Submitted:
20 July 2023
Posted:
20 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
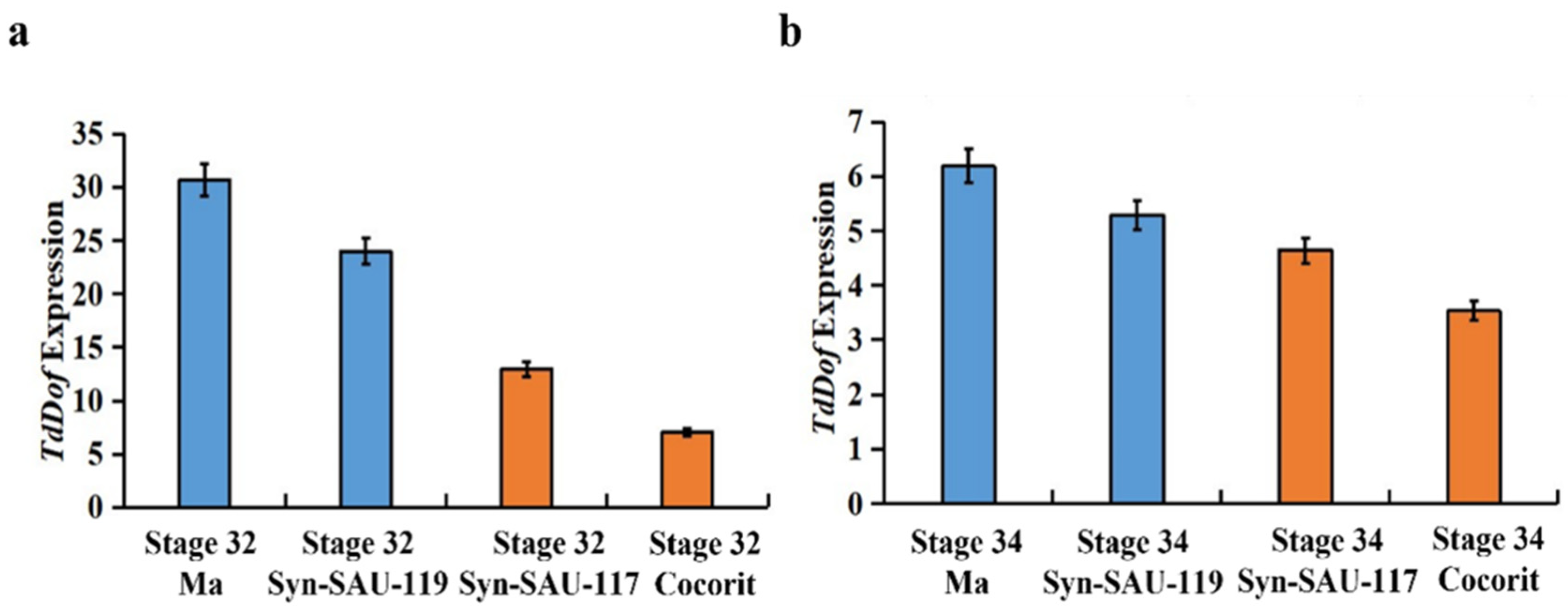
2.1. Differential Expression of the TdDof Gene in Different Materials
2.2. Genetic Analysis of the Solid Stem Suppressor Gene Su-TdDof
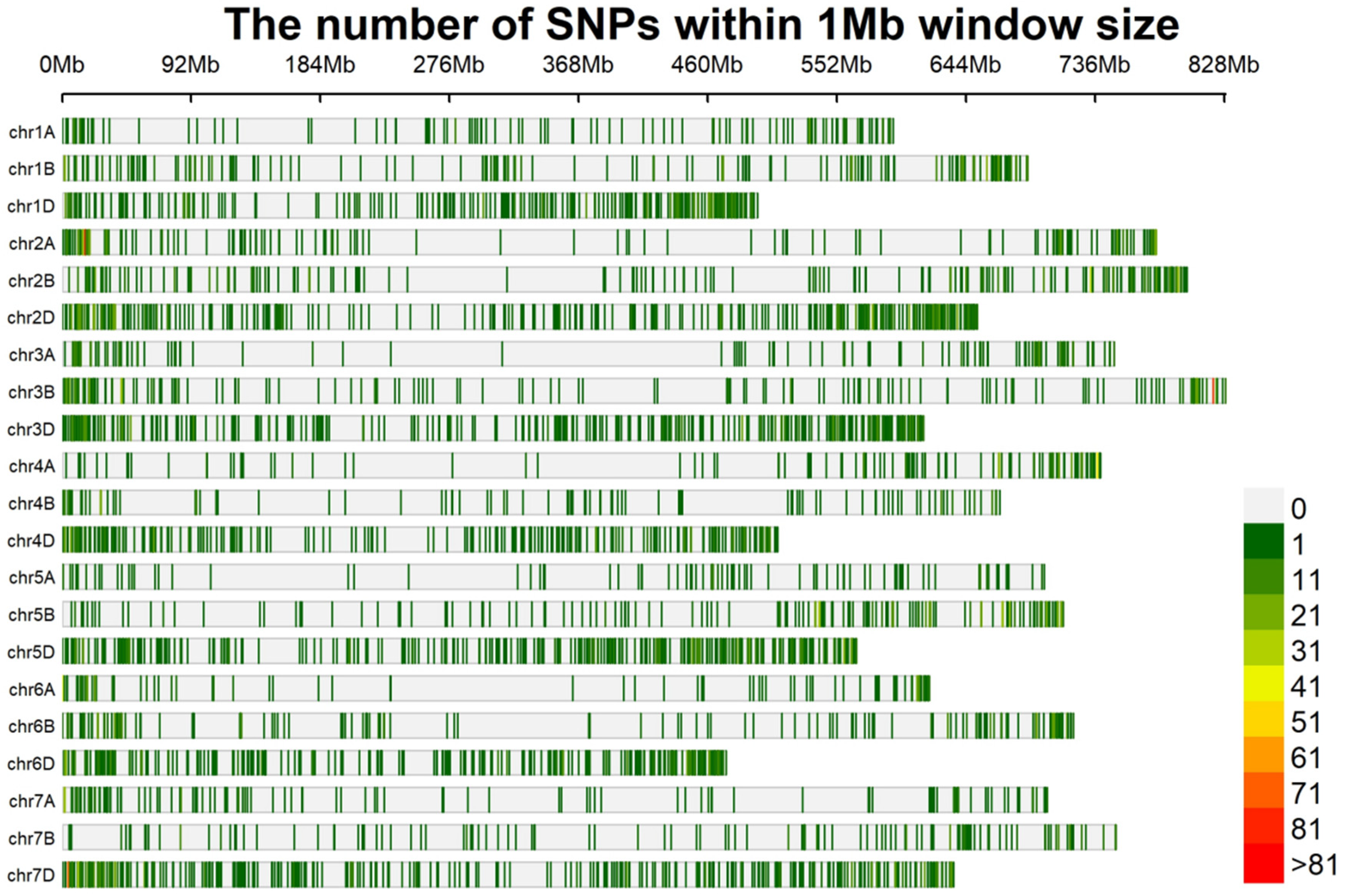
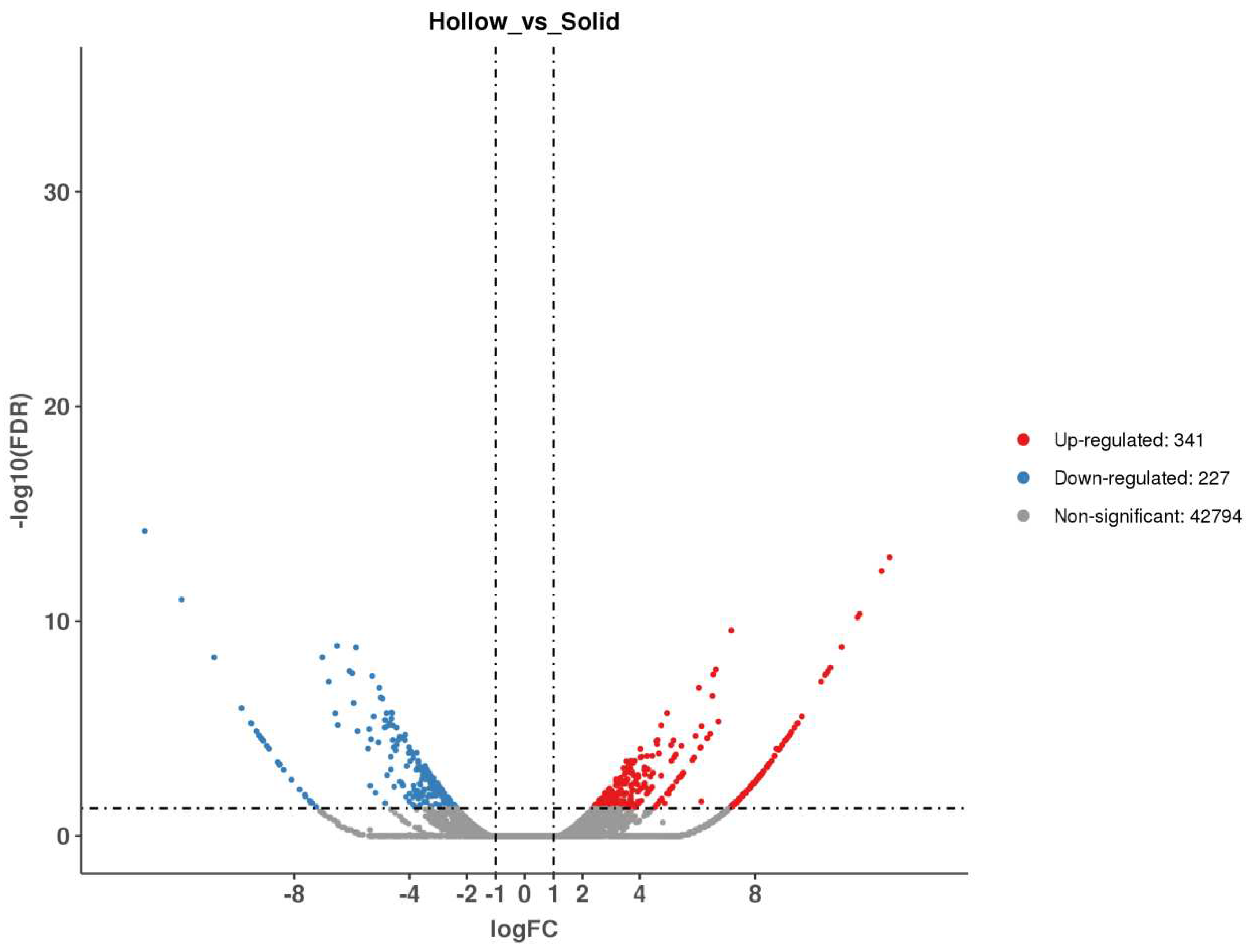
2.3. BSR-Seq Analysis of the RNA of Bulks with Contrasting Stem Solidity
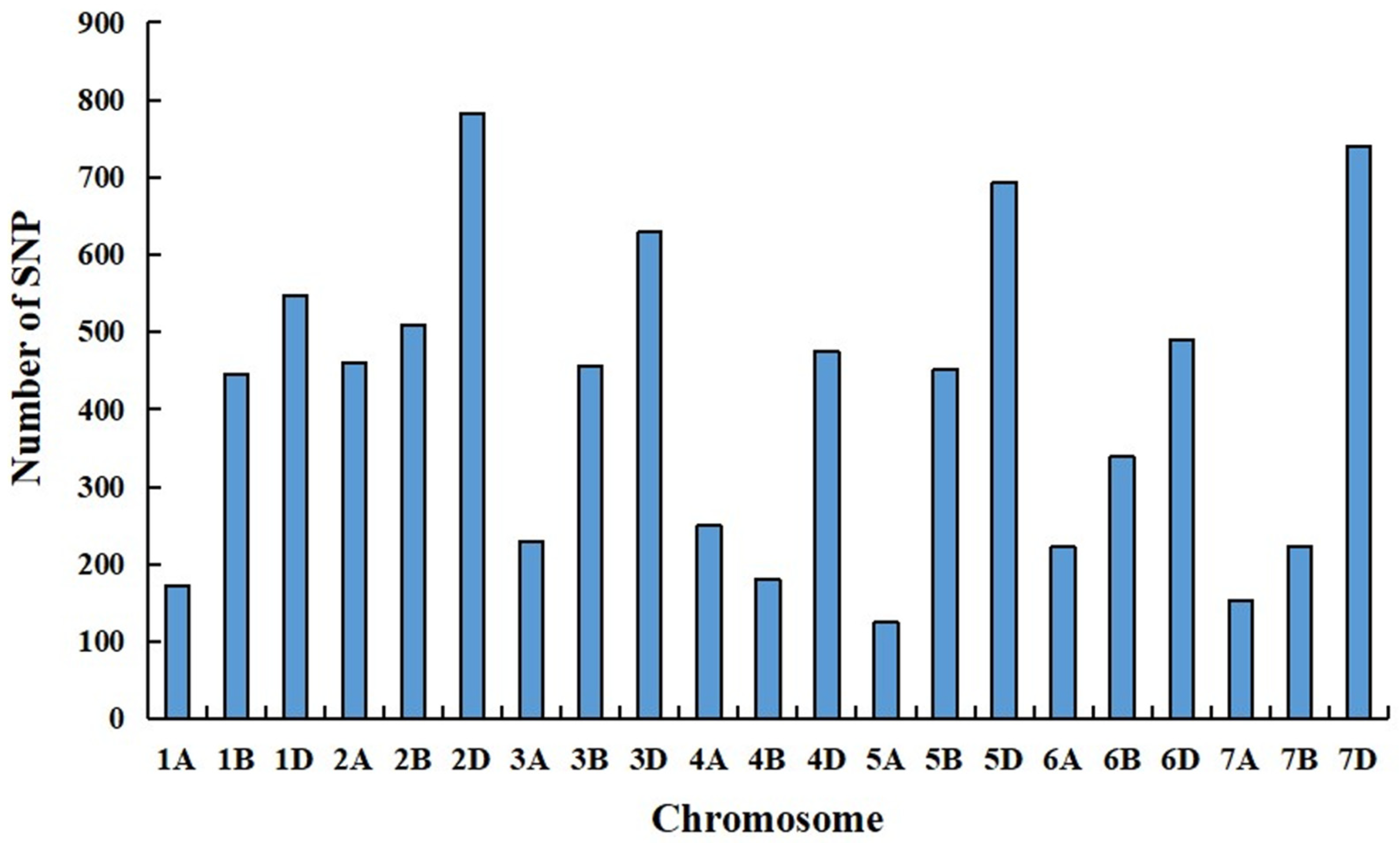
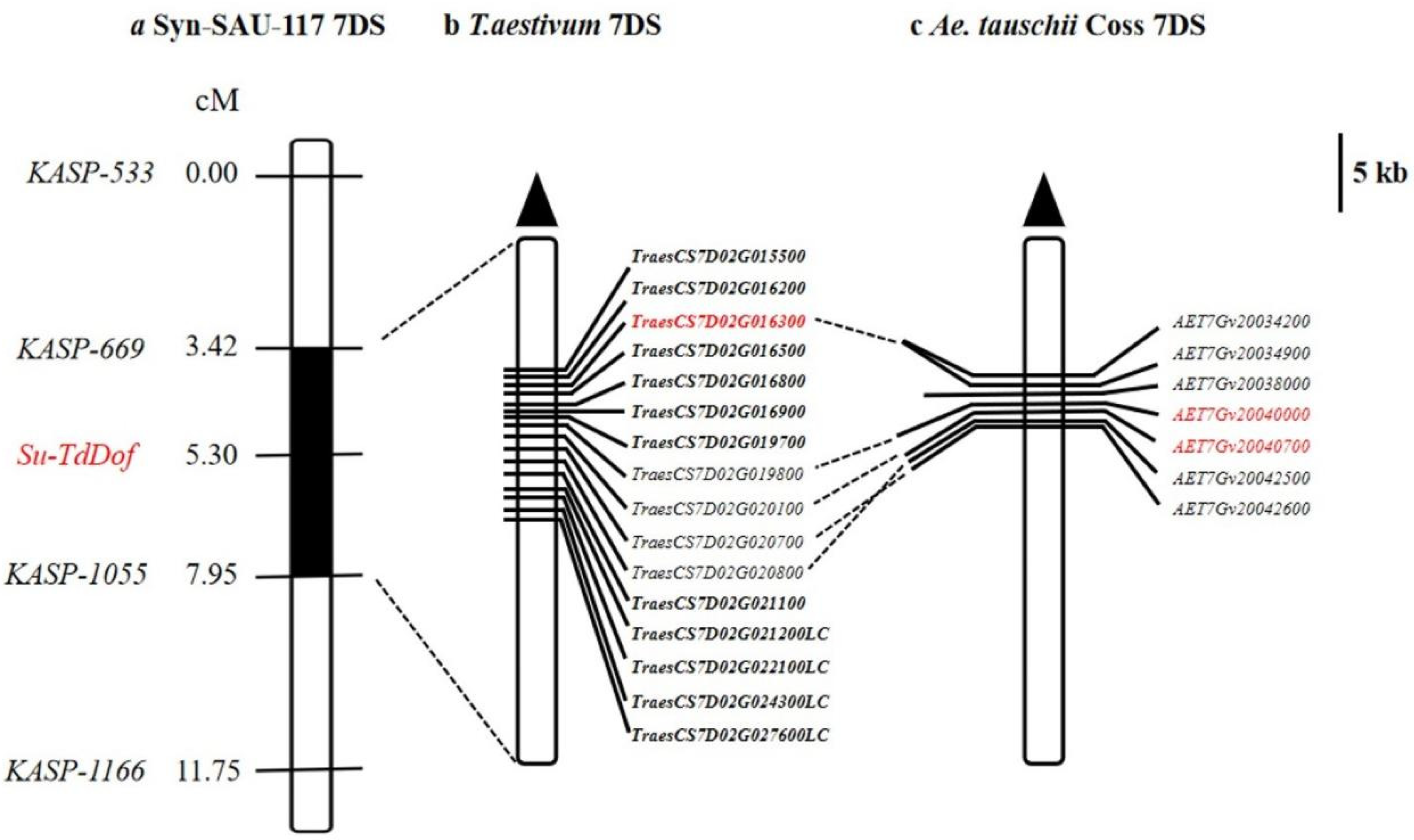
2.4. Molecular Mapping of Su-TdDof
2.5. Gene Analysis of the Su-TdDof Genomic Region
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Population Construction and Phenotypic Investigation
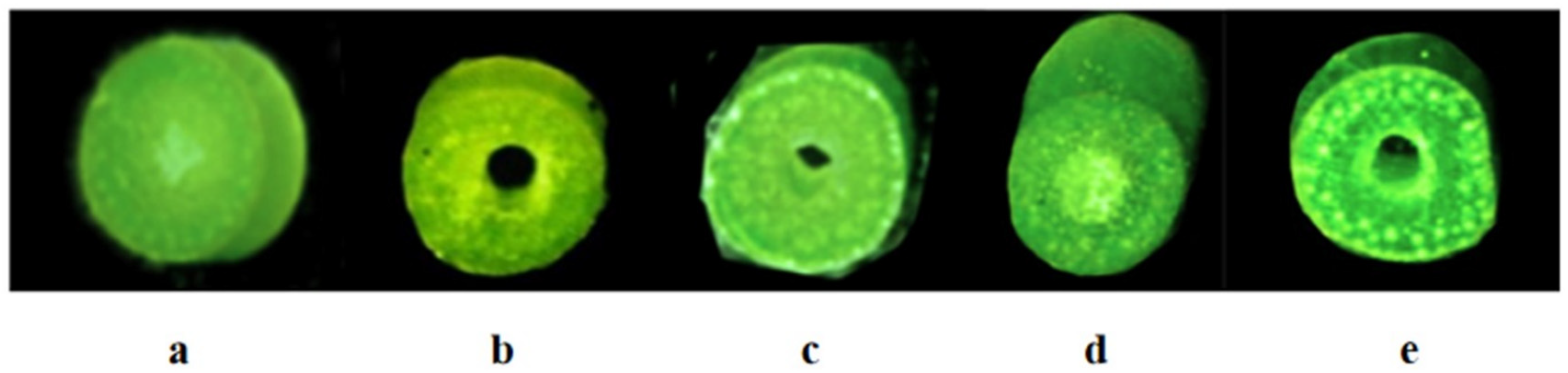
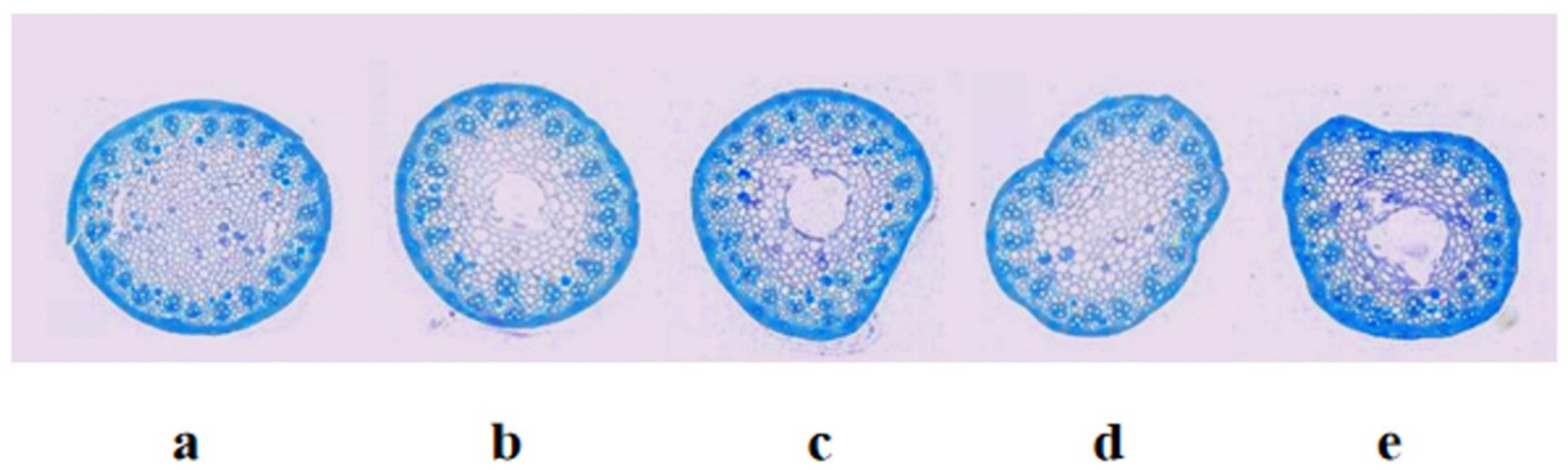
4.3. Observation of the Anatomical Structures of Stems
4.4. Solid Stem Gene Expression Analysis
4.5. Bulked Segregant RNA-Seq (BSR-Seq)
4.6. Kompetitive Allele-Specific PCR (KASP) Assays
4.7. Data Analysis
4.8. Candidate Gene Analysis
Supplementary Materials
Author Contributions statement
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Food and agricultural organization of the United Nations, FAOSTAT https:// www. fao. org/ faost at/en/. Accessed 28 January 2022.
- UN (United Nations, Department of Economic and Social Affairs, Population Division). World Population Prospects: The 2015 Revision, Key Findings and Advance Tables. 2015.
- Zhang, C.Z.; Huang, L.; Zhang, H.F.; Hao, Q.Q.; Lyu, B.; Wang, M.N.; Lynn, E.; Liu, M.; Kou, C.L.; Qi, J.; et al. An ancestral NB-LRR with duplicated 3’ UTRs confers stripe rust resistance in wheat and barley. Nat. Commun. 2019, 10, 4023. [CrossRef]
- Keller M, Karutz C, Schmid J E, et al. Quantitative trait loci for lodging resistance in a segregating wheat× spelt population. Theor. Appl. Genet. 1999, 98, 1171–1182. [CrossRef]
- Islam, M.S; Peng, S.B.; Visperas, R.; Ereful, N.; Bhuiya, M.S. Julfiquar, A.W. Lodging-related morphological traits of hybrid rice in a tropical irrigated ecosystem. Field Crop Res. 2007, 101, 240–248. [CrossRef]
- Acreche, M.M.; Slafer, G.A. Lodging yield penalties as affected by breeding in Mediterranean wheats. Field Crop Res. 2011, 122, 40–48. [CrossRef]
- Berry, P.M.; Spink, J. Predicting yield losses caused by lodging in wheat. Field Crop Res. 2012, 137, 19–26. [CrossRef]
- Peake, A.S.; Huth, N.I.; Carberry, P.S.; Raine, S.R.; Smith, R.J. Quantifying potential yield and lodging-related yield gaps for irrigated spring wheat in sub-tropical Australia. Field Crop Res. 2014, 158, 1–14. [CrossRef]
- Flintham, J.E.; Börner, A.; Worland, A.J.; GALE, M.D. Optimizing wheat grain yield: effects of Rht (gibberellin-insensitive) dwarfing genes. J. Agr. Sci. 1997, 128, 11–25. [CrossRef]
- Hirano, K.; Ordonio, R.L.; Matsuoka, M. Engineering the lodging resistance mechanism of post-Green Revolution rice to meet future demands. P. Jpn. Acad. B-Phys. 2017, 93(4), 220–233. [CrossRef]
- Keller, M.; Karutz, C.; Schmid, J.E.; Stamp, P.; Winzeler, M.; Keller, B. Messmer, M.M. Quantitative trait loci for lodging resistance in a segregating wheat × spelt population. Theor. Appl. Genet. 1999, 98, 1171–1182. [CrossRef]
- Larson, R.I.; MacDonald, M.D.; Inheritance of the type of solid stem in Golden Ball (Triticum durum). III. The effect of selection for solid stem beyond F5 in hexaploid segregates of the hybrid Rescue (T. aestivum) × Golden Ball. Can. J. Genet. Cytol. 1963, 5, 437–444.
- Kong, E.Y.; Liu, D.C.; Guo, X.L.; Yang, W.L.; Sun, J.Z.; Li, X.; Zhan, K.H.; Cui, D.Q.; Lin, J.X.; Zhang, A.M. Anatomical and chemical characteristics associated with lodging resistance in wheat. Crop J., 2013, 1, 43–49. [CrossRef]
- Beres, B.L.; Cárcamo, H.A.; Byers, J.R.; Clarke, F. R.; Pozniak C.J.; Basu, S.K.; DePauw, R.M. Host plant interactions between wheat germplasm source and wheat stem sawfly Cephus cinctus Norton (Hymenoptera: Cephidae) I. Commercial cultivars. Can. J. Plant Sci. 2013, 93, 607–617. [CrossRef]
- International Wheat Genome Sequencing Consortium (IWGSC).; Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science, 2018, 361, eaar7191. [CrossRef]
- Nilsen, K.T.; Walkowiak, S.; Xiang D.Q.; Gao, P.; Quilichini, T.D.; Willick, L.R.; Byrns, B.; Diaye, A.N.; Ens, J.; Wiebe, K.; et al. Copy number variation of TdDof controls solid-stemmed architecture in wheat. Proc. Natl. Acad. Sci. 2020, 117, 28708–28718. [CrossRef]
- Liu, Q.; Zhao, Y.; Rahman, S.J.; She, M.; Zhang, J.J.; Yang, R.C.; Islam, S.; O’Hara, G.; Varshney, R.; Liu, H.; et al. The putative vacuolar processing enzyme gene TaVPE3cB is a candidate gene for wheat stem pith-thickness. Theor. Appl. Genet. 2023, 136(6): 138. [CrossRef]
- Damania, A.B.; Pecetti, L.; Qualset, C.O.; Humeid, B. Diversity and geographic distribution of stem solidness and environmental stress tolerance in a collection of durum wheat landraces from Turkey. Genet. Resour. Crop Evol. 1997, 44, 101–108. [CrossRef]
- Platt, A.; Farstad, C. W.; Breeding spring wheats for resistance to wheat stem sawfly attack. Proc. 7th Pac. Sci. Cong. 1949, 4, 215–220.
- Clarke, F.R.; Clarke, J.M.; Knox, R.E.; Inheritance of stem solidness in eight durum wheat crosses. Can. J. Plant Sci. 2002, 82, 661–664. [CrossRef]
- Singh, A.K.; Clarke, J.M.; Knox, R.E.; Depauw, R.M.; McCaig, T.N.; Cuthbert, R.D.; Clarke, F.R.; Fernandez, M.R. AAC Raymore durum wheat. Can. J. Plant Sci. 2014, 94, 1289–1296. [CrossRef]
- Larson, R.I. Inheritance of the type of solid stem in Golden Ball (Triticum durum): I. early generations of a hybrid with Rescue (T. aestivum). Can. J. Bot. 1959, 37, 889–896. [CrossRef]
- Larson, R.I. Inheritance of the type of solid stem in Golden Ball (Triticum durum): II. Cytogenetics of the relation between solid stem and other morphological characters in hexaploid F5 lines of a hybrid with Rescue (T. aestivum). Can. J. Bot. 1959, 37, 1207–1216.
- Baker, E.P. Basic studies relating to the transference of genetic characters from Triticum monococcum L. to hexaploid wheat. Aust. J. Biol. Sci. 1975, 28, 189–200. [CrossRef]
- Bai, D.; Knott, D.R. Suppression of rust resistance in bread wheat (Triticum aestivum L.) by D-genome chromosomes. Genome 1992, 35, 276–282.
- Innes, R.L.; Kerber, E.R. Resistance to wheat leaf rust and stem rust in Triticum tauschii and inheritance in hexaploid wheat of resistance transferred from T. tauschii. Genome 1994, 37, 813–822.
- Kema, G.H.J.; Lange, W.; Van Silfhout, C.H. Differential suppression of stripe rust resistance in synthetic wheat hexaploids derived from Triticum turgidum subsp. dicoccoides and Aegilops squarrosa. Phytopathology. 1995, 85, 425–429.
- Hanušová, R.; Hsam, S.L.K.; Bartoš, P.; Zeller, F.J. Suppression of powdery mildew resistance gene Pm8 in Triticum aestivum L. (common wheat) cultivars carrying wheat-rye translocation T1BL·1RS. Heredity 1996, 77, 383–387. [CrossRef]
- Nelson, J.C.; Singh, R.P.; Autrique, J.E.; Sorrells, M.E. Mapping genes conferring and suppressing leaf rust resistance in wheat. Crop Sci. 1997, 37, 1928–1935. [CrossRef]
- Hiebert, C.W.; Moscou, M.J.; Hewitt, T.; Steuernagel, B.; Hernández-Pinzón, I.; Green, P.; Pujol, V.; Zhang, P.; Rouse, M.N.; Jin, Y.; et al. Stem rust resistance in wheat is suppressed by a subunit of the mediator complex. Nat. Commun. 2020, 11, 1123. [CrossRef]
- Assefa, S.; Fehrmann, H. Evaluation of Aegilops tauschii Coss. for resistance to wheat stem rust and inheritance of resistance genes in hexaploid wheat. Genet. Resour. Crop Evol. 2004, 51, 663–669. [CrossRef]
- Liu, W.; Danilova, T.V.; Rouse, M.N.; Bowden, B.L.; Friebe, B.; Gill, B.S.; Pumphrey, M.O. Development and characterization of a compensating wheat-Thinopyrum intermedium Robertsonian translocation with Sr44 resistance to stem rust (Ug99). Theor. Appl. Genet. 2013, 126, 1167–1177. [CrossRef]
- Liang, D.Y.; Zhang, M.H.; Liu, X.; Li, H.; Jia, Z.J.; Wang, D.H.; Peng, T.; Hao, M.; Liu, D.C.; Jiang, B.; et al. Development and identification of four new synthetic hexaploid wheat lines with solid stems. Sci. Rep. 2022, 12, 4898. [CrossRef]
- Pauw, R.M.D.; Read, D.W.L. The effect of nitrogen and phosphorus on the expression of stem solidness in Canuck wheat at four locations in southwestern Saskatchewan. Can. J. Plant Sci. 1982, 62, 593–598. [CrossRef]
- Chen, H.H.; Li, J.; Wan, H.S.; Wang, L.L.; Peng, Z.S.; Yang, W.Y. Microsatellite markers for culm wall thickness and anatomical features of solid stem wheat 86-741. Acta Agron. Sin. 2008, 34, 1381–1385.
- Li, Y.H.; Shi, X.H.; Hu, J.H.; Wu, P.P.; Qiu, D.; Qu, Y.F.; Xie, J.Z.; Wu, Q.H.; Zhang, H.J.; Yang, L.; et al. Identification of a recessive gene PmQ conferring resistance to powdery mildew in wheat landrace Qingxinmai using BSR-Seq analysis. Plant Dis. 2020, 104, 743–751. [CrossRef]
- Chen, S.; Zhou, Y.; Chen,Y, Gu, J Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890.
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics 2013, 29, 15–21. [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K,; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabrielet, S.; Daly, M.; et al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Re. 2010, 20, 1297–1303. [CrossRef]
- Lander, E.S.; Green, P.; Abrahamson, J.; Barlow, A.; Daly, M.J.; Lincoln, S.E.; Newburg, L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1, 174–181. [CrossRef]
- Kosambi, D.D. The estimation of map distances from recombination values. In Selected Works in Mathematics and Statistics; Springer: New Delhi, India, 2016; pp. 125–130.
- Liu, R.H.; Meng, J.L. MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 2003, 25, 317–321.
- Luo, M.C.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.V.; Deal, K.R.; Huo, N.X.; Zhu, T.T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [CrossRef]
- Oiestad, A.J.; Martin, J.M.; Cook, J.; Giroux, M.J. Identification of candidate genes responsible for stem pith production using expression analysis in solid-stemmed wheat. Plant Genome 2017, 10, plantgenome2017.2002.0008. [CrossRef]
- Rutjens, B.; Bao, D.; Van, Eck-Stouten E.V.; Brand, M.; Smeekens, S.; Proveniers, M. Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J. 2009, 58, 641–654. [CrossRef]
- Tao, Y.; Chen, M.; Shu, Y.J.; Zhu, Y.J.; Wang, S.; Huang, L.Y.; Yu, X.W.; Wang, Z.K.; Qian, P.P.; Gu, W.H.; Ma, H. Identification and functional characterization of a novel BEL1-LIKE homeobox transcription factor GmBLH4 in soybean. Plant Cell Tiss. Org. 2018, 134, 331–344. [CrossRef]
- Smith, H.M.S.; Ung, N.; Lal, S.; Courtier, J. Specification of reproductive meristems requires the combined function of SHOOT MERISTEMLESS and floral integrators FLOWERING LOCUS T and FD during Arabidopsis inflorescence development. J Exp. Bot. 2011, 62, 583–593. [CrossRef]
- Fujimoto, M.; Sazuka, T.; Oda, Y.; Kawahigashi, H.; Wu, J.Z.; Takanashi, H.; Ohnishi, T.; Yoneda, J.; Ishimori, M.; Kajiya-Kanegae, H.; et al. Transcriptional switch for programmed cell death in pith parenchyma of sorghum stems. Proc. Natl. Acad. Sci. 2018, 115, E8783–E8792. [CrossRef]
- Ma, Q.H. The expression of caffeic acid 3-O-methyltransferase in two wheat genotypes differing in lodging resistance. J Exp. Bot. 2009, 60, 2763–2771. [CrossRef]
- Tu, Y.; Rochfort, S.; Liu, Z.Q.; Ran, Y.D.; Griffith, M.; Badenhorst P.; Louie, G.V.; Bowman, M.E.; Smith, K.F.; Noel, J.P.; et al. Functional analyses of caffeic acid O-methyltransferase and cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 2010, 22, 3357–3373.
- Miller, C.N.; Harper, A.L.; Trick, M.; Werner, P.; Waldron, K.; Bancroft, I. Elucidation of the genetic basis of variation for stem strength characteristics in bread wheat by Associative Transcriptomics. BMC Genomics. 2016, 17, 500. [CrossRef]
- Knott, D.R.; Ramanujam, S. Transfer of genes for rust resistance to wheat from related species. Therm Sci. 1978, 16, 513–525.
- Hurni, S.; Brunner, S.; Stirnweis, D.; Herren, G.; Peditto, D.; McIntosh, R.A.; Keller, B. The powdery mildew resistance gene Pm8 derived from rye is suppressed by its wheat ortholog Pm3. Plant J. 2014, 79, 904–913. [CrossRef]
- Wu, L.; Xia, X.C.; Rosewarne, G.M.; Zhu, H.Z.; Li, S.Z.; Zhang, Z.Y.; He, Z.H. Stripe rust resistance gene Yr18 and its suppressor gene in Chinese wheat landraces. Plant Breed. 2015, 134, 634–640. [CrossRef]
- Athiyannan, N.; Long, Y.; Kang, H.; Chandramohan, S.; Bhatt, D.; Zhang, Q.J.; Klindworth, D.L.; Rouse, M.N.; Friesen, T.L.; McIntosh, R.; et al. Haplotype variants of Sr46 in Aegilops tauschii, the diploid D genome progenitor of wheat. Theor. Appl. Genet. 2022, 135, 2627–2639. [CrossRef]
- Jin, H.L.; Zhang, H.P.; Zhao, X.Y.; Long, L.; Guan, F.N.; Wang, Y.P.; Huang, L.Y.; Zhang, X.Y.; Wang, Y.Q.; Li, H.; et al. Identification of a suppressor for the wheat stripe rust resistance gene Yr81 in Chinese wheat landrace Dahongpao. Theor. Appl. Genet. 2023, 136, 67. [CrossRef]
- Huang, L.; Brooks, S.A.; Li, W.L.; Feller, J.P.; Trick, H.N.; Gill, B.S. Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 2003, 164, 655–664. [CrossRef]
- Arora, S.; Steuernagel, B.; Gaurav, K.; Chandramohan, S.; Long, Y.M.; Matny, O.; Johnson, R.; Enk, J.; Periyannan, S.; Singh, N.; et al. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 2019, 37, 139–143. [CrossRef]
- Dyck, P.L. Genetic inhibition of expression of resistance gene Lr23 in wheat to Puccinia recondita. Can. J. Plant Sci. 1982, 62, 219–220. [CrossRef]
- Kerber, E.R.; Green, G.J. Suppression of stem rust resistance in the hexaploid wheat cv. Canthatch by chromosome 7DL. Can. J. Bot. 1980, 58, 1347–1350. [CrossRef]
- Kerber, E.R.; Stem-rust resistance in 'Canthatch' hexaploid wheat induced by a nonsuppressor mutation on chromosome 7DL. Genome 1991, 34, 935–939. [CrossRef]
- Williams, N.D.; Miller, J.D.; Klindworth, D.L. Induced mutations of a genetic suppressor of resistance to wheat stem rust. Crop Sci. 1992, 32, 612–616. [CrossRef]
- Talajoor, M.; Jin, Y.; Wan, A.; Chen, X.M.; Bhavani, S.; Tabe, L.; Lagudah, E.; Huang, L. Specificity of a rust resistance suppressor on 7DL in the spring wheat cultivar Canthatch. Phytopathology 2015, 105, 477–481. [CrossRef]
- Mclntosh, R.A. Genetics of resistance to pathogens and pests: Recent developments. Proc. 8th Int. Wheat Genet. Symp. 1995, 2, 889–896.
- Hao, M.; Zhang, L.Q.; Zhao, L.B.; Dai, S.F.; Li, A.L.; Yang, W.Y.; Xie, D.; Li, Q.C.; Ning, S.Z.; Yan, Z.H.; et al. A breeding strategy targeting the secondary gene pool of bread wheat: introgression from a synthetic hexaploid wheat. Theor. Appl. Genet. 2019, 132, 2285–2294. [CrossRef]
| Parents and cross | Generationa | No. of plants/families | Observed ratiob | Actual ratio | Expected ratio | χ2 | P-value | ||
| S | Seg | Ss | |||||||
| Syn-SAU-117 | PS | 20 | 20 | ||||||
| Syn-SAU-119 | PH | 20 | 20 | ||||||
| PSs × PS | F1 | 20 | 20 | ||||||
| F2 | 156 | 36 | 120 | 0.9:3 | 1:3 | 0.308 | 0.579 | ||
| F2:3 | 134 | 35 | 67 | 32 | 1.04:2:0.96 | 1:2:1 | 0.134 | 0.935 | |
| Marker | Physical position (bp) | Allele 1 primera | Allele 2 primerb | Common/reverse primer |
|---|---|---|---|---|
| KASP-533 | 5336907 | TCAGCTTCAATTTCGGCAGC | TCAGCTTCAATTTCGGCAGT | AGAAGCTGAACGTGCGGAAG |
| KASP-669 | 6695986 | GTCGGATTCGGTTACTTTGAC | GTCGGATTCGGTTACTTTGAT | AGAGGTGCATGGTGTCGT |
| KASP-1055 | 10558194 | TCTTTCTCCTTCAGCCTCTTA | TCTTTCTCCTTCAGCCTCTTG | GCCTGATTGTAGTACATTATG |
| KASP-1166 | 11664145 | AACGAGGTCCCGCGCTCCTCCC | AACGAGGTCCCGCGCTCCTCCG | GTGTGAAGAGCGCTTCTGC |
| Parents | Marker genotypea | |||
|---|---|---|---|---|
| KASP-533 | KASP-669 | KASP-1055 | KASP-1166 | |
| AS92 | CC | CC | AA | CC |
| Syn-SAU-117 | CC | CC | AA | CC |
| Syn-SAU-119 | TT | TT | GG | GG |
| AS96 | TT | TT | GG | GG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





