Submitted:
07 July 2023
Posted:
10 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Selection of transcription factors responsive to drought and salinity stress
2.2. Selection of transcription factors responsive to drought and salinity stress
2.3. Structural and functional annotation of the coding region
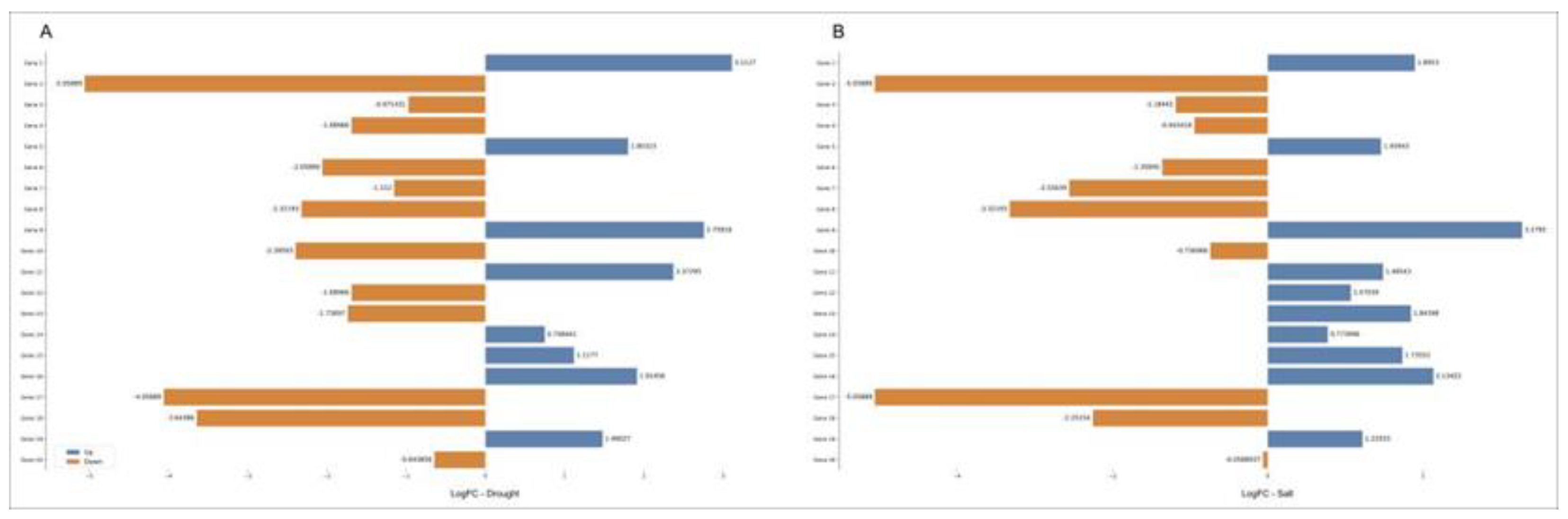
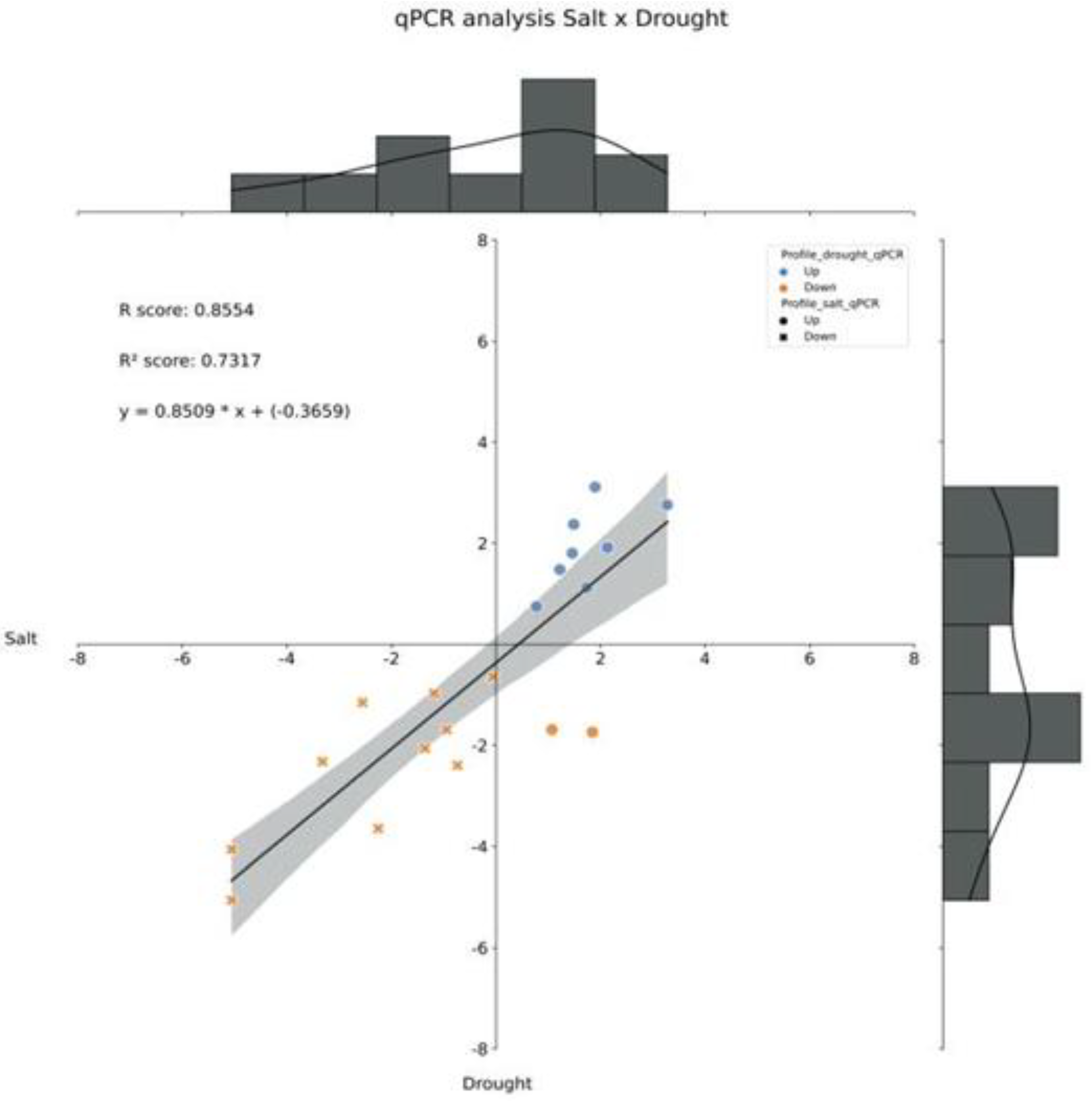
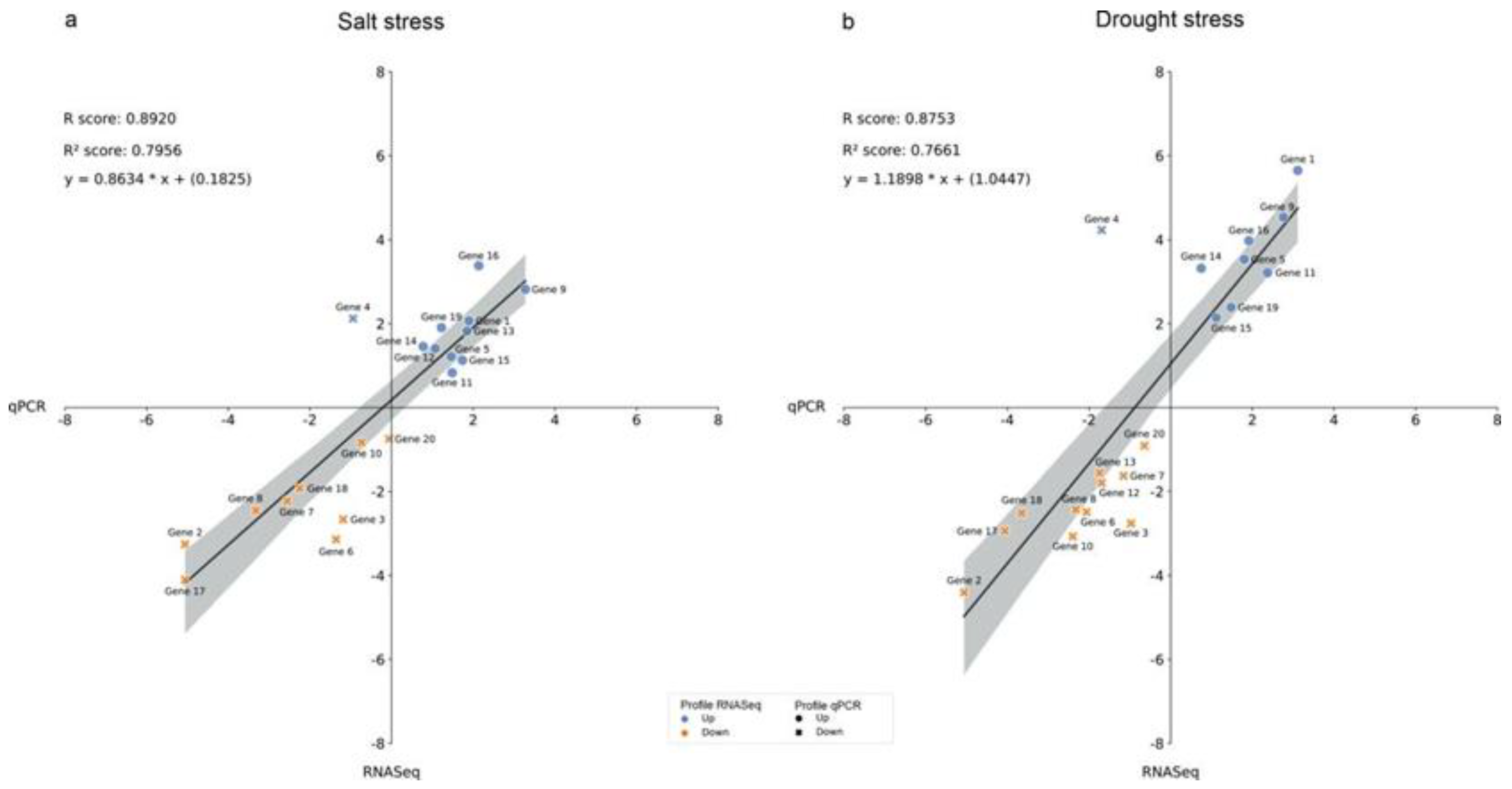
2.4. Differential expression and correlation analyses
3. Discussion
4. Materials & Methods
4.1. RNA-Seq data generation
4.2. RNA-Seq data analysis
4.3. Genes selection and annotation
4.4. RNA extraction, reverse transcriptase-PCR and quantitative real-time PCR analyses
4.5. Pair-by-pair comparison and correlation analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Statista, 2023. Available online: www.statista.com (accessed on June 30, 2023).
- Brasil. Ministério da Agricultura, Pecuária e Abastecimento. 2028. Diagnóstico da Produção Sustentável da Palma de Óleo no Brasil / Ministério da Agricultura, Pecuária e Abastecimento - MAPA. Available online: http://www.abrapalma.org/pt/wp-content/uploads/2014/12/DIAGNOSTICO_PALMA.pdf.
- Corley, R.H.V.; Rao, V.; Palat, T.; Praiwan, T. Breeding for drought tolerance in oil palm. J. Oil Palm Res. 2018, 30, 26–35. [Google Scholar]
- Zaman, M.; Shahid, S.A.; Heng, L. Guideline for salinity assessment, mitigation and adaptation using nuclear and related techniques. Springer International Publishing 2018. [CrossRef]
- Machado, R.; Serralheiro, R. Soil salinity: Effect on vegetable crop growth. Manag. Pract. Prev. Mitigate Soil Salinization. Hortic. 2017, 3, 30. [Google Scholar] [CrossRef]
- Shahid, S.A.; Zaman, M.; Heng, L. Soil salinity: Historical perspectives and a world overview of the problem; Guideline for Salinity Assessment, Mitigation and Adaptation Using Nuclear and Related Techniques; Zaman, M., Shahid, S.A., Heng, L., Eds.; Springer International Publishing, 2018; pp. 43–53. [Google Scholar] [CrossRef]
- Ivushkin, K.; Bartholomeus, H.; Bregt, A.K.; Pulatov, A.; Kempen, B.; De Sousa, L. Global mapping of soil salinity change. Remote Sens. Environ. 2019, 231, 111260. [Google Scholar] [CrossRef]
- Angon, P.B.; Tahjib-Ul-Arif, Md. , Samin, S.I.; Habiba, U.; Hossain, M.A.; Brestic, M. How do plants respond to combined drought and salinity stress?—A systematic review. Plants 2022, 11, 2884. [Google Scholar] [CrossRef]
- Wang, W.; Pang, J.; Zhang, F.; Sun, L.; Yang, L.; Fu, T.; Guo, L.; Siddique, K.H. M. Salt-responsive transcriptome analysis of canola roots reveals candidate genes involved in the key metabolic pathway in response to salt stress. Sci. Rep. 2022, 12, 1666. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Wang, J.; Ye, Y.; Xu, M.; Feng, L.; Xu, L. Roles of the SPL gene family and miR156 in the salt stress responses of tamarisk (Tamarix chinensis). BMC Plant Biol. 2019, 19, 370. [Google Scholar] [CrossRef]
- Javed, T.; Shabbir, R.; Ali, A.; Afzal, I.; Zaheer, U.; Gao, S.-J. Transcription factors in plant stress responses: Challenges and potential for sugarcane improvement. Plants 2020, 9, 491. [Google Scholar] [CrossRef]
- He, M.; He, C.-Q.; Ding, N.-Z. Abiotic stresses: General defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 2018, 9, 1771. [Google Scholar] [CrossRef]
- Hajyzadeh, M.; Turktas, M.; Khawar, K.M.; Unver, T. Mir408 overexpression causes increased drought tolerance in chickpea. Gene 2015, 555, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, R.S.; Jangale, B.L.; Krishna, B.; Sane, P.V. Improved abiotic stress tolerance in Arabidopsis by constitutive active form of a banana DREB2 type transcription factor, MaDREB20.CA, than its native form, MaDREB20. Protoplasma 2023, 260, 671–690. [Google Scholar] [CrossRef] [PubMed]
- Belamkar, V.; Weeks, N.T.; Bharti, A.K.; Farmer, A.D.; Graham, M.A.; Cannon, S.B. Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genom. 2014, 15, 950. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Khurana, J.P.; Jain, M. Characterization of rice homeobox genes, oshox22 and oshox24, and over-expression of oshox24 in transgenic arabidopsis suggest their role in abiotic stress response. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.; Ding, Y.; Hu, S.; Ding, L.; Chen, Z.; Zhu, C. The role of HD-Zip class I transcription factors in plant response to abiotic stresses. Physiol. Plant. 2019, 167, 516–525. [Google Scholar] [CrossRef]
- Sharif, R.; Raza, A.; Chen, P.; Li, Y.; El-Ballat, E.M.; Rauf, A.; Hano, C.; El-Esawi, M.A. Hd-zip gene family: Potential roles in improving plant growth and regulating stress-responsive mechanisms in plants. Genes 2021, 12, 1256. [Google Scholar] [CrossRef] [PubMed]
- Dezar, C.A.; Gago, G.M.; González, D.H.; Chan, R.L. Hahb-4, a sunflower homeobox-leucine zipper gene, is a developmental regulator and confers drought tolerance to Arabidopsis thaliana plants. Transgenic Res. 2005, 14, 429–440. [Google Scholar] [CrossRef]
- Jiao, P.; Jiang, Z.; Wei, X.; Liu, S.; Qu, J.; Guan, S.; Ma, Y. Overexpression of the homeobox-leucine zipper protein ATHB-6 improves the drought tolerance of maize (Zea mays L.). Plant Sci. 2022, 316, 111159. [Google Scholar] [CrossRef]
- Zhao, Y.; Ma, Q.; Jin, X.; Peng, X.; Liu, J.; Deng, L.; Yan, H.; Sheng, L.; Jiang, H.; Cheng, B. A novel maize homeodomain–leucine zipper (Hd-zip) i gene, zmhdz10, positively regulates drought and salt tolerance in both rice and arabidopsis. Plant Cell Physiol. 2014, 55, 1142–1156. [Google Scholar] [CrossRef]
- Salgado, F.F.; Vieira, L.R.; Silva, V.N. B.; Leão, A.P.; Grynberg, P.; Do Carmo Costa, M.M.; Togawa, R.C.; De Sousa, C.A. F.; Júnior, M.T. S. Expression analysis of miRNAs and their putative target genes confirm a preponderant role of transcription factors in the early response of oil palm plants to salinity stress. BMC Plant Biol. 2021, 21, 518. [Google Scholar] [CrossRef]
- Salgado, F.F.; Da Silva, T.L. C.; Vieira, L.R.; Silva, V.N. B.; Leão, A.P.; Costa, M.M. D. C.; Togawa, R.C.; De Sousa, C.A. F.; Grynberg, P.; Souza, M.T. The early response of oil palm (Elaeis guineensis Jacq.) plants to water deprivation: Expression analysis of miRNAs and their putative target genes, and similarities with the response to salinity stress. Front. Plant Sci. 2022, 13, 970113. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, Z.; Zhao, H.; Zhao, Y.; Cheng, B.; Xiang, Y. Genome-wide analysis of soybean hd-zip gene family and expression profiling under salinity and drought treatments. PLoS ONE 2014, 9, e87156. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, T.J.; McIntyre, C.L.; Collet, C.; Xue, G.-P. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol. Biol. 2007, 65, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Roelofs, D.; Morgan, J.; Stürzenbaum, S. The significance of genome-wide transcriptional regulation in the evolution of stress tolerance. Evol. Ecol. 2010, 24, 527–539. [Google Scholar] [CrossRef]
- Qin, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011, 52, 1569–1582. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. (2007). Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Em J. A. Goode & D. Chadwick (Orgs.), Novartis Foundation Symposia (p. 176–189). John Wiley & Sons, Ltd. [CrossRef]
- Ni, Z.; Hu, Z.; Jiang, Q.; Zhang, H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 2013, 82, 113–129. [Google Scholar] [CrossRef]
- Xu, Z.; Gongbuzhaxi, *!!! REPLACE !!!*; Wang, C.; Xue, F.; Zhang, H.; Ji, W. Wheat NAC transcription factor TaNAC29 is involved in response to salt stress. Plant Physiol. Biochem. 2015, 96, 356–363. [Google Scholar] [CrossRef]
- Song, S.; Xu, Y.; Huang, D.; Miao, H.; Liu, J.; Jia, C.; Hu, W.; Valarezo, A.V.; Xu, B.; Jin, Z. Identification of a novel promoter from banana aquaporin family gene ( MaTIP1;2 ) which responses to drought and salt-stress in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2018, 128, 163–169. [Google Scholar] [CrossRef]
- Yu, Y.; Xu, W.; Wang, J.; Wang, L.; Yao, W.; Xu, Y.; Ding, J.; Wang, Y. A core functional region of the RFP1 promoter from Chinese wild grapevine is activated by powdery mildew pathogen and heat stress. Planta 2013, 237, 293–303. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Roy, S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 2016, 11, e1117723. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple functions of myb transcription factors in abiotic stress responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Gao, S.; Zhang, Y.L.; Yang, L.; Song, J.B.; Yang, Z.M. AtMYB20 is negatively involved in plant adaptive response to drought stress. Plant Soil. 2014, 376, 433–443. [Google Scholar] [CrossRef]
- Gong, Q.; Li, S.; Zheng, Y.; Duan, H.; Xiao, F.; Zhuang, Y.; He, J.; Wu, G.; Zhao, S.; Zhou, H.; Lin, H. SUMOylation of MYB30 enhances salt tolerance by elevating alternative respiration via transcriptionally upregulating AOX1a in Arabidopsis. Plant J. 2020, 102, 1157–1171. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Suh, M.C. Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 2015, 34, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.E.; Repetti, P.P.; Adams, T.R.; Creelman, R.A.; Wu, J.; Warner, D.C.; Anstrom, D.C.; Bensen, R.J.; Castiglioni, P.P.; Donnarummo, M.G.; Hinchey, B.S.; Kumimoto, R.W.; Maszle, D.R.; Canales, R.D.; Krolikowski, K.A.; Dotson, S.B.; Gutterson, N.; Ratcliffe, O.J.; Heard, J.E. Plant nuclear factor Y (Nf-y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 2007, 104, 16450–16455. [Google Scholar] [CrossRef]
- Leyva-González, M.A.; Ibarra-Laclette, E.; Cruz-Ramírez, A.; Herrera-Estrella, L. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis nf-ya family members. PLoS ONE 2012, 7, e48138. [Google Scholar] [CrossRef]
- Li, L.; Yu, Y.; Wei, J.; Huang, G.; Zhang, D.; Liu, Y.; Zhang, L. Homologous HAP5 subunit from Picea wilsonii improved tolerance to salt and decreased sensitivity to ABA in transformed Arabidopsis. Planta 2013, 238, 345–356. [Google Scholar] [CrossRef]
- Kumar, R. Role of micrornas in biotic and abiotic stress responses in crop plants. Appl. Biochem. Biotechnol. 2014, 174, 93–115. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, D.; Kong, F.; Lin, K.; Zhang, H.; Li, G. The Arabidopsis thaliana nuclear factor y transcription factors. Frontiers in Plant Science 2017, 07. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Z.; Zhang, Y.; Guo, J.; Liu, L.; Wang, C.; Wang, B.; Han, G. The roles of HD-ZIP proteins in plant abiotic stress tolerance. Front. Plant Sci. 2022, 13, 1027071. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, J.-H.; Ma, X.; Luo, D.-X.; Gong, Z.-H.; Lu, M.-H. The plant heat stress transcription factors (Hsfs): Structure, regulation, and function in response to abiotic stresses. Frontiers in Plant Science 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wan, X.-L.; Yu, J.-Y.; Wang, K.-L.; Zhang, J. Genome-wide identification, classification, and expression analysis of the hsf gene family in carnation (Dianthus caryophyllus). Int. J. Mol. Sci. 2019, 20, 5233. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liang, J.; Wang, C.; Zhao, X.; Zhong, X.; Cao, X.; Li, G.; He, J.; Yi, M. Overexpression of lily HsfA3s in Arabidopsis confers increased thermotolerance and salt sensitivity via alterations in proline catabolism. J. Exp. Bot. 2018, 69, 2005–2021. [Google Scholar] [CrossRef]
- Pérez-Salamó, I.; Papdi, C.; Rigó, G.; Zsigmond, L.; Vilela, B.; Lumbreras, V.; Nagy, I.; Horváth, B.; Domoki, M.; Darula, Z.; Medzihradszky, K.; Bögre, L.; Koncz, C.; Szabados, L. The heat shock factor a4a confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases mpk3 and mpk6. Plant Physiology 2014, 165, 319–334. [Google Scholar] [CrossRef]
- Zhu, J.-K. SALT AND DROUGHT STRESS SIGNAL TRANSDUCTION IN PLANTS. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Rasmussen, S.; Barah, P.; Suarez-Rodriguez, M.C.; Bressendorff, S.; Friis, P.; Costantino, P.; Bones, A.M.; Nielsen, H.B.; Mundy, J. Transcriptome responses to combinations of stresses in arabidopsis. Plant Physiol. 2013, 161, 1783–1794. [Google Scholar] [CrossRef]
- Hazzouri, K.M.; Flowers, J.M.; Nelson, D.; Lemansour, A.; Masmoudi, K.; Amiri, K.M. A. Prospects for the study and improvement of abiotic stress tolerance in date palms in the post-genomics era. Front. Plant Sci. 2020, 11, 293. [Google Scholar] [CrossRef]
- McCoy, R.M.; Julian, R.; Kumar, S.R.V.; Ranjan, R.; Varala, K.; Li, Y. A Systems Biology Approach to Identify Essential Epigenetic Regulators for Specific Biological Processes in Plants. Plants 2021, 10, 364. [Google Scholar] [CrossRef]
- Carvalho da Silva, T.L.; Grynberg, P.; Togawa, R.C.; Souza, M., Jr. T. The Brazilian Agricultural Research Corporation, Embrapa Agroenergy, Brasília, DF, Brazil. 2023, status (manuscript in preparation; to be submitted).
- OmicsBox – Bioinformatics Made Easy, BioBam Bioinformatics, 2019. 3 March. Available online: https://www.biobam.com/omicsbox.
- Singh, R.; Ong-Abdullah, M.; Low, E.-T. L.; Manaf, M.A. A.; Rosli, R.; Nookiah, R.; Ooi, L.C.-L.; Ooi, S.; Chan, K.-L.; Halim, M.A.; et al. Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 2013, 500, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; von Mering, C.; Bork, P. Eggnog 5. 0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. Mega11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
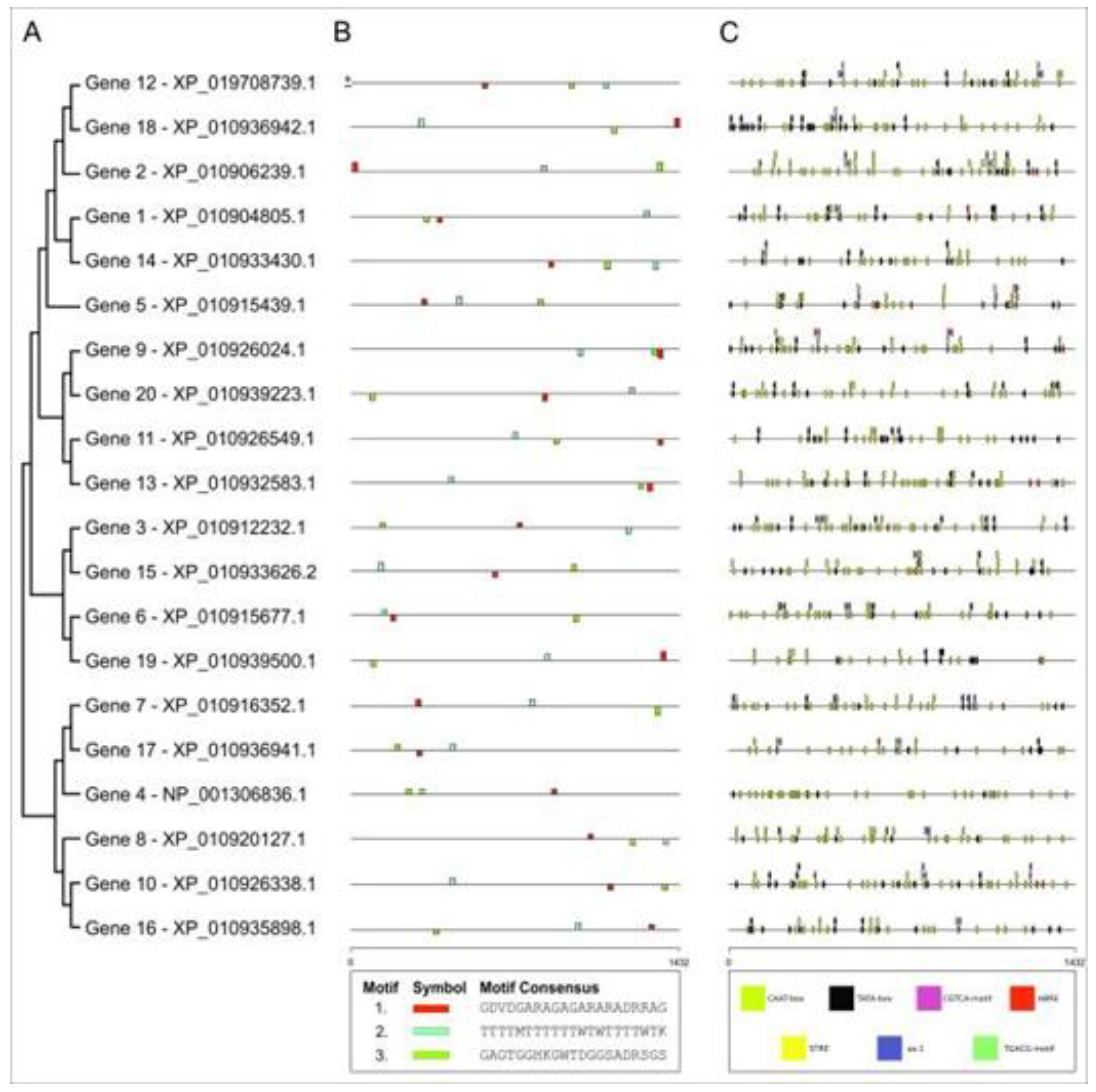
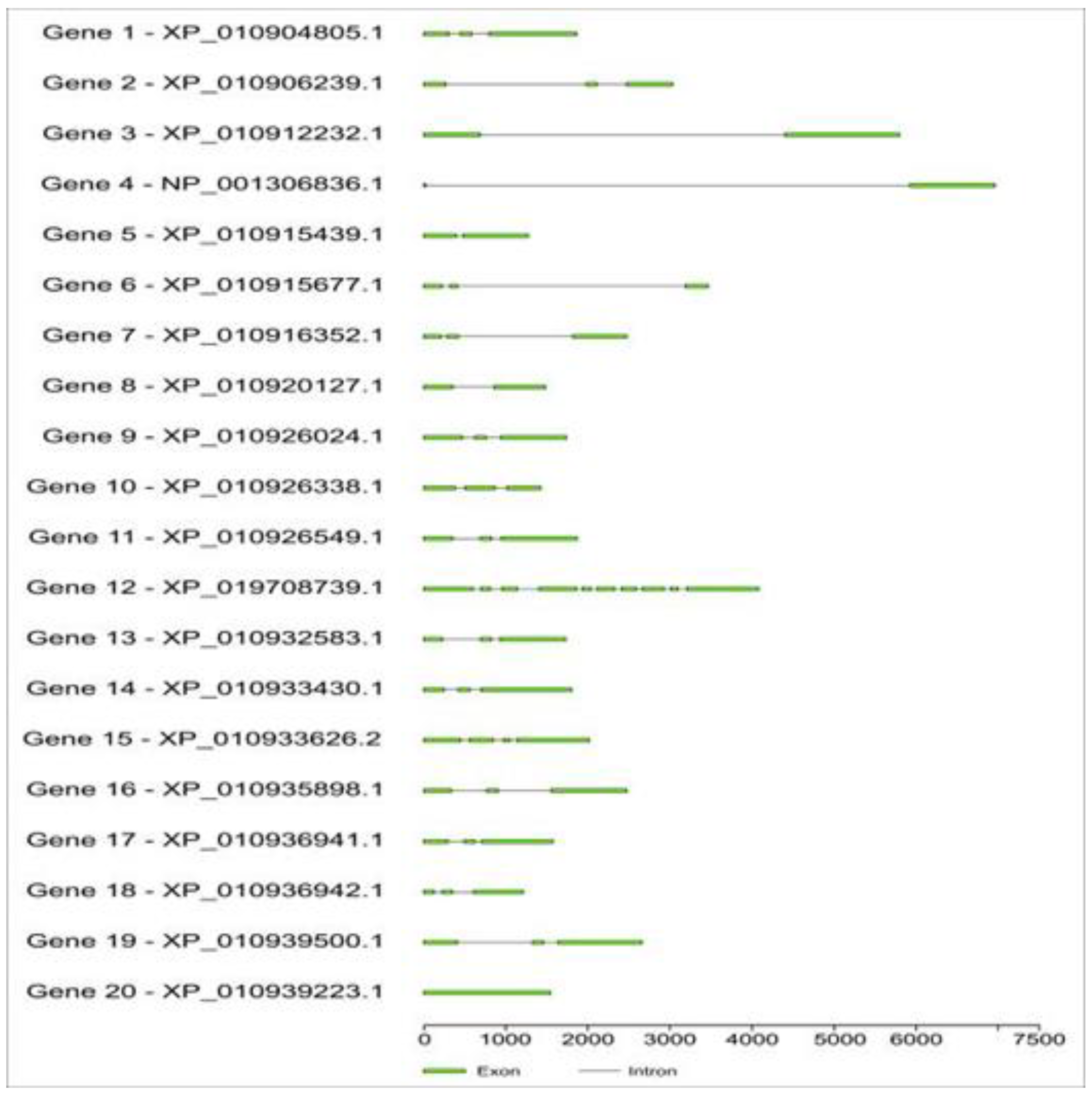
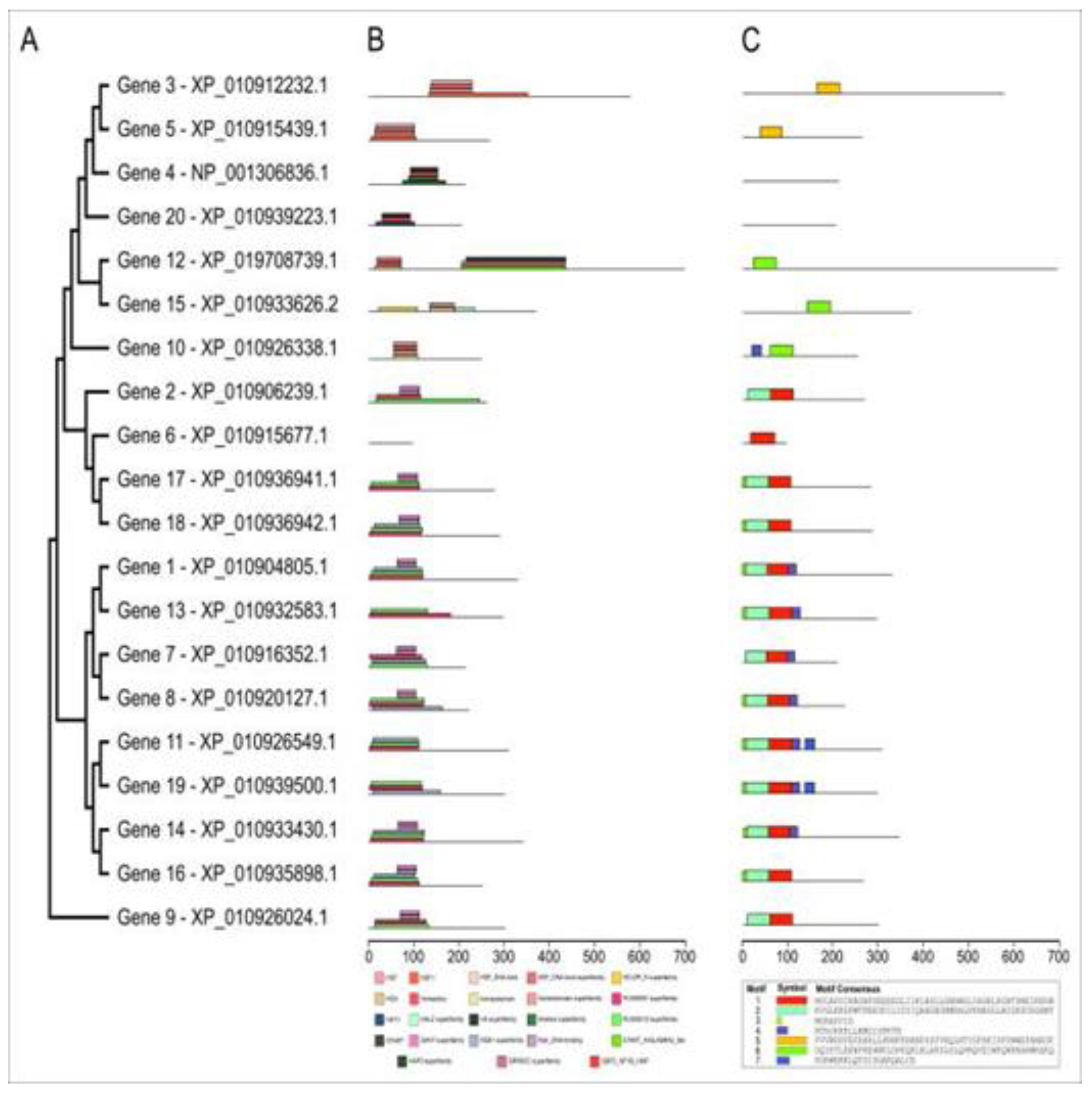
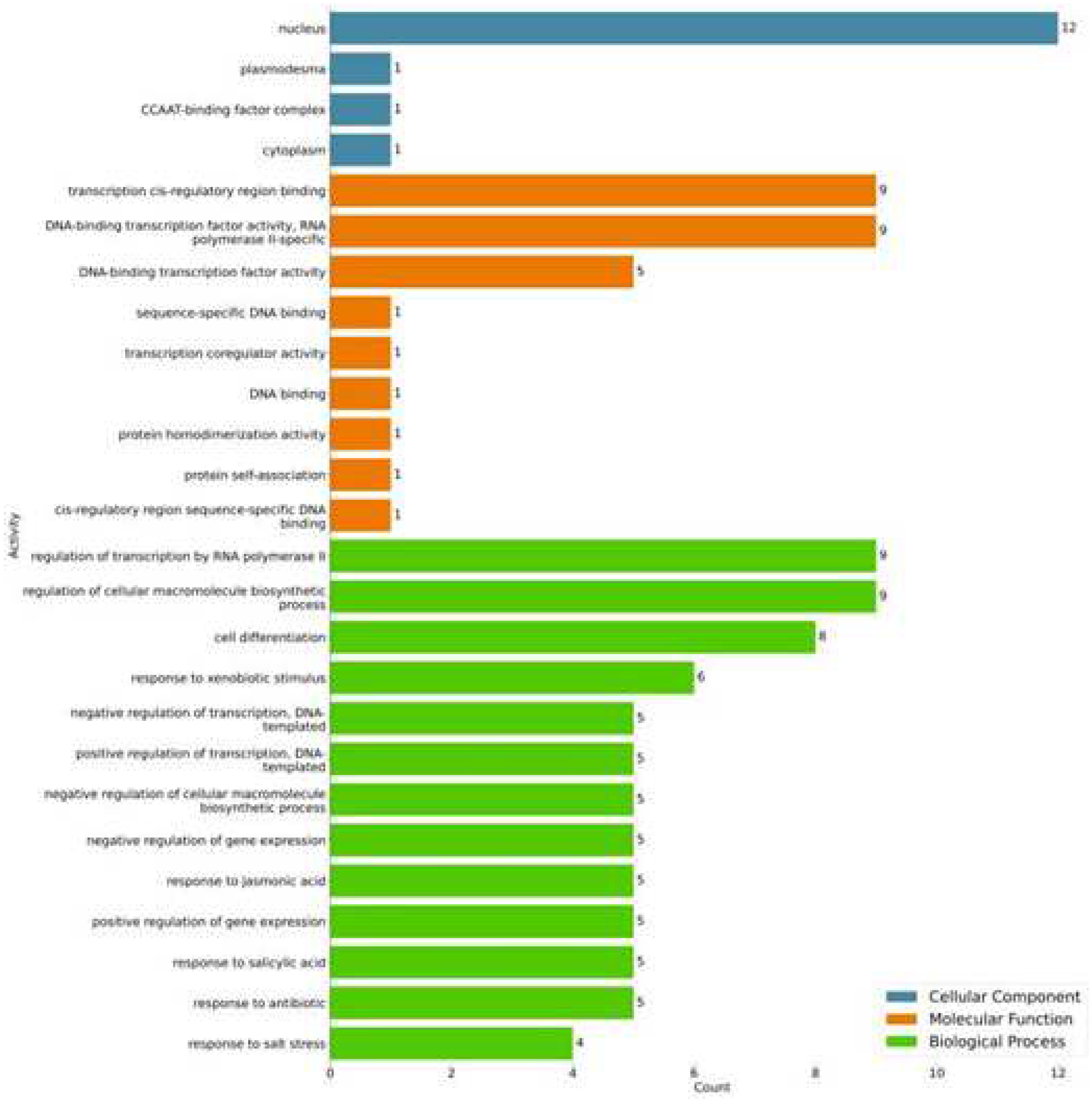
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Gene 01 | GATAAGAATGGTCTGAAGAAGGG | CAATTGCTGACCACTTGTTCC |
| Gene 02 | GGAGGACCAGAAACTTGTAGAC | ATTAGAGACCATCTGTTGCCC |
| Gene 03 | TTTTCGCAAGATTGATGCTG | TCATGAGATGCTCCTGTTGC |
| Gene 04 | GGTAGACCTCTCTGCATCG | CCTACCACCAGCTGAAAATACC |
| Gene 05 | GCTCAACACCTATGGGTTTCG | ACCTTCTCTTCGATCTTCCTCTG |
| Gene 06 | CACCACTTAAGATCCGTTGCC | GCTATCAAATCCCACCTGTCAC |
| Gene 07 | ACGAGAAGCTCATTAACTACATCC | CTATCAATGACCACCGATTGCC |
| Gene 08 | GGACCAAGGAGGAAGATGAC | AGAGACCATTTGTTGCCGAG |
| Gene 09 | CTAAGTGCTCAGGCTTGAAGAG | GTCCTCCAGTAGTTCTTTATCTCG |
| Gene 10 | TCGAGCGTGGTGTTTAATG | CGGAGCAGTACCAAGAGAGG |
| Gene 11 | TTTAGGCAATAGGTGGGCAG | TATTGAGCTTCTCCACAGACAG |
| Gene 12 | AGTCACAGTTCACAAGAGCAC | CATTCAGAGCACGAAGGAGAG |
| Gene 13 | AGCTAGCAGGATTGCTTAGG | CTGGTCAACAGTTTCAGCG |
| Gene 14 | CAAGGGTTCATGGACACCAG | TTCGACCATTTGTTTCCAAGC |
| Gene 15 | GACACAAGCCATGATTGAAG | TAGCGAGCAACCCTTTTATCTC |
| Gene 16 | CAAGCAAGCTGGTCTATTGAGG | CGGCTGGTTTGGATTCACTC |
| Gene 17 | CTTCACTCTAGCGATATCTACCG | AACGTTTCAACCCTGCTCTC |
| Gene 18 | CCATGTTGTTCAAAGGAGGG | GCAATTAGAGACCATCTGTTTCC |
| Gene 19 | CCTACTAATACTGGGCTGATGAG | GGTGTTCCAATAGTTCTTGATGTC |
| Gene 20 | GGTGGGAATTCTTCCTCTTC | TAATTCAGAGACCAACCCAACC |
| Control Gene* | CCAGGGTTCAGCTGATTAAG | TCGTCCAAATCCAGCAATC |
| Gene | Family | Protein ID | Description | Salt Stress | Drought Stress | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EP | FC | Log2(FC) | FDR | EP | FC | Log2(FC) | FDR | ||||
| 1 | MYBP | XP_010904805.1 | transcription factor MYB102 | Up | 4.2 | 2.1 | 0 | Up | 50.2 | 5.6 | 0 |
| 2 | MYBP | XP_010906239.1 | transcription repressor MYB5-like | Down | -9.6 | -3.3 | 0 | Down | -21.2 | -4.4 | 0 |
| 3 | HSFF | XP_010912232.1 | heat stress transcription factor A-3 | Down | -6.3 | -2.7 | 0 | Down | -6.8 | -2.8 | 0 |
| 4 | NFYC, HAP5 | NP_001306836.1 | nuclear transcription factor Y subunit C-2-like | Up | 4.3 | 2.1 | 0 | Up | 18.7 | 4.2 | 0 |
| 5 | HSFF | XP_010915439.1 | heat stress transcription factor C-2b | Up | 2.3 | 1.2 | 0 | Up | 11.6 | 3.5 | 0 |
| 6 | MYBP | XP_010915677.1 | transcription factor TRY | Down | -8.8 | -3.1 | 0 | Down | -5.6 | -2.5 | 0 |
| 7 | MYBP | XP_010916352.1 | myb-related protein 308 | Down | -4.7 | -2.2 | 0 | Down | -3.1 | -1.6 | 0 |
| 8 | MYBP | XP_010920127.1 | myb-related protein 308 | Down | -5.5 | -2.5 | 0 | Down | -5.4 | -2.4 | 0 |
| 9 | MYBP | XP_010926024.1 | transcription factor MYB62 | Up | 7 | 2.8 | 0 | Up | 23.2 | 4.5 | 0 |
| 10 | HD-ZIP | XP_010926338.1 | homeobox-leucine zipper protein HOX8 | Down | -1.8 | -0.8 | 0.016 | Down | -8.4 | -3.1 | 0 |
| 11 | MYBP | XP_010926549.1 | myb-related protein 306 | Up | 1.8 | 0.8 | 0.029 | Up | 9.3 | 3.2 | 0 |
| 12 | HD-ZIP | XP_019708739.1 | homeobox-leucine zipper protein ROC8 | Up | 2.6 | 1.4 | 0.027 | Down | -3.5 | -1.8 | 0.007 |
| 13 | MYBP | XP_010932583.1 | transcription factor MYB20 | Up | 3.5 | 1.8 | 0.001 | Down | -2.9 | -1.6 | 0.001 |
| 14 | MYBP | XP_010933430.1 | myb-related protein Zm1-like | Up | 2.7 | 1.5 | 0.022 | Up | 10 | 3.3 | 0 |
| 15 | HD-ZIP | XP_010933626.2 | homeobox-leucine zipper protein HAT4 | Up | 2.2 | 1.1 | 0.024 | Up | 4.4 | 2.1 | 0 |
| 16 | MYBP | XP_010935898.1 | transcription factor MYB30 | Up | 10.4 | 3.4 | 0 | Up | 15.7 | 4 | 0 |
| 17 | MYBP | XP_010936941.1 | transcription factor MYB4 | Down | -17.2 | -4.1 | 0 | Down | -7.7 | -2.9 | 0 |
| 18 | MYBP | XP_010936942.1 | anthocyanin regulatory C1 protein-like | Down | -3.8 | -1.9 | 0.002 | Down | -5.7 | -2.5 | 0 |
| 19 | MYBP | XP_010939500.1 | myb-related protein 306 | Up | 3.7 | 1.9 | 0 | Up | 5.2 | 2.4 | 0 |
| 20 | NFYB, HAP3 | XP_010939223.1 | nuclear transcription factor Y subunit B-3 | Down | -1.7 | -0.7 | 0.002 | Down | -1.9 | -0.9 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





