Submitted:
14 August 2023
Posted:
18 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant material
2.2. Extraction and immunodetection of S-nitrosylated wheat proteins
2.3. In-gel trypsin digestion
2.4. Solid-phase extraction
2.5. Nano LC-MS/MS
2.6. Data analysis
2.7. Bioinformatic analysis of the availability of S-nitrosylation sites of wheat proteins
2.8. Protein-ligand docking and protein-protein interactions
2.9. Statistical analysis
3. Results
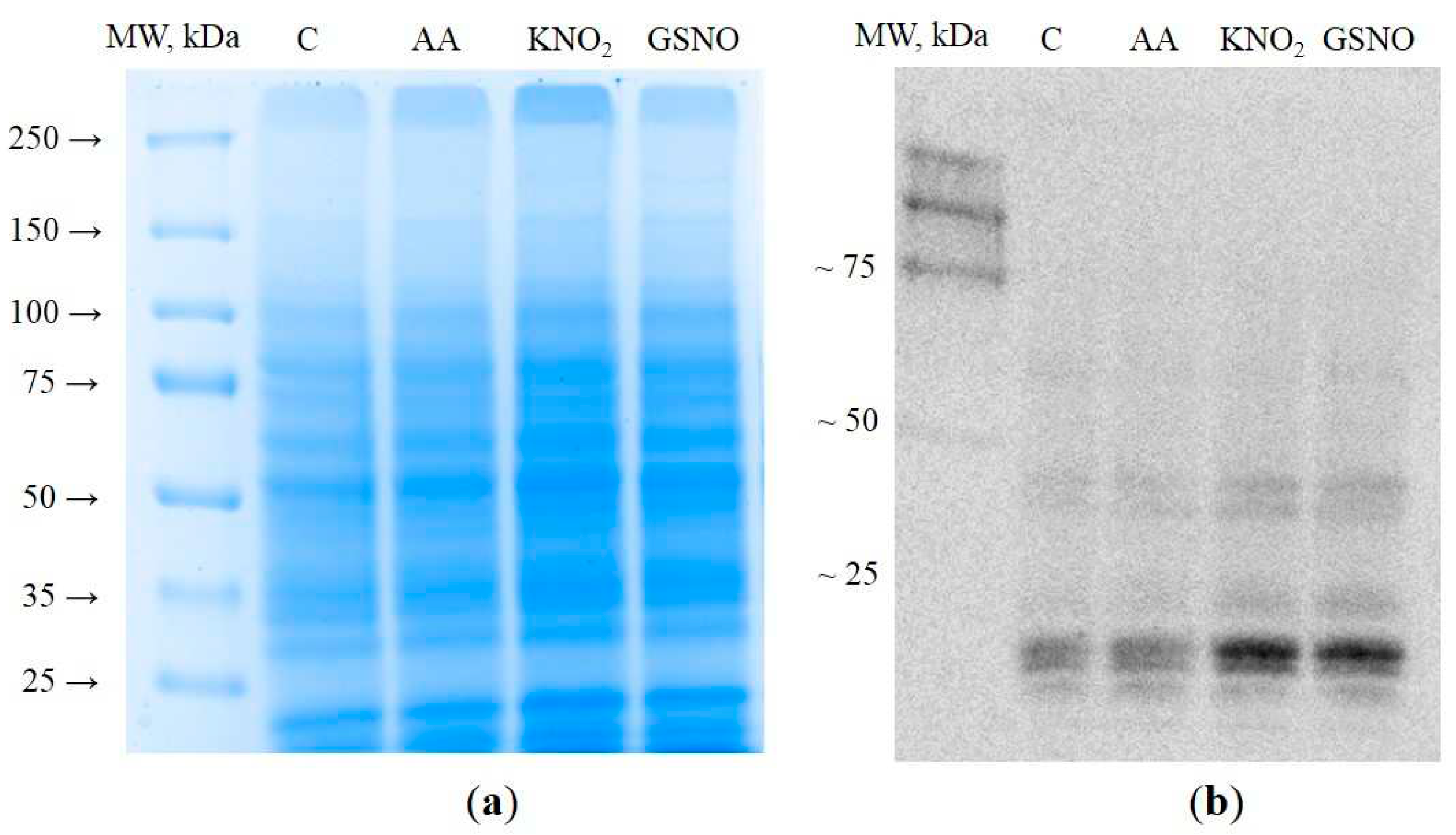
3.1. Extraction and visualization of S-nitrosylated proteins during induction of autophagy
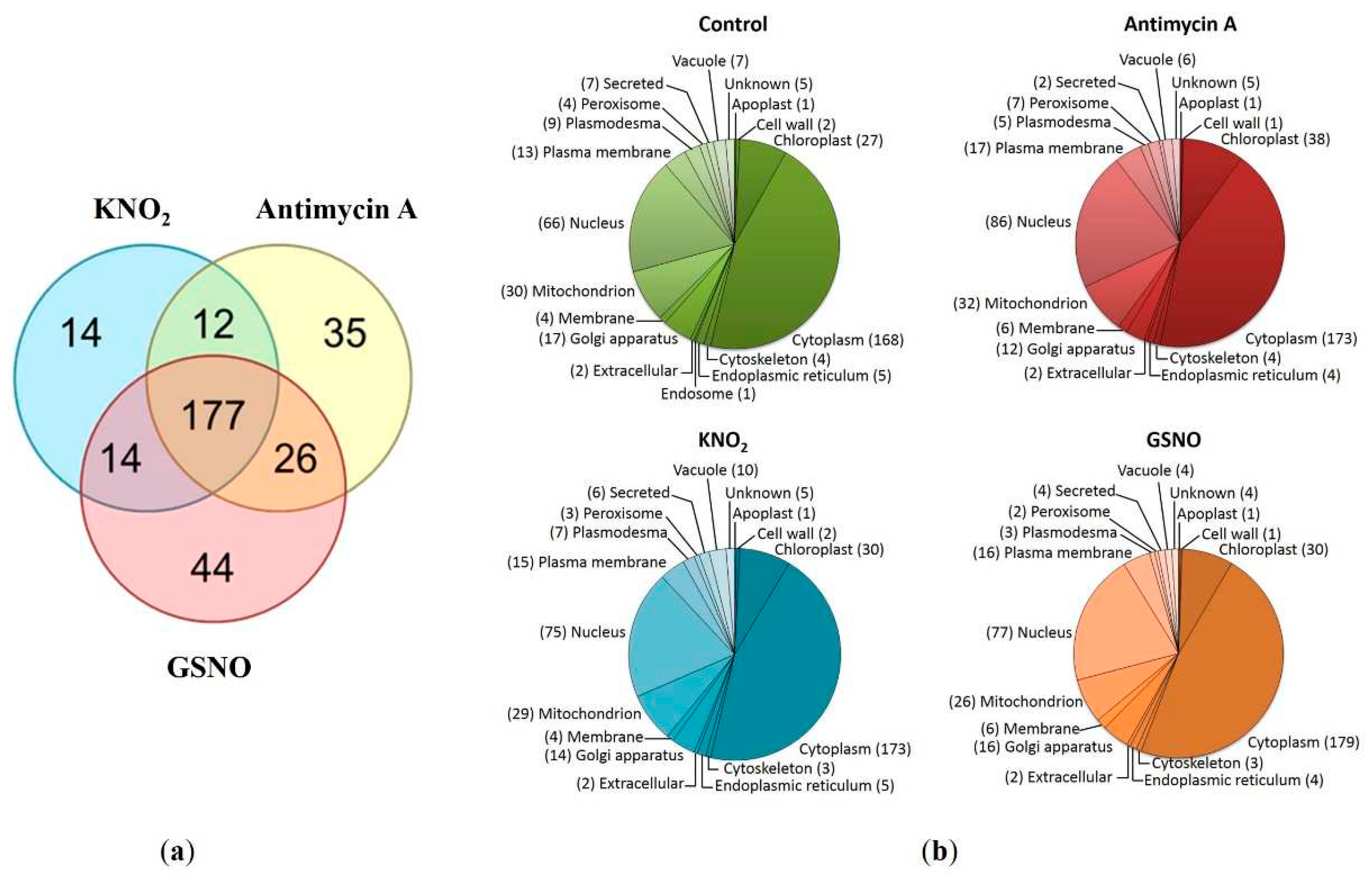
3.2. Protein identification and search for S-nitrosylation sites
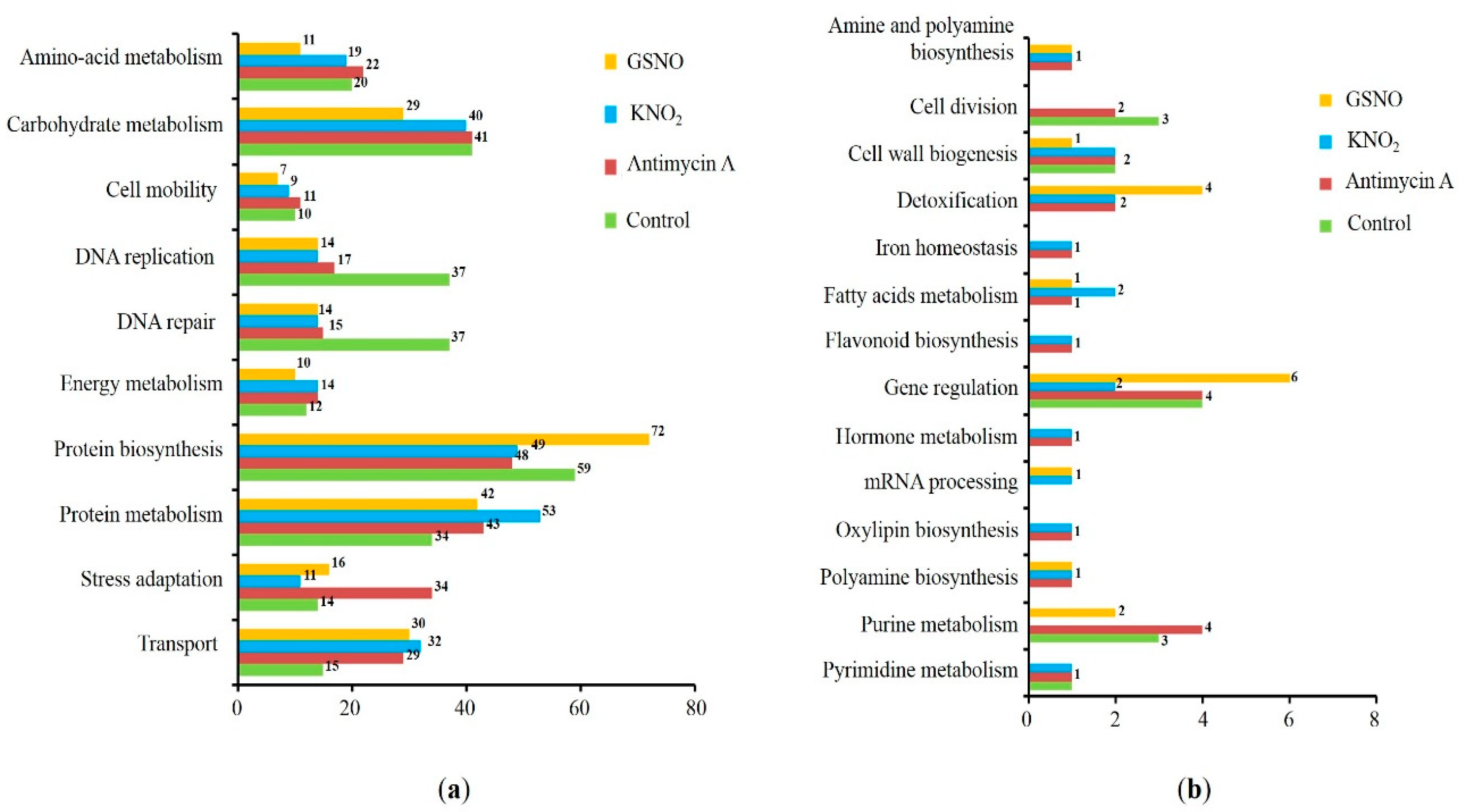
3.3. Prediction of S-nitrosylation sites, localization, and functional annotation of S-nitrosylated proteins
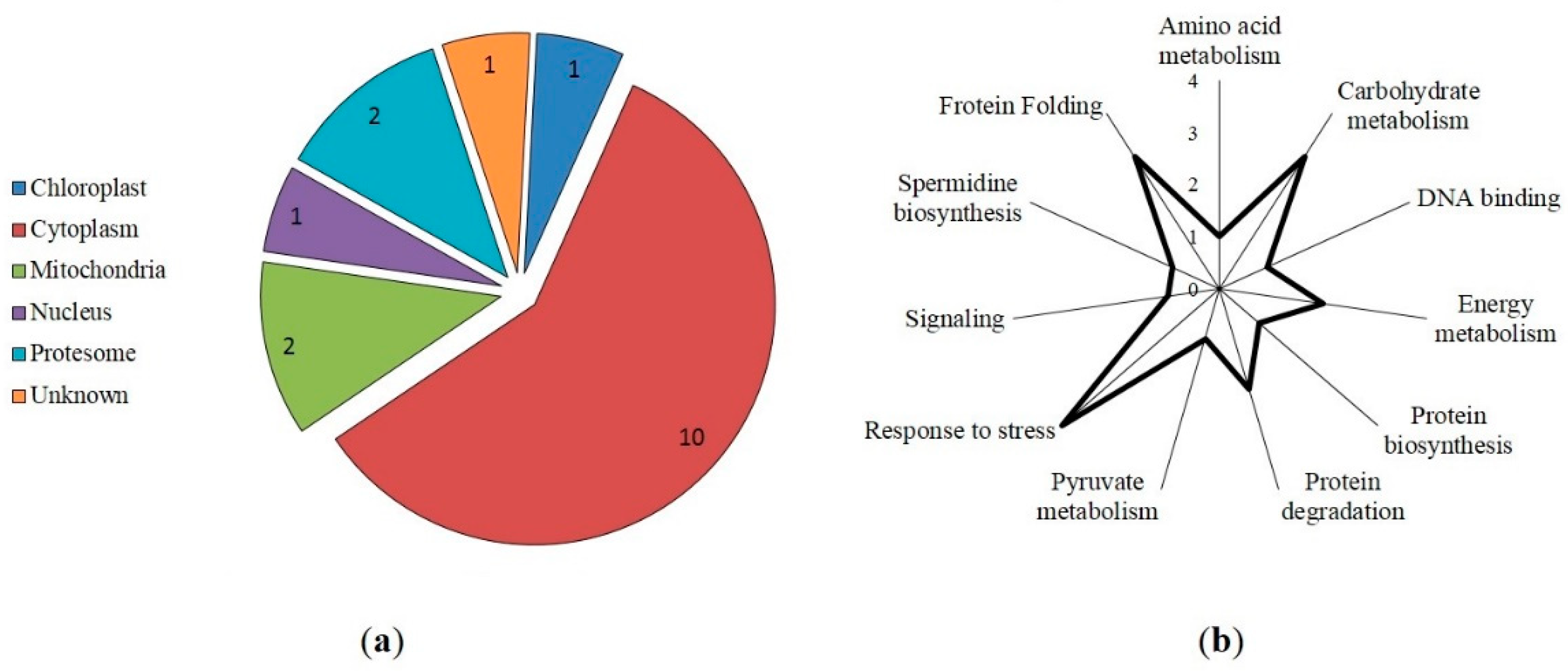
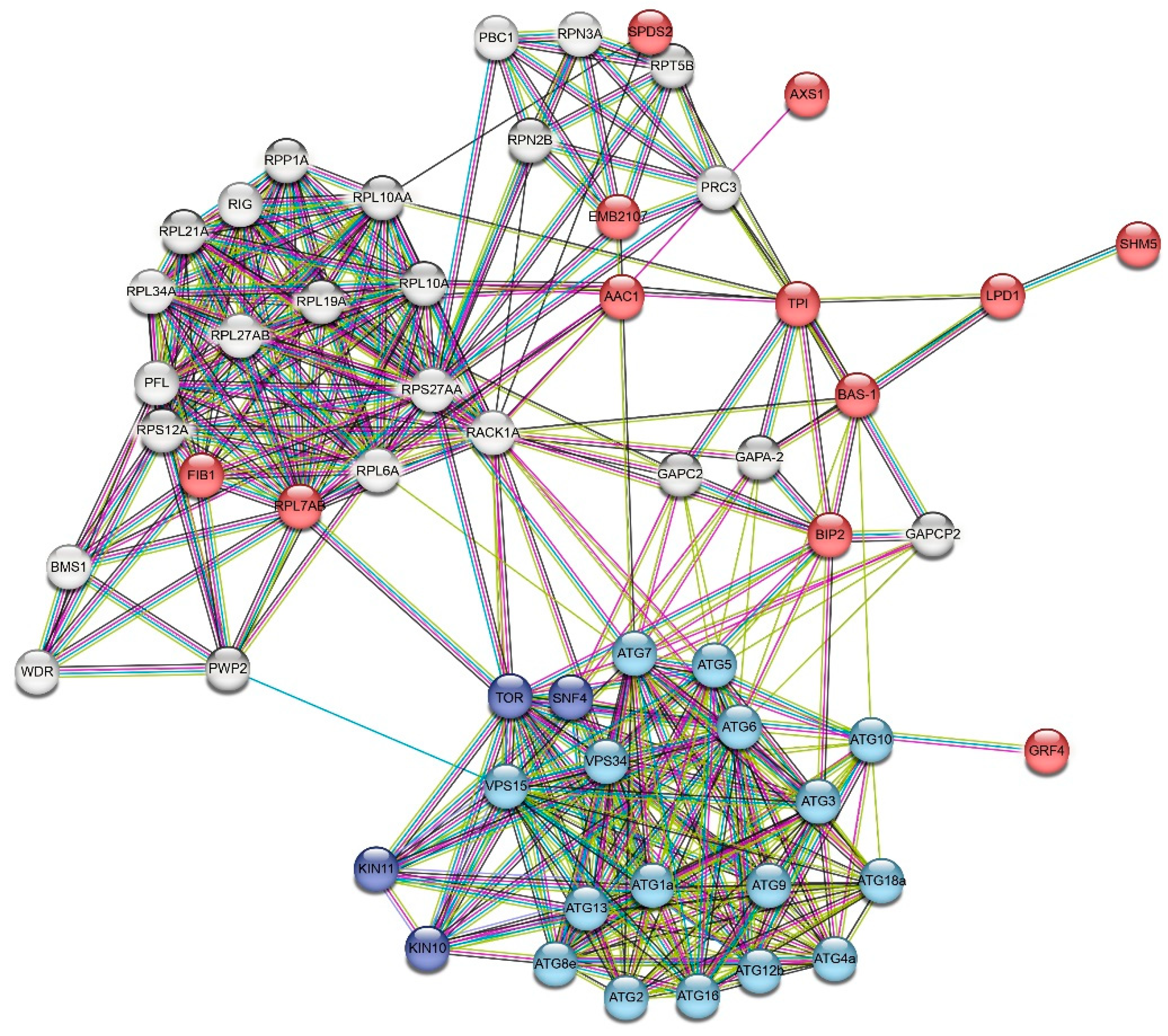
3.4. Protein-ligand docking and protein-protein interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siyiannis, V.F.; Protonotarios, V.E.; Zechmann, B.; Chorianopoulou, S.N.; Müller, M.; Hawkesford, M.J.; Bouranis, D.L. Comparative spatiotemporal analysis of root aerenchyma formation processes in maize due to sulphate, nitrate or phosphate deprivation. Protoplasma 2012, 249, 671–686. [Google Scholar] [CrossRef] [PubMed]
- Hanamata, S.; Kurusu, T.; Okada, M.; Suda, A.; Kawamura, K.; Tsukada, E.; Kuchitsu, K. In vivo imaging and quantitative monitoring of autophagic flux in tobacco BY-2 cells. Plant Signal. Behav. 2013, 8, e22510. [Google Scholar] [CrossRef] [PubMed]
- Filomeni, G.; De Zio, D.; Cecconi, F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015, 22, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Sláviková, S.; Shy, G.; Yao, Y.; Glozman, R.; Levanony, H.; Pietrokovski, S.; Elazar, Z.; Galili, G. The autophagy-associated atg8 gene family operates both under favourable growth conditions and under starvation stresses in arabidopsis plants. J. Exp. Bot. 2005, 56, 2839–2849. [Google Scholar] [CrossRef]
- Shi, J.; Feng, H.; Lee, J.; Ning Chen, W. Comparative proteomics profile of lipid-cumulating oleaginous yeast: an ITRAQ-coupled 2-D LC-MS/MS analysis. PLoS ONE 2013, 8, e85532. [Google Scholar] [CrossRef] [PubMed]
- Melia, T.J.; Lystad, A.H.; Simonsen, A. Autophagosome biogenesis: from membrane growth to closure. J. Cell Biol. 2020, 219, e202002085. [Google Scholar] [CrossRef]
- Liu, B.; Huang, X.; Li, Y.; Liao, W.; Li, M.; Liu, Y.; He, R.; Feng, D.; Zhu, R.; Kurihara, H. JS-K, a nitric oxide donor, induces autophagy as a complementary mechanism inhibiting ovarian cancer. BMC Cancer 2019, 19, 645. [Google Scholar] [CrossRef]
- Sarkar, S.; Korolchuk, V.I.; Renna, M.; Imarisio, S.; Fleming, A.; Williams, A.; Garcia-Arencibia, M.; Rose, C.; Luo, S.; Underwood, B.R.; et al. Complex inhibitory effects of nitric oxide on autophagy. Mol. Cell 2011, 43, 19–32. [Google Scholar] [CrossRef]
- Zhang, X.; Jin, L.; Tian, Z.; Wang, J.; Yang, Y.; Liu, J.; Chen, Y.; Hu, C.; Chen, T.; Zhao, Y.; et al. Nitric oxide inhibits autophagy and promotes apoptosis in hepatocellular carcinoma. Cancer Sci. 2019, 110, 1054–1063. [Google Scholar] [CrossRef]
- Dmitrieva, S.A.; Ponomareva, A.A.; Gurjanov, O.P.; Mazina, A.B.; Andrianov, V.V.; Iyudin, V.S.; Minibayeva, F.V. Spermine induces autophagy in plants: possible role of NO and reactive oxygen species. Dokl. Biochem. Biophys. 2018, 483, 341–343. [Google Scholar] [CrossRef]
- Kuo, E.Y.; Chang, H.-L.; Lin, S.-T.; Lee, T.-M. High light-induced nitric oxide production induces autophagy and cell death in Chlamydomonas reinhardtii. Front. Plant Sci. 2020, 11, 772. [Google Scholar] [CrossRef]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem. J. 2012, 441, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Bignon, E.; Allega, M.F.; Lucchetta, M.; Tiberti, M.; Papaleo, E. Computational structural biology of S-nitrosylation of cancer targets. Front. Oncol. 2018, 8, 272. [Google Scholar] [CrossRef]
- Astier, J.; Kulik, A.; Koen, E.; Besson-Bard, A.; Bourque, S.; Jeandroz, S.; Lamotte, O.; Wendehenne, D. Protein S-nitrosylation: what’s going on in plants? Free Radic. Biol. Med. 2012, 53, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- León, J. Protein tyrosine nitration in plant nitric oxide signaling. Front. Plant Sci. 2022, 13, 859374. [Google Scholar] [CrossRef]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Protein nitration: a connecting bridge between nitric oxide (NO) and plant stress. Plant Stress 2021, 2, 100026. [Google Scholar] [CrossRef]
- Montagna, C.; Rizza, S.; Maiani, E.; Piredda, L.; Filomeni, G.; Cecconi, F. To eat, or NOt to eat: S -nitrosylation signaling in autophagy. FEBS J. 2016, 283, 3857–3869. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Wang, L.; Wang, P.; Xue, Y.; Li, X.; Qiao, X.; Zhang, X.; Xu, T.; Liu, G.; et al. Autophagy impairment mediated by S-nitrosation of ATG4B leads to neurotoxicity in response to hyperglycemia. Autophagy 2017, 13, 1145–1160. [Google Scholar] [CrossRef]
- Oh, C.; Dolatabadi, N.; Cieplak, P.; Diaz-Meco, M.T.; Moscat, J.; Nolan, J.P.; Nakamura, T.; Lipton, S.A. S-nitrosylation of p62 inhibits autophagic flux to promote α-synuclein secretion and spread in Parkinson’s disease and Lewy body dementia. J. Neurosci. 2022, 42, 3011–3024. [Google Scholar] [CrossRef]
- Wright, C.; Iyer, A.K.V.; Kulkarni, Y.; Azad, N. S -nitrosylation of Bcl-2 negatively affects autophagy in lung epithelial cells. J. Cell. Biochem. 2016, 117, 521–532. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, C.; Liu, Q. PTEN S-nitrosylation by NOS1 inhibits autophagy in NPC cells. Cell Death Dis. 2019, 10, 306. [Google Scholar] [CrossRef]
- Nagarajan, N.; Oka, S.; Nah, J.; Wu, C.; Zhai, P.; Mukai, R.; Xu, X.; Kashyap, S.; Huang, C.-Y.; Sung, E.-A.; et al. Thioredoxin 1 promotes autophagy through transnitrosylation of Atg7 during myocardial ischemia. J. Clin. Investig. 2023, 133, e162326. [Google Scholar] [CrossRef] [PubMed]
- Guha, P.; Harraz, M.M.; Snyder, S.H. Cocaine elicits autophagic cytotoxicity via a nitric oxide-GAPDH signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, 1417–1422. [Google Scholar] [CrossRef]
- Zhan, N.; Wang, C.; Chen, L.; Yang, H.; Feng, J.; Gong, X.; Ren, B.; Wu, R.; Mu, J.; Li, Y.; et al. S-nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol. Cell 2018, 71, 142–154. [Google Scholar] [CrossRef]
- Minibayeva, F.; Dmitrieva, S.; Ponomareva, A.; Ryabovol, V. Oxidative stress-induced autophagy in plants: the role of mitochondria. Plant Physiol. Biochem. 2012, 59, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Mazina, A.B.; Dmitrieva, S.A.; Minibayeva, F.V. (2017). Donors of nitric oxide as autophagy indicators in wheat cells. In Proceedings of Molecular Aspects of Plant Redox Metabolism, Ufa, Russia, 351-354., June 2017. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970, 227, 680. [Google Scholar] [CrossRef]
- Bassal, M.; Abukhalaf, M.; Majovsky, P.; Thieme, D.; Herr, T.; Ayash, M.; Tabassum, N.; Al Shweiki, M.R.; Proksch, C.; Hmedat, A.; et al. Reshaping of the Arabidopsis thaliana proteome landscape and co-regulation of proteins in development and immunity. Mol. Plant 2020, 13, 1709–1732. [Google Scholar] [CrossRef] [PubMed]
- Spiller, S.; Frolov, A.; Hoffmann, R. Quantification of specific glycation sites in human serum albumin as prospective type 2 diabetes mellitus biomarkers. Protein Pept. Lett. 2018, 24, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Shumilina, J.; Kiryushkin, A.S.; Frolova, N.; Mashkina, V.; Ilina, E.L.; Puchkova, V.A.; Danko, K.; Silinskaya, S.; Serebryakov, E.B.; Soboleva, A.; et al. Integrative proteomics and metabolomics analysis reveals the role of small signaling peptide rapid alkalinization factor 34 (RALF34) in cucumber roots. Int. J. Mol. Sci. 2023, 24, 7654. [Google Scholar] [CrossRef]
- Chen, S.-J.; Liao, D.-L.; Chen, C.-H.; Wang, T.-Y.; Chen, K.-C. Construction and analysis of protein-protein interaction network of heroin use disorder. Sci. Rep. 2019, 9, 4980. [Google Scholar] [CrossRef]
- Begara-Morales, J.C.; Chaki, M.; Valderrama, R.; Sánchez-Calvo, B.; Mata-Pérez, C.; Padilla, M.N.; Corpas, F.J.; Barroso, J.B. Nitric oxide buffering and conditional nitric oxide release in stress response. J. Exp. Bot. 2018, 69, 3425–3438. [Google Scholar] [CrossRef] [PubMed]
- Galeeva, E.I.; Trifonova, T.V.; Ponomareva, A.A.; Viktorova, L.V.; Minibayeva, F.V. Nitrate reductase from Triticum aestivum leaves: regulation of activity and possible role in production of nitric oxide. Biochem. (Mosc.). 2012, 77, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Kumari, A.; Florez-Sarasa, I.; Fernie, A.R.; Igamberdiev, A.U. Interaction of nitric oxide with the components of the plant mitochondrial electron transport chain. J. Exp. Bot. 2018, 69, 3413–3424. [Google Scholar] [CrossRef] [PubMed]
- Antonova, K.; Vikhnina, M.; Soboleva, A.; Mehmood, T.; Heymich, M.-L.; Leonova, T.; Bankin, M.; Lukasheva, E.; Gensberger-Reigl, S.; Medvedev, S.; et al. Analysis of chemically labile glycation adducts in seed proteins: case study of methylglyoxal-derived hydroimidazolone 1 (MG-H1). Int. J. Mol. Sci. 2019, 20, 3659. [Google Scholar] [CrossRef] [PubMed]
- Benhar, M.; Forrester, M.T.; Stamler, J.S. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 721–732. [Google Scholar] [CrossRef]
- Leiper, J.; Murray-Rust, J.; McDonald, N.; Vallance, P. S -nitrosylation of dimethylarginine dimethylaminohydrolase regulates enzyme activity: further interactions between nitric oxide synthase and dimethylarginine dimethylaminohydrolase. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 13527–13532. [Google Scholar] [CrossRef]
- Sisti, G.; Kanninen, T.T.; Ramer, I.; Witkin, S.S. Interaction between the inducible 70-KDa heat shock protein and autophagy: effects on fertility and pregnancy. Cell Stress and Chaperones 2015, 20, 753–758. [Google Scholar] [CrossRef]
- Hurley, J.H.; Young, L.N. Mechanisms of autophagy initiation. Annu. Rev. Biochem. 2017, 86, 225–244. [Google Scholar] [CrossRef]
- Jia, H.; Liang, Z.; Zhang, X.; Wang, J.; Xu, W.; Qian, H. 14-3-3 proteins: an important regulator of autophagy in diseases. Am. J. Transl. Res. 2017, 9, 4738–4746. [Google Scholar]
- Chodasiewicz, M.; Kerber, O.; Gorka, M.; Moreno, J.C.; Maruri-Lopez, I.; Minen, R.I.; Sampathkumar, A.; Nelson, A.D.L.; Skirycz, A. 2′,3′-CAMP treatment mimics the stress molecular response in Arabidopsis thaliana. Plant Physiol. 2022, 188, 1966–1978. [Google Scholar] [CrossRef]
- Pereira, C.; Chaves, S.; Alves, S.; Salin, B.; Camougrand, N.; Manon, S.; Sousa, M.J.; Côrte-Real, M. Mitochondrial degradation in acetic acid-induced yeast apoptosis: the role of pep4 and the ADP/ATP carrier: mitochondria degradation in apoptosis. Mol. Microbiol. 2010, 76, 1398–1410. [Google Scholar] [CrossRef] [PubMed]
- Ryder, L.; Arendrup, F.S.; Martínez, J.F.; Snieckute, G.; Pecorari, C.; Shah, R.A.; Lund, A.H.; Blasius, M.; Bekker-Jensen, S. Nitric oxide-induced ribosome collision activates ribosomal surveillance mechanisms. Cell Death Dis. 2023, 14, 467. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, S.; Zhang, H.; Xu, J. Identification of nitric oxide as an endogenous inhibitor of 26S proteasomes in vascular endothelial cells. PLoS ONE 2014, 9, e98486. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Li, F.; Gemperline, D.C.; Book, A.J.; Vierstra, R.D. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell 2015, 58, 1053–1066. [Google Scholar] [CrossRef]
- Cheng, F.; Yin, L.-L.; Zhou, J.; Xia, X.-J.; Shi, K.; Yu, J.-Q.; Zhou, Y.-H.; Foyer, C.H. Interactions between 2-Cys peroxiredoxins and ascorbate in autophagosome formation during the heat stress response in Solanum lycopersicum. J. Exp. Bot. 2016, 67, 1919–1933. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.-M.; Lee, M.-Y. Airborne nanoparticles (PM 0.1) induce autophagic cell death of human neuronal cells: autophagic cell death by PM 0.1. J. Appl. Toxicol. 2016, 36, 1332–1342. [Google Scholar] [CrossRef]

| Uniprot ID | Protein | Hypothetical S-nitrosylation sites |
|---|---|---|
| P46077 | 14-3-3-like protein GF14 phi | 106 |
| Q9FIB6 | 26S proteasome non-ATPase regulatory subunit 12 homolog A | 397 |
| Q9LNU4 | 26S proteasome non-ATPase regulatory subunit 3 homolog A | 141 |
| P80602 | 2-Cys peroxiredoxin BAS1 chloroplastic (Fragment) | 64, 185 |
| Q9LZH9 | 60S ribosomal protein L7a-2 | 193 |
| P31167 | ADP ATP carrier protein 1 mitochondrial | 130 |
| Q41629 | ADP ATP carrier protein 1 mitochondrial | 81, 206 |
| A8MS68 | Dihydrolipoyl dehydrogenase 1 chloroplastic | 400 |
| Q9S7C0 | Heat shock 70 kDa protein 14 | 268 |
| F4HQD4 | Heat shock 70 kDa protein 15 | 268 |
| Q39043 | Heat shock 70 kDa protein BIP2 | 298 |
| Q9FEF8 | rRNA 2'-O-methyltransferase fibrillarin 1 | 252 |
| Q9SVM4 | Serine hydroxymethyltransferase 5 | 324 |
| O48661 | Spermidine synthase 2 | 43 |
| P48491 | Triosephosphate isomerase cytosolic | 13, 127 |
| Q9ZUY6 | UDP-D-apiose/UDP-D-xylose synthase 1 | 187 |
| Q9SGE0 | UDP-D-apiose/UDP-D-xylose synthase 2 | 187 |
| Name | Description (UNIPROT) | UNIPROT ID |
|---|---|---|
| GRF4 | 14-3-3-like protein GF14 phi | P46077 |
| EMB2107 | 26S proteasome non-ATPase regulatory subunit 12 homolog A | Q9FIB6 |
| BAS-1 | 2-Cys peroxiredoxin BAS1 chloroplastic (Fragment) | Q9C5R8 |
| RPL7AB | 60S ribosomal protein L7a-2 | Q9LZH9 |
| AAC1 | ADP ATP carrier protein 1 mitochondrial | P31167 |
| LPD1 | Dihydrolipoyl dehydrogenase 1 chloroplastic | A8MS68 |
| BIP2 | Heat shock 70 kDa protein BIP2 | Q39043 |
| FIB1 | rRNA 2'-O-methyltransferase fibrillarin 1 | Q9FEF8 |
| SHM5 | Serine hydroxymethyltransferase 5 | Q9SVM4 |
| SDS2 | Spermidine synthase 2 | O48661 |
| TPI | Triosephosphate isomerase cytosolic | P48491 |
| AXS1 | UDP-D-apiose/UDP-D-xylose synthase 1 | Q9ZUY6 |
| ATG1a | Serine/threonine-protein kinase ATG1a | Q94C95 |
| ATG2 | Autophagy-related protein 2 | F8S296 |
| ATG3 | Autophagy-related protein 3 | Q0WWQ1 |
| ATG4a | Cysteine protease ATG4a | Q8S929 |
| ATG5 | Autophagy protein 5 | Q9FFI2 |
| ATG6 | Beclin-1-like protein | Q9M367 |
| ATG7 | Ubiquitin-like modifier-activating enzyme atg7 | Q94CD5 |
| ATG8e | Autophagy-related protein 8e | Q8S926 |
| ATG9 | Autophagy-related protein 9 | Q8RUS5 |
| ATG10 | Ubiquitin-like-conjugating enzyme ATG10 | Q8VZ52 |
| ATG12b | Ubiquitin-like protein ATG12B | Q9LVK3 |
| ATG13 | Autophagy-related protein 13a | Q9SCK0 |
| ATG16 | Autophagy-related protein 16 | Q6NNP0 |
| ATG18a | Autophagy-related protein 18a | Q93VB2 |
| TOR | Serine/threonine-protein kinase TOR | Q9FR53 |
| VPS15 | Serine/threonine-protein kinase VPS15 | Q9M0E5 |
| VPS34 | Phosphatidylinositol 3-kinase VPS34 | P42339 |
| KIN10 | SNF1-related protein kinase catalytic subunit alpha KIN10 | Q38997 |
| KIN11 | SNF1-related protein kinase catalytic subunit alpha KIN11 | P92958 |
| SNF4 | Sucrose nonfermenting 4-like protein | Q944A6 |
| GAPC2 | Glyceraldehyde-3-phosphate dehydrogenase GAPC2, cytosolic | Q9FX54 |
| GAPA-2 | Glyceraldehyde-3-phosphate dehydrogenase GAPA2, chloroplastic | Q9LPW0 |
| GAPCP2 | Glyceraldehyde-3-phosphate dehydrogenase GAPC2, cytosolic | Q9FX54 |
| PRC3 | Proteasome subunit alpha type-2-A | O23708 |
| RPT5B | 26S proteasome regulatory subunit 6A homolog B | O04019 |
| RPN3A | 26S proteasome non-ATPase regulatory subunit 3 homolog A | Q9LNU4 |
| PBC1 | Proteasome subunit beta type-3-A | Q9XI05 |
| RPN2B | 26S proteasome non-ATPase regulatory subunit 1 homolog B | Q9MAT0 |
| RPL10AA | Large ribosomal subunit protein uL1z | Q8VZB9 |
| RPL10A | Large ribosomal subunit protein uL16z | Q93VT9 |
| RPS27AA | Ubiquitin-ribosomal protein eS31z fusion protein | P59271 |
| RACK1A | Small ribosomal subunit protein RACK1z | O24456 |
| RPL6A | Large ribosomal subunit protein eL6z | Q9FZ76 |
| RPL19A | Large ribosomal subunit protein eL19x | Q9SRX2 |
| RPP1A | Large ribosomal subunit protein P1w | Q8LCW9 |
| RIG | Small ribosomal subunit protein uS19u | Q08112 |
| RPL21A | Large ribosomal subunit protein eL21z/eL21y | Q43291 |
| RPL27AB | Large ribosomal subunit protein uL15y | Q9LR33 |
| RPL34A | Large ribosomal subunit protein eL34z | Q42351 |
| PFL | Small ribosomal subunit protein uS13z/uS13y/uS13x | P34788 |
| RPS12A | Small ribosomal subunit protein eS12z | Q9S9P1 |
| BMS1 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | F4IDR3 |
| WDR | Uncharacterized protein At1g15425 | Q8L403 |
| PWP2 | Periodic tryptophan protein 2 | Q8VYZ5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





