Submitted:
10 July 2023
Posted:
11 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Test materials
2.2. Methodology
2.2.1. Extraction of total DNA
2.2.2. Synthesis and screening of ISSR primers
2.2.3. ISSR reaction system and procedure
2.2.4. Data processing
3. Results
3.1. Detection of total DNA quality
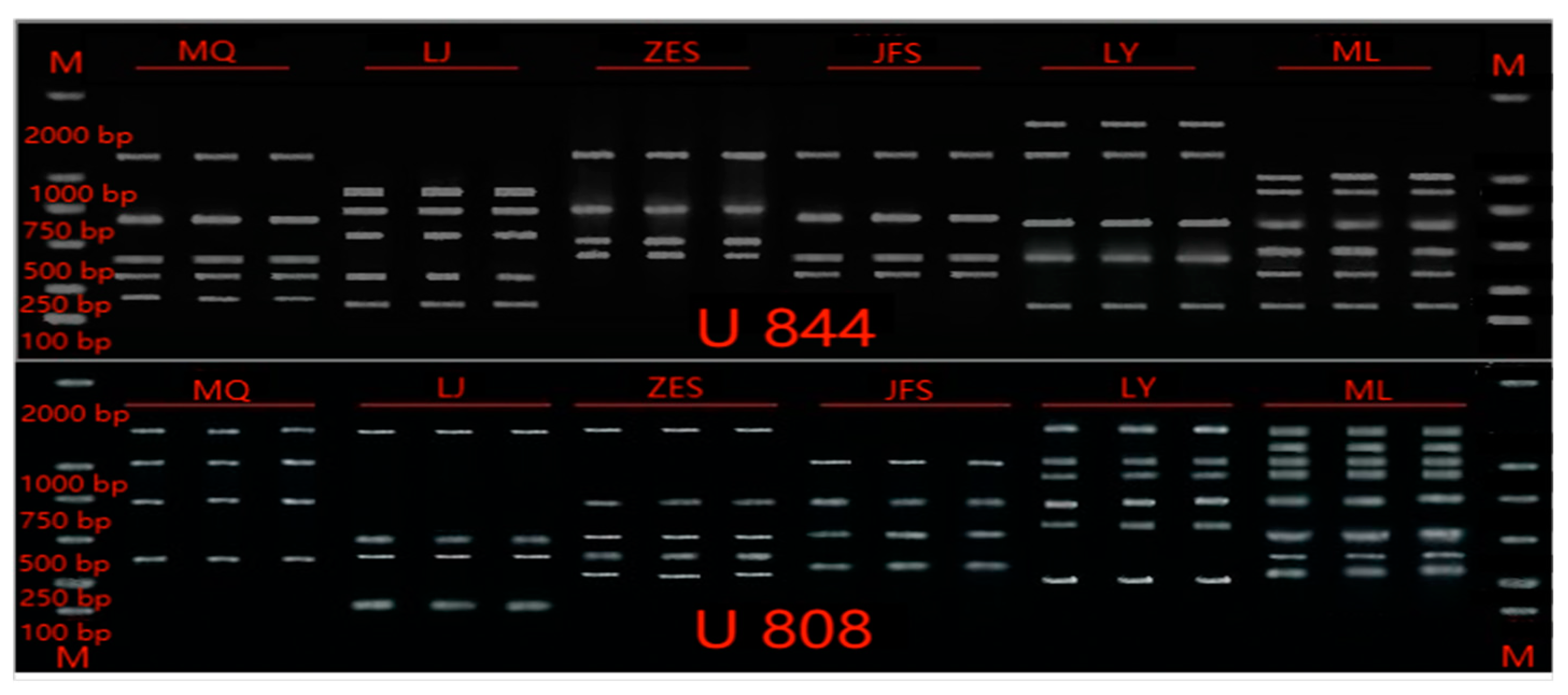
3.2. Primer screening
3.3. ISSR genetic diversity analysis
3.3.1. Analysis of genetic diversity parameters
3.3.2. Population genetic structure analysis
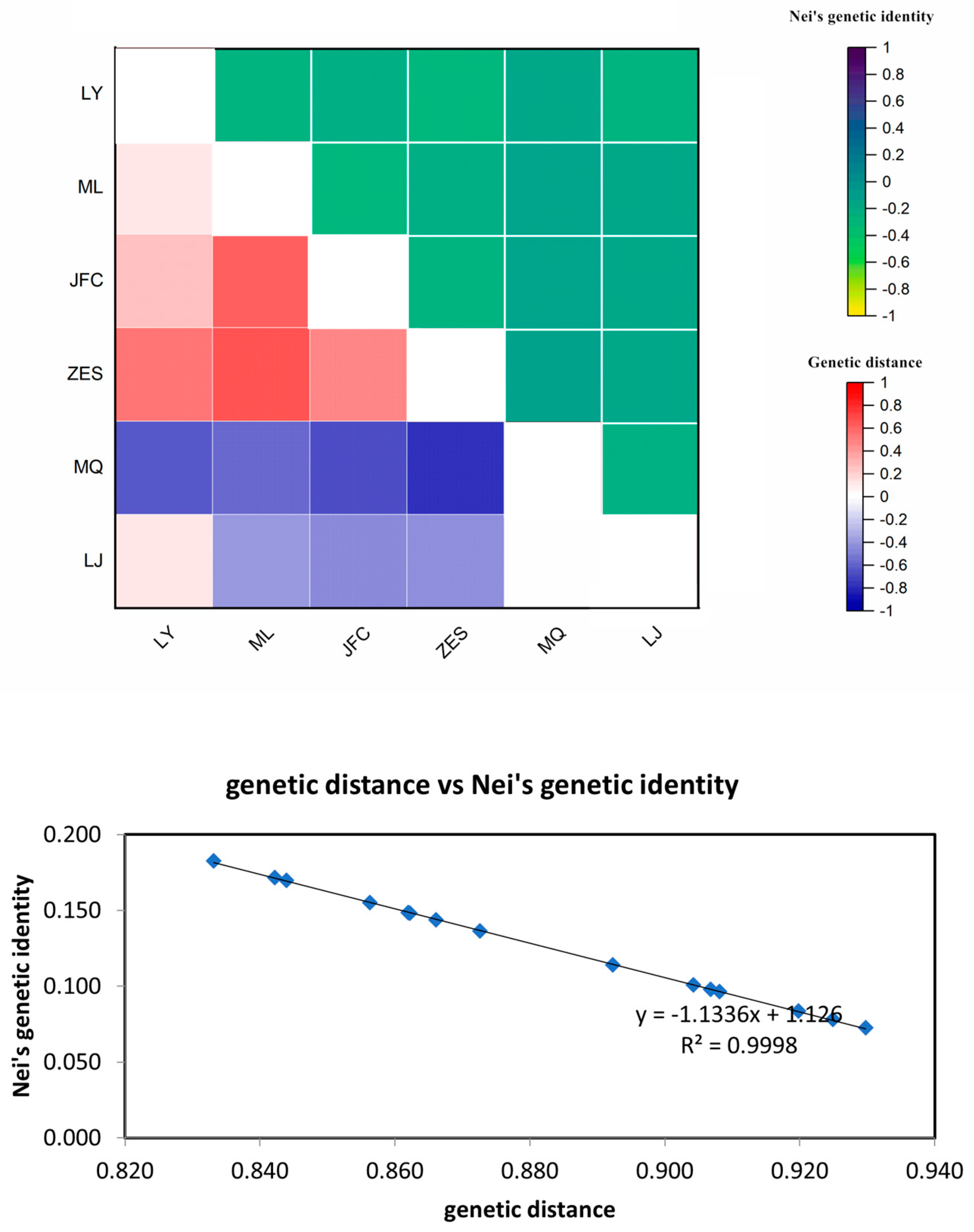
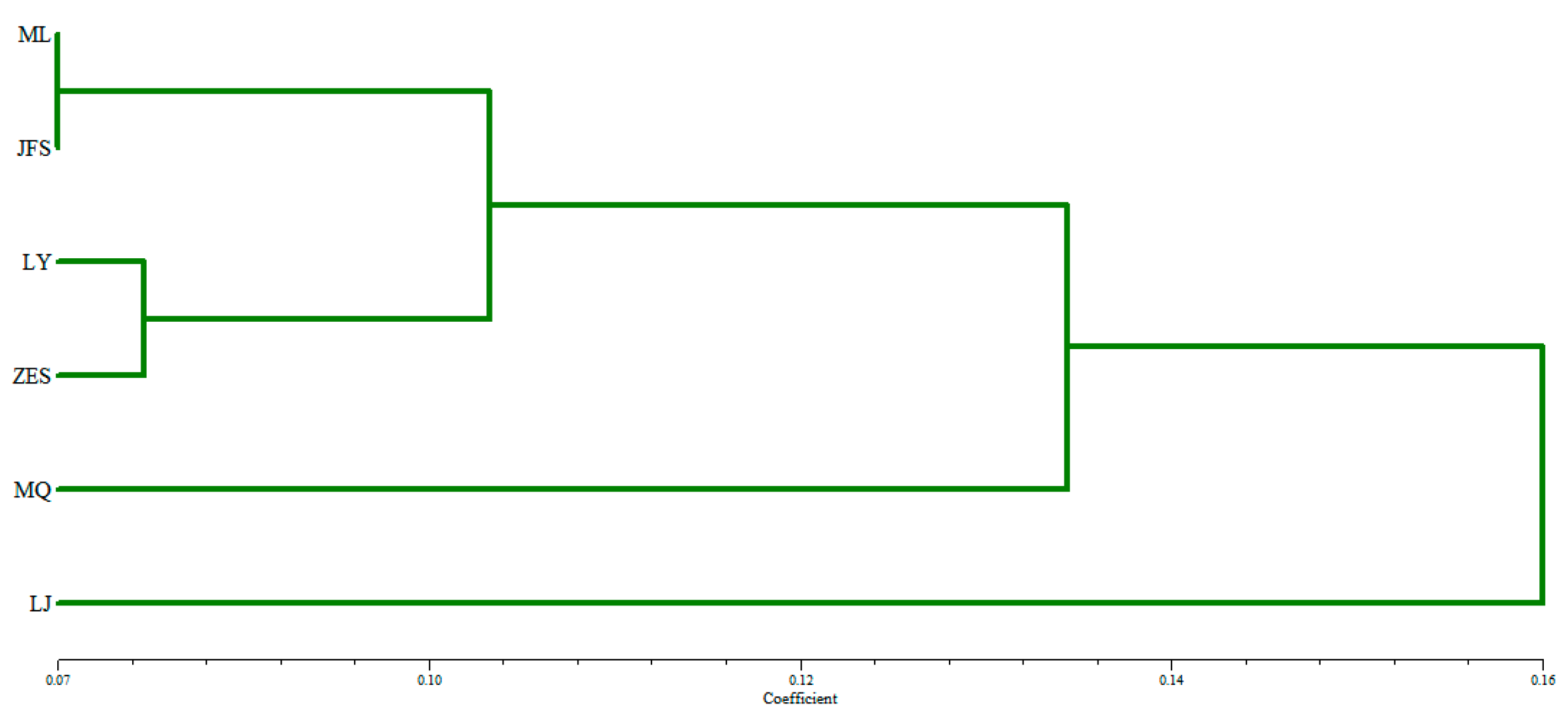
3.3.3. Genetic distance and clustering analysis of H.dentata.
3.3.4. Cluster analysis of wild populations
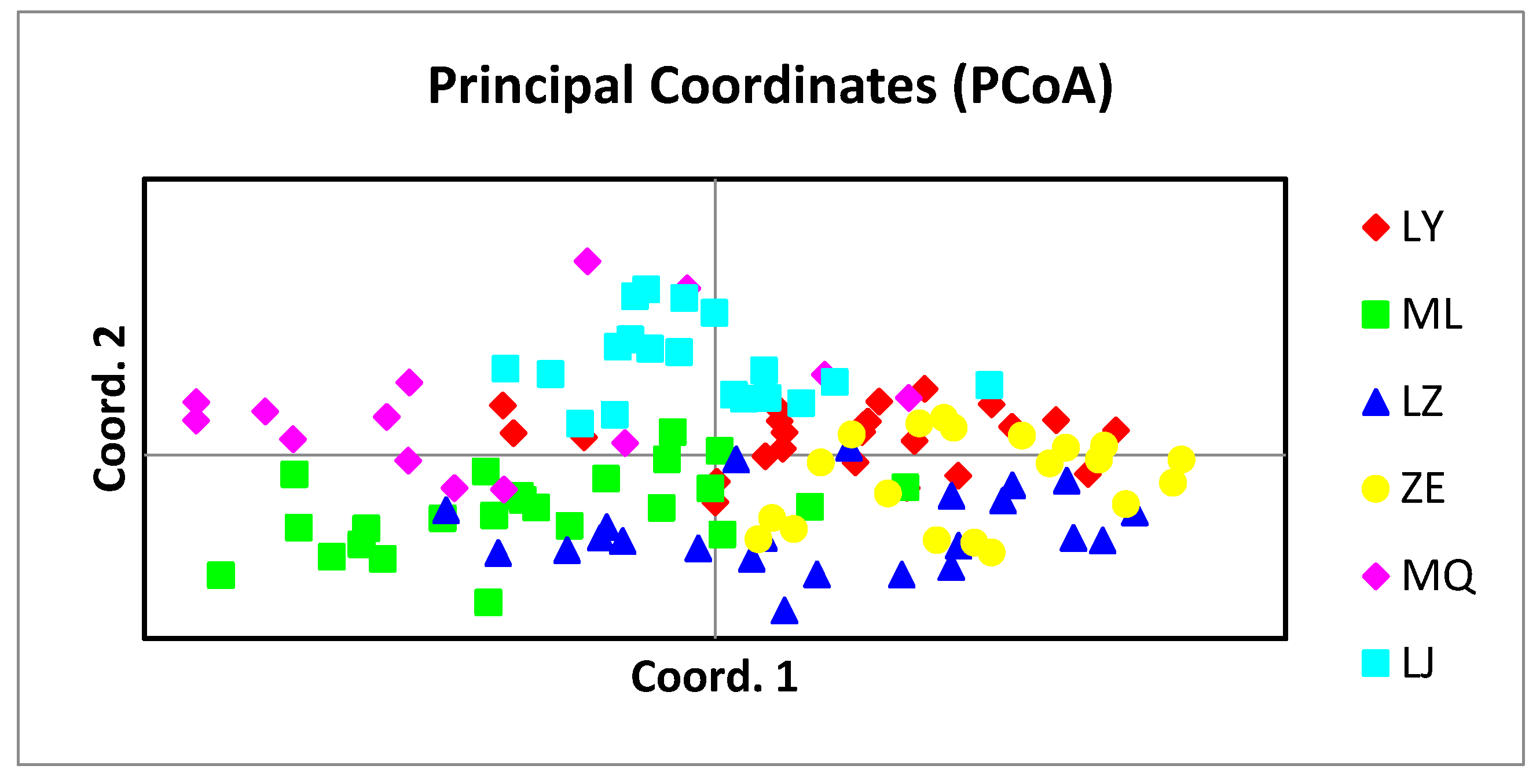
3.3.5. Principal component analysis of the populations
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Azani, N. ; Babi[1]Luo Yajin, Tang Jianmin, Jiang Qiang, et al. Progress in Conservation Studies of Orchids in Plant in Guangxi Yachang Reserve[J]. Journal of Guangxi Academy of Sciences, 2020(1): 5-16.
- Editorial Committee of Flora of China, Chinese Academy of Sciences. Flora of China[M]. Beijing: Science Press, 1999, (17), 472.
- Jiangxi Provincial Health Bureau Revolutionary Committee. Jiangxi herbal medicine[M]. Nanchang: Xinhua Bookstore Jiangxi Province, 1970: 420-421.
- Liu Yitao, Long Chunlin, Dao Zhiling, et al. Ethnobotanical survey on medicinal roots eaten by the local people in Simao, Yunnan Province, during the Dragon-boat Festival [J]. Journal of Plant Resources and Environment, 2003, (2): 33-38. [CrossRef]
- Kunming Municipal Health Bureau. Commonly used folk herbal medicine in Kunming[M]. Kunming: Kunming Municipal Health Bureau, 1970: 392.
- Compilation group of ' National Chinese Herbal Medicine Compilation ' : National Chinese Herbal Medicine Compilation ( Vol II ) [M]. Beijing: People's health publishing Press, 1975.
- Wu Fuqin, Ma Licui, Zheng Jinxuan, et al. Diversity of Medicinal Vascular Plant Resources from Wetland in Yunnan Province[J], 2021, 40(9): 75-80. [CrossRef]
- Chen Yaya, Mao Tangfen, Li Qike et al. Tissue Culture and Plantlet Regeneration of Habenaria dentata[J]. Plant Physiology Letters, 2007, 244 (6): 1136.
- Shao, Li. Effects of different cytokinins on one-step seedling formation of aseptic seedlings of Habenaria dentata[J]. Contemporary Horticulture, 2022, 45(13): 15-17.
- Chen Yaya, Zhu Guosheng, Mao Tangfen et al. A Preliminary Study on Habenaria dentate Endophytic Fungi[J]. Guizhou Agricultural Sciences, 2008, 218 (3): 12-13,3.
- Chen Yaya, Yang Lin, Zhu Guosheng, et al. ISolation and Identification of Mycorhiza Fungi on Habenaria dentate[J]. Guizhou Agricultural Sciences, 2010, 38 (8): 84-86.
- Chen Yaya, Zhu Guosheng, Mao Tangfen et al. The Research on Habenaria dentate Mycorrhizal Fungi Acclerating the Growth of Habenaria dentate in vitro Seedling[J]. Guizhou Agricultural Sciences, 2009, 37 (12): 33-34, 39.
- Wang Caiyun, Hou Jun, Zhou Maochang, et al. Study in genetic diversity of Gastrodia elata[J]. Hubei Agricultural Sciences, 2022, 61(18): 131-138.
- Ge Song, Hong Deyuan. Genetic diversity and its detection methods[A]. Qian Yingqian, Ma Keping. Principles and Methods of Biological Genetic Diversity Research[M]. Beijing: Science Press, 1994, 123-140.
- Qiu Guojun, Cheng Min, Guo Jihua. Application of ISSR Molecular Marker Technology in Plants and Its Research Progress[J]. Journal of Minzu Normal University of Xingyi, 2020,(01):117-120.
- Luo Mingxin, Liu Fengmin, Zhang Weili, et al. Application of SRAP and CDDP Markers in Genetic Diversity Analysis of Morinda officinalis How.[J]. Molecular Plant Breeding, 2021, 19(11): 3661-3669. [CrossRef]
- Yang Shuting, Ma Xiaona, Bai Xiaolin,et al. CDDP genetic diversity analysis of a very small population of wild plant Cypripedium palangshanense[J]. Journal of Sichuan University(Natural Science Edition), 2022,59(06):155-163. [CrossRef]
- Elena E K, Irina Y G N,Vladimir V Z. Using ISSR-Markers for Genetic Diversity of some Representatives of Orchidaceae[J]. Biosciences Biotechnology Research Asia, 2016, 13(1). [CrossRef]
- Sun Feifei. Research on Genetic Diversity and Development and Utilization of Germplasm Resource of Medicinal Plant Tetrapanax papyrifer[D]. Guilin: Guilin Medical University.
- Lu Jiashi, Bu Chaoyang, Lv Weili, et al. Analysis on Genetic Diversity of 20 Plants of Orchidaceae by ISSR Molecular Marker[J]. Southwest China Journal of Agricultural Sciences, 2012, 25(6): 2252-2257.
- Zhang Qing, Wang Hanchen, Cheng Zhuo, et al. Current Status of Wild Orchid Resource in China, Focusing on Their Conservation and Utilization[J]. China Biotechnology, 2022, 42(11): 59-72. [CrossRef]
- Zhao Zeyu, Liu Na, Xing Xiaoke. Research advances in mechanisms of interaction between mycorrhizal fungi and Orchidaceae plants by using omics techniques[J]. Mycosystema,, 2021, 40(3): 423-435.
- Tang Yanjing, Guo Shunxing, Chen Juan. Advances in the specificity of Orchid-mycorrhizal fungi[J]. Journal of Capital Normal University(Natural Science Edition),2021, 42(3): 63-74.
- Zhang Z, Yan Y, Tian Y, et al. Distribution and conservation of orchid species richness in China[J]. Biological Conservation, 2015, 181: 64-72. [CrossRef]
- Su Ziying, Li Yue, Zhang Xiying, et al. Ornamental Evaluation of Endemic Orchids from Guangdong Province[J]. Chinese Journal of Tropical Crops, 2020, 41(8): 1560-1565. [CrossRef]
- Tang Huan. DNA barcoding identification and ecological suitability of important medicinal plants of Orchidaceae[D].Yaan: Sichuan Agricultural University.
- Wei Xiao, Tang Jianmin,Chai Shengfeng. Study on the Current Situation and Sustainable Development Strategy of Orchidaceas Resources in Guangxi[J]. Journal of Guangxi Academy of Sciences, 2022, 38(2): 99-107, 117.
- Wang Tiejuan, Li Weiqiong, Zhang Shuyan, et al. Genetic Diversity and Differentiation of Five Natural Populations of Artemisia halodendron[J]. Scientia Silvae Sinicae, 2010, 46(12): 171-175.
- Hutchison D W, Templeton A R. Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability[J]. Evolution, 1999, 53(6): 1898-1914. [CrossRef]
- Huang Jinchun, Wan Siqi, Chen Yang, et al. Genetic diversity of Anoectochilus roxburghii based on ISSR and SRAP molecular markers[J]. Journal of Zhejiang A & F University, 2023, 40(1): 22-29. [CrossRef]
- Li Zongyan, Guan Mingyuan, Li Jing, et al. Genetic Diversity of Paphiopedilum micranthum Detected by ISSR Data[J]. Acta Botanica Boreali-Occidentalia Sinica,, 2016, 36(7): 1351-1356.
- Jiang Yawen, Sun Xiaoqin, Luo Huolin, et al. Studies on Genetic Diversity of Cymbidium kanran Populations from the Main Mountains in Jiangxi Province Based on ISSR Marker[J]. Acta Horticulturae Sinica, 2017, 44(10): 1993-2000. [CrossRef]
- Govindaraju D, R. Relationship between dispersal ability and levels of gene flow in plants[J]. Oikos, 1988: 31-35. [CrossRef]
- Wright, S. Evolution in Mendelian populations[J]. Genetics, 1931, 16(2): 97-159. [CrossRef]
- Nybom H, Bartish I V. Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants[J]. Perspectives in Plant Ecology, Evolution and Systematics, 2000, 3(2): 93-114. [CrossRef]
- Fischer M, Matthies D. RAPD variation in relation to population size and plant fitness in the rare Gentianella germanica (Gentianaceae) [J]. American Journal of Botany, 1998, 85(6): 811-819. [CrossRef]
- Qin Huizhen, Pan Bo, Zhao Jian, et al. Genetic Diversity Analysis by ISSR of Paphiopedilum emersonill, a plant Species with Extremely Small Population[J]. Guangxi Sciences, 2022, 29(6): 1134-1140.

| Area | North latitude | East latitude | Number of samples | Biotope |
| Minqiang Village, Shanglong Township, Longzhou County, Chongzuo City (MQ) | 22°25′23″ | 106°54′43″ | 22 | Hilltop Grass |
| Jianfeng Mountain, Xinzhuangxiong Village, Jiangzhou District, Chongzuo City (JFS) | 22°29′30″ | 106°55′2″ | 23 | Mountain slopes |
| Jingxi City, Lu Dong Township, by the curse on the village (ZES) | 23°8′00″ | 106°19′45″ | 20 | Roadside slopes |
| Longji Village, Luodong Township, Jingxi City (LJ) | 23°8′22″ | 106°21′26″ | 20 | Roadside slopes |
| Huaping Village, Huaping Township, Leye County, Baise City (LY) | 24°50′55″ | 106°21′50″ | 24 | Roadside grasses |
| Huanjiang Maonan Autonomous County, Hechi City, Chuanshan Town, Xizaitun Mu Lun Nature Reserve (ML) | 24°55′33″ | 106°33′42″ | 24 | Grass by the river |
| Primer | Sequence(5'-3') | Annealing temperature(℃) | Number of total amplified bands | Number of polymorphic bands | Percentage of polymorphic(%) |
| U808 | AGAGAGAGAGAGAGAGGC | 48.7 | 8 | 8 | 100% |
| U823 | TCTCTCTCTCTCTCTCTCC | 48.0 | 5 | 4 | 80.0% |
| U844 | CTCTCTCTCTCTCTCTCTRC | 48.6 | 6 | 6 | 100% |
| U855 | ACACACACACACACACACACYT | 52.7 | 7 | 5 | 71.4% |
| U876 | GATAGATAGACAGACA | 38.4 | 6 | 4 | 66.7% |
| U878 | GGATGGATGGATGGAT | 47.0 | 7 | 6 | 85.7% |
| U880 | GGAGAGGAGAGGAGA | 47.9 | 6 | 5 | 83.3% |
| U885 | BHBGAGAGAGAGAGAGAGA | 48.0 | 5 | 4 | 80.0% |
| Population | Number of polymorphic loci | Number of alleles | Effective number of alleles | Nei's genetic diversity | Shannon's information index | Percentage of polymorphic loci (PPL) |
| ML | 33 | 1.846±0.366 | 1.506±0.362 | 0.294±0.184 | 0.436±0.256 | 84.621 |
| LY | 31 | 1.795±0.409 | 1.463±0.350 | 0.275±0.1807 | 0.414±0.251 | 79.492 |
| JFS | 27 | 1.692±0.468 | 1.353±0.361 | 0.211±0.195 | 0.321±0.275 | 69.232 |
| ZES | 30 | 1.769±0.427 | 1.387±0.356 | 0.235±0.1806 | 0.361±0.252 | 76.922 |
| LJ | 31 | 1.795±0.409 | 1.430±0.353 | 0.257±0.1828 | 0.393±0.250 | 79.493 |
| MQ | 27 | 1.692±0.468 | 1.406±0.365 | 0.240±0.1949 | 0.360±0.277 | 69.234 |
| Mean | 29.8 | 1.765 | 1.424 | 0.252 | 0.381 | 76.499 |
| P value | 0.168 | 0.101 | 0.087 | 0.089 | ||
| Species level | 2.000 | 1.539 | 0.324 | 0.493 | 100 |
| Total genetic diversity (Ht ) | Population genetic diversity (Hs ) | Gene differentiation factor (Gst ) | Gene flow (Nm ) | |
| Mean | 0.326 | 0.252 | 0.227 | 1.706 |
| Standard deviation | 0.019 | 0.010 |
| Source of variation | df | Sum of squares | Variance components | Percentage of variation | P value |
| Among population | 5 | 250.856 | 2.014 | 27% | <0.01 |
| Within population | 127 | 708.618 | 5.580 | 73% | <0.01 |
| Total | 132 | 959.474 | 7.594 | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




