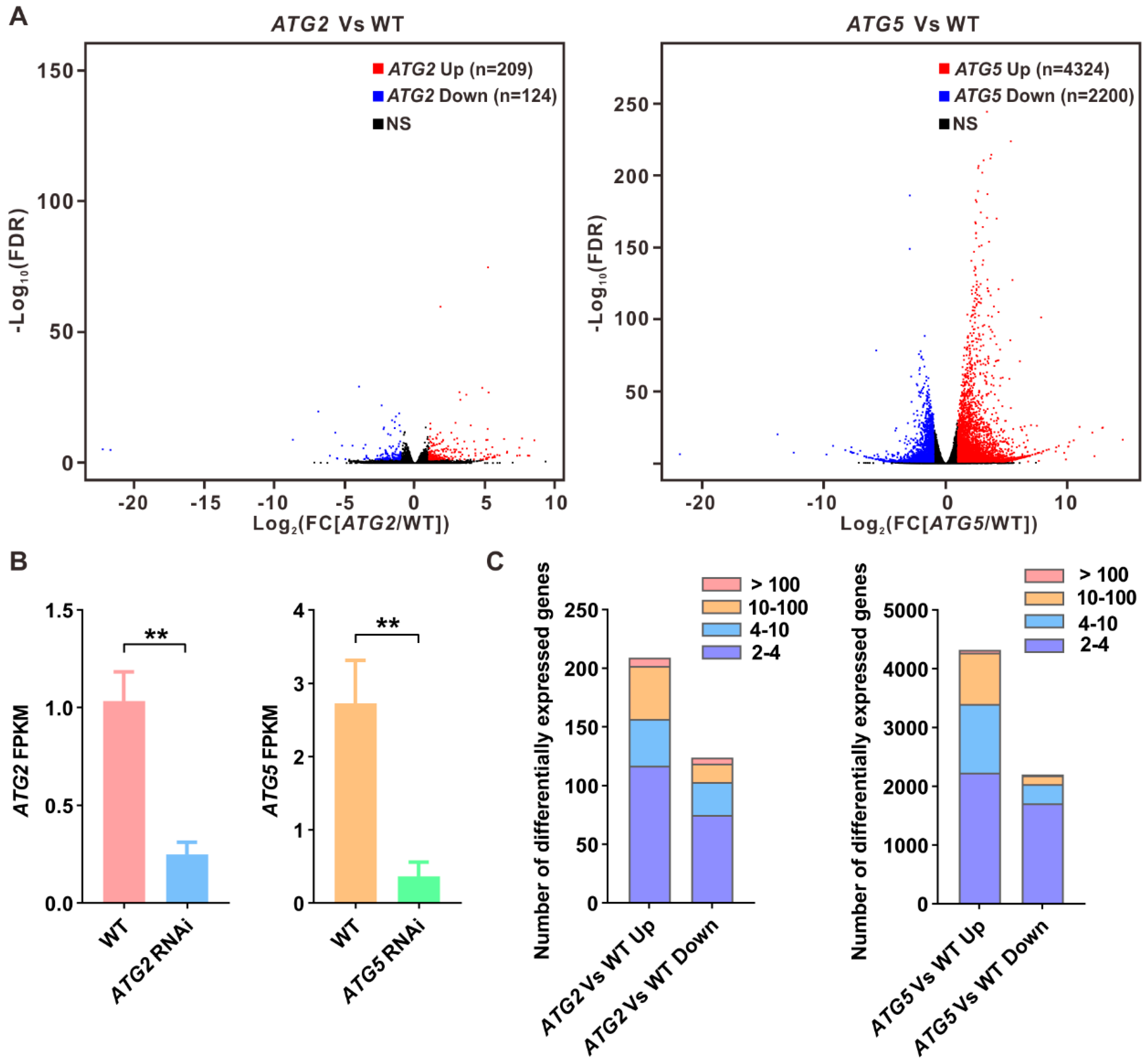
2.2. Different impact of silencing of ATG2 and ATG5 on pollen transcriptome
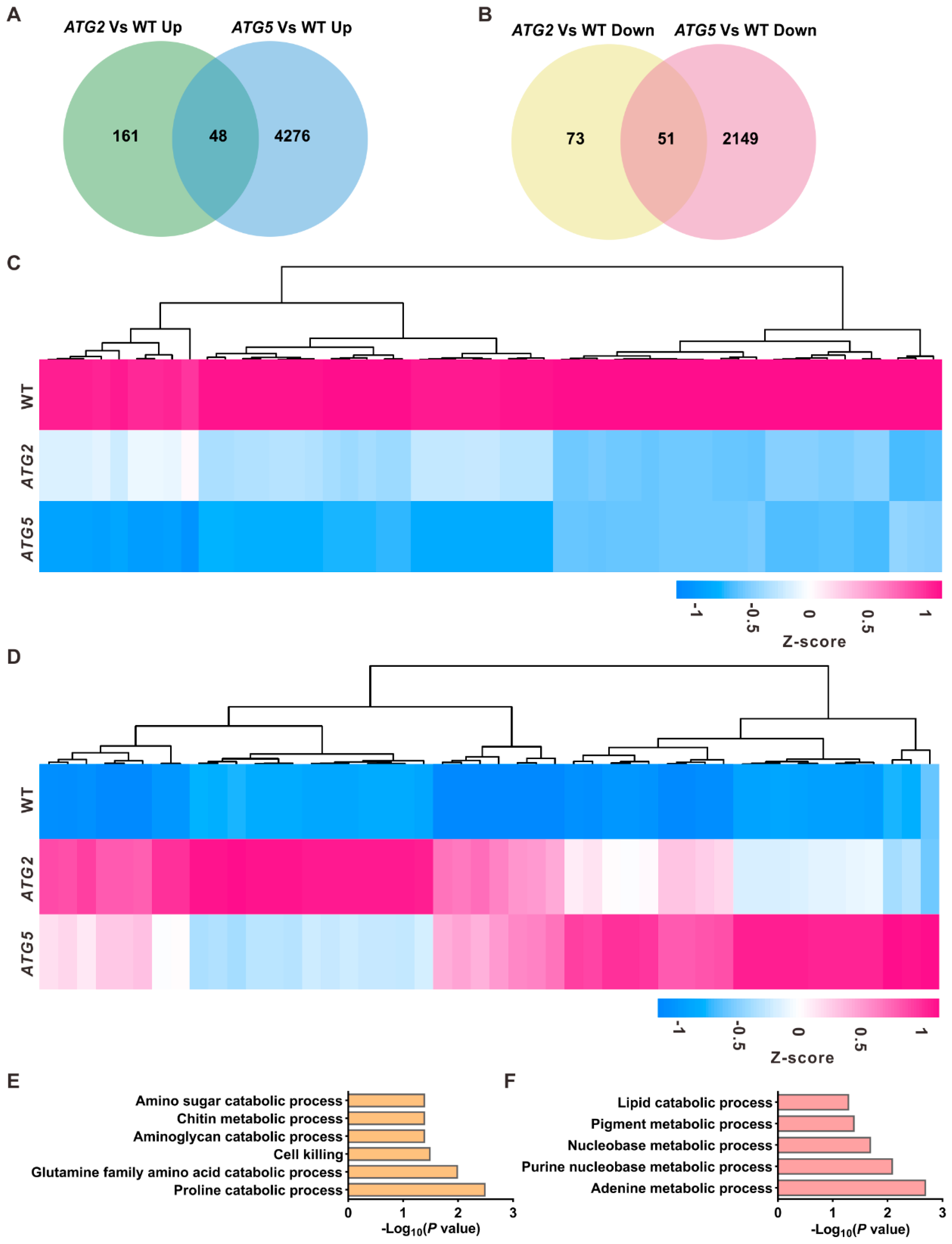
2.3. The levels of transcripts linked to metabolic process changed in ATG2- and ATG5-silenced pollen
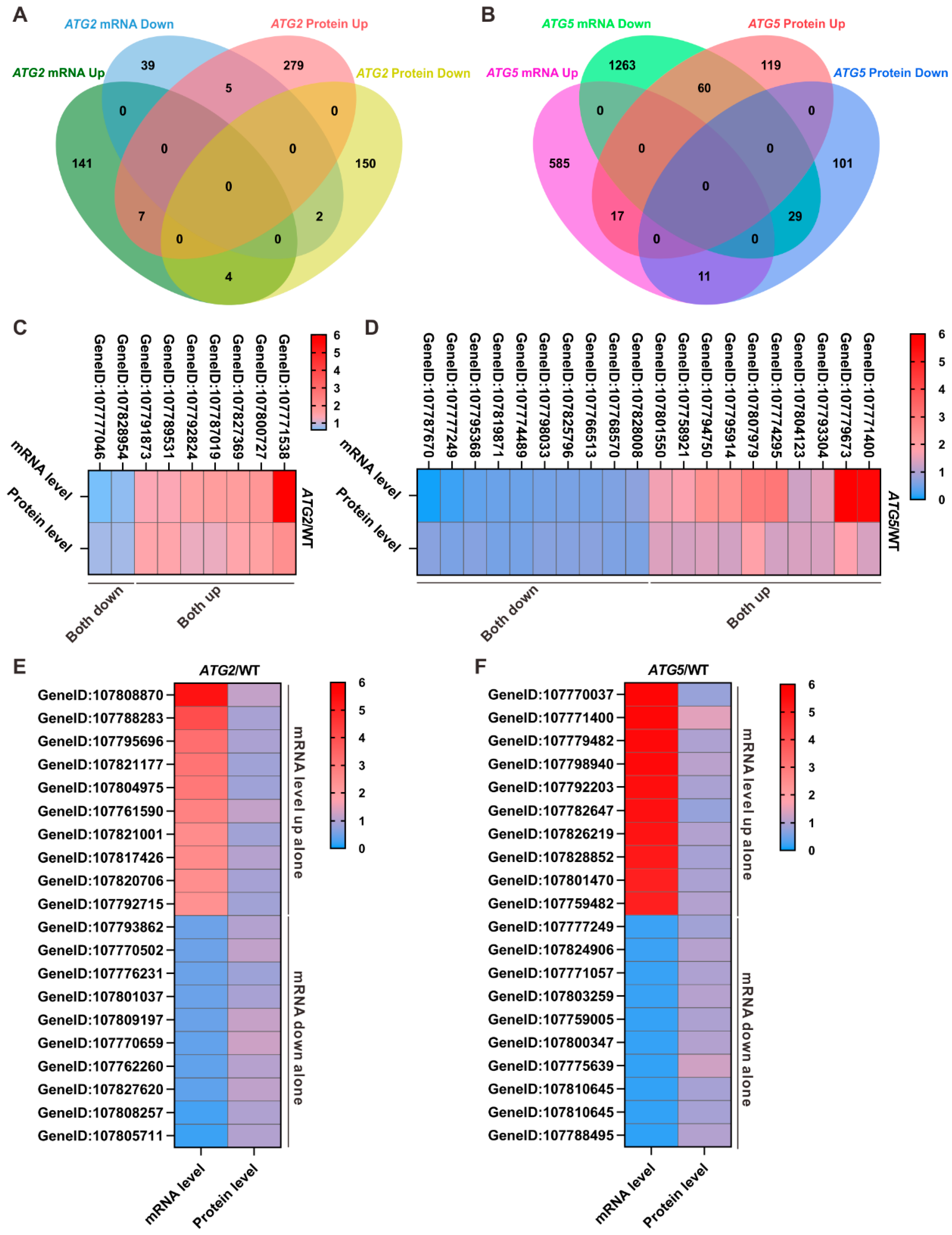
2.4. The expressions at mRNA level and protein level are differentially affected by autophagy
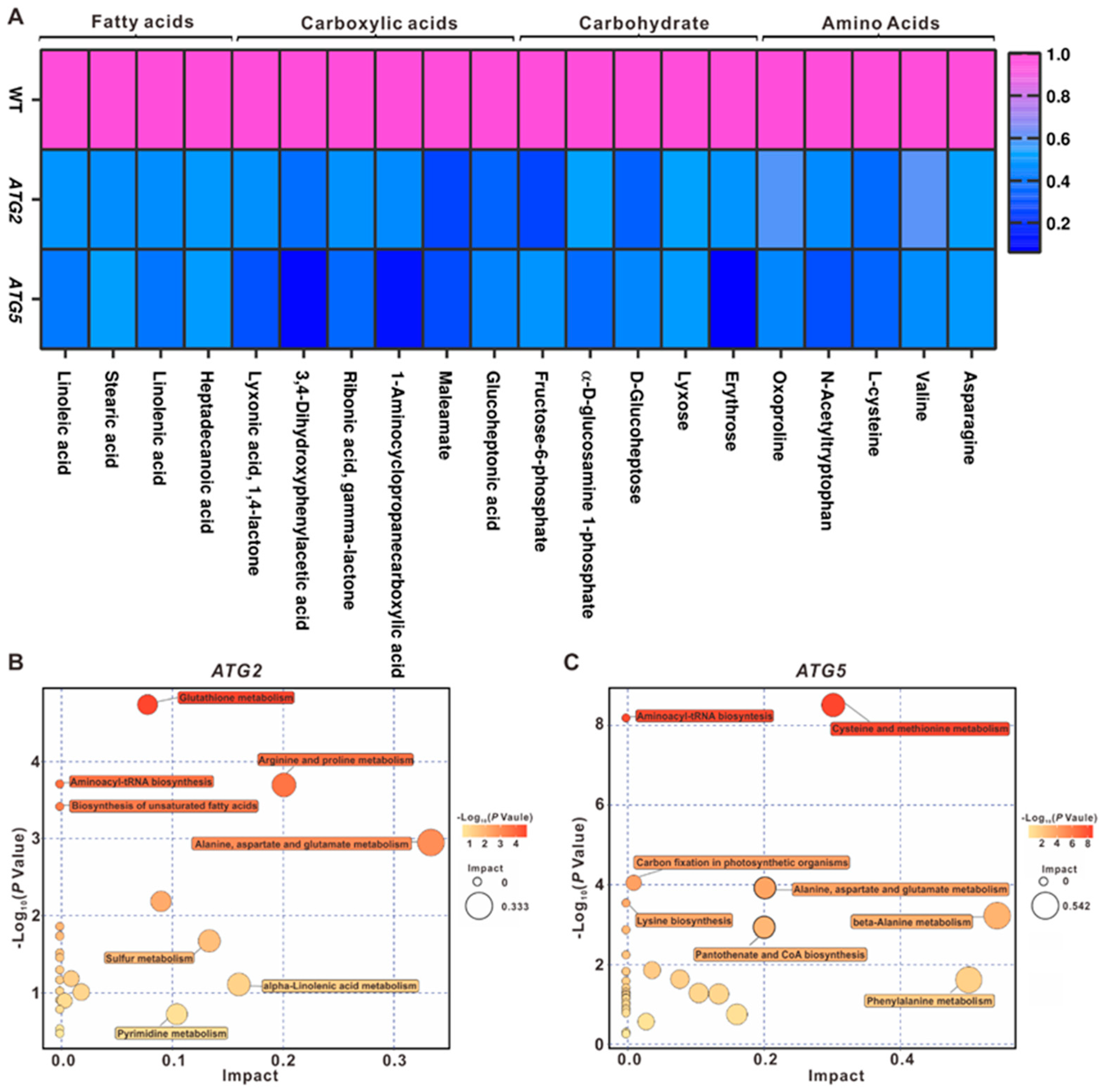
2.5. Down-regulation of ATGs leads to the decrease of metabolic level in pollen
2.6. Metabolic pathways impacted by autophagy
3. Discussion
3.1. Differential regulation of downstream genes of autophagy at mRNA and protein levels
3.2. Autophagy-dependent cellular metabolism is critical for promoting pollen germination
3.3. Differences and similarities between tobacco and Arabidopsis atg5 transcriptomes
4. Materials and Methods
4.1. Plant materials
4.2. RNA isolation and RNA-seq
4.3. RNA-seq data analysis
4.4. GO analysis
4.5. KEGG analysis
4.6. High-throughput analysis of primary metabolites in pollen using GC-TOF/MS
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Han, S.; Yu, B.; Wang, Y.; Liu, Y. , Role of plant autophagy in stress response. Protein & cell 2011, 2, 784–791. [Google Scholar]
- Kim, J.; Lee, H.; Lee, H. N.; Kim, S. H.; Shin, K. D.; Chung, T. , Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 2013, 25, 4956–4966. [Google Scholar] [CrossRef] [PubMed]
- Wada, S.; Ishida, H.; Izumi, M.; Yoshimoto, K.; Ohsumi, Y.; Mae, T.; Makino, A. , Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol 2009, 149, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, B.; Zhao, J.; Guo, J.; Li, Y.; Han, S.; Huang, L.; Du, Y.; Hong, Y.; Tang, D.; Liu, Y. , Autophagy contributes to leaf starch degradation. Plant Cell 2013, 25, 1383–1399. [Google Scholar] [CrossRef] [PubMed]
- Avin-Wittenberg, T.; Bajdzienko, K.; Wittenberg, G.; Alseekh, S.; Tohge, T.; Bock, R.; Giavalisco, P.; Fernie, A. R. , Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell 2015, 27, 306–322. [Google Scholar] [CrossRef]
- Have, M.; Luo, J.; Tellier, F.; Balliau, T.; Cueff, G.; Chardon, F.; Zivy, M.; Rajjou, L.; Cacas, J. L.; Masclaux-Daubresse, C. , Proteomic and lipidomic analyses of the Arabidopsis atg5 autophagy mutant reveal major changes in endoplasmic reticulum and peroxisome metabolisms and in lipid composition. New Phytol 2019, 223, 1461–1477. [Google Scholar] [CrossRef]
- McLoughlin, F.; Augustine, R. C.; Marshall, R. S.; Li, F.; Kirkpatrick, L. D.; Otegui, M. S.; Vierstra, R. D. , Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nat Plants 2018, 4, 1056–1070. [Google Scholar] [CrossRef]
- Zhao, P.; Zhou, X. M.; Zhao, L. L.; Cheung, A. Y.; Sun, M. X. , Autophagy-mediated compartmental cytoplasmic deletion is essential for tobacco pollen germination and male fertility. Autophagy 2020, 1–13. [Google Scholar] [CrossRef]
- Meijer, W. H.; van der Klei, I. J.; Veenhuis, M.; Kiel, J. A. , ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 2007, 3, 106–116. [Google Scholar] [CrossRef]
- Xia, K.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M. , Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res 2011, 18, 363–377. [Google Scholar] [CrossRef]
- Zhou, X. M.; Zhao, P.; Wang, W.; Zou, J.; Cheng, T. H.; Peng, X. B.; Sun, M. X. , A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res 2015, 22, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Doelling, J. H.; Walker, J. M.; Friedman, E. M.; Thompson, A. R.; Vierstra, R. D. , The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 2002, 277, 33105–33114. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A. R.; Doelling, J. H.; Suttangkakul, A.; Vierstra, R. D. , Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 2005, 138, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Contento, A. L.; Bassham, D. C. , AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J 2005, 42, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Fan, J.; Taylor, D. C.; Ohlrogge, J. B. , DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 2009, 21, 3885–3901. [Google Scholar] [CrossRef]
- Abildgaard, M. H.; Brynjolfsdottir, S. H.; Frankel, L. B. , The Autophagy-RNA Interplay: Degradation and Beyond. Trends Biochem Sci 2020, 45, 845–857. [Google Scholar] [CrossRef]
- Varshavsky, A. , The Ubiquitin System, Autophagy, and Regulated Protein Degradation. Annu Rev Biochem 2017, 86, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, W. Z.; Song, L. F.; Zou, J. J.; Su, Z.; Wu, W. H. , Transcriptome Analyses Show Changes in Gene Expression to Accompany Pollen Germination and Tube Growth in Arabidopsis. Plant Physiol 2008, 148, 1201–1211. [Google Scholar] [CrossRef]
- Obermeyer, G.; Fragner, L.; Lang, V.; Weckwerth, W. , Dynamic adaption of metabolic pathways during germination and growth of lily pollen tubes after inhibition of the electron transport chain. Plant Physiol 2013, 162, 1822–1833. [Google Scholar] [CrossRef]
- Rotsch, A. H.; Kopka, J.; Feussner, I.; Ischebeck, T. , Central metabolite and sterol profiling divides tobacco male gametophyte development and pollen tube growth into eight metabolic phases. Plant J 2017, 92, 129–146. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Clement, G.; Anne, P.; Routaboul, J. M.; Guiboileau, A.; Soulay, F.; Shirasu, K.; Yoshimoto, K. , Stitching together the Multiple Dimensions of Autophagy Using Metabolomics and Transcriptomics Reveals Impacts on Metabolism, Development, and Plant Responses to the Environment in Arabidopsis. Plant Cell 2014, 26, 1857–1877. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; Zhang, X.; Wang, J.; Yang, H.; Fang, L.; Chen, Q. , SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S. L. , HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics 2010, 11, 117. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C. N. , RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. Bmc Bioinformatics 2011, 12. [Google Scholar] [CrossRef]
- Love, M. I.; Huber, W.; Anders, S. , Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15, 550. [Google Scholar] [CrossRef]
- Guo, R.; Shi, L.; Yang, C.; Yan, C.; Zhong, X.; Liu, Q.; Xia, X.; Li, H. , Comparison of Ionomic and Metabolites Response under Alkali Stress in Old and Young Leaves of Cotton (Gossypium hirsutum L.) Seedlings. Frontiers in plant science 2016, 7, 1785. [Google Scholar] [CrossRef]
- Dunn, W. B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J. D.; Halsall, A.; Haselden, J. N.; Nicholls, A. W.; Wilson, I. D.; Kell, D. B.; Goodacre, R.; Human Serum Metabolome, C. , Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature protocols 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




