Submitted:
05 July 2023
Posted:
06 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Nine Shisa Subfamilies Identified in Zebrafish
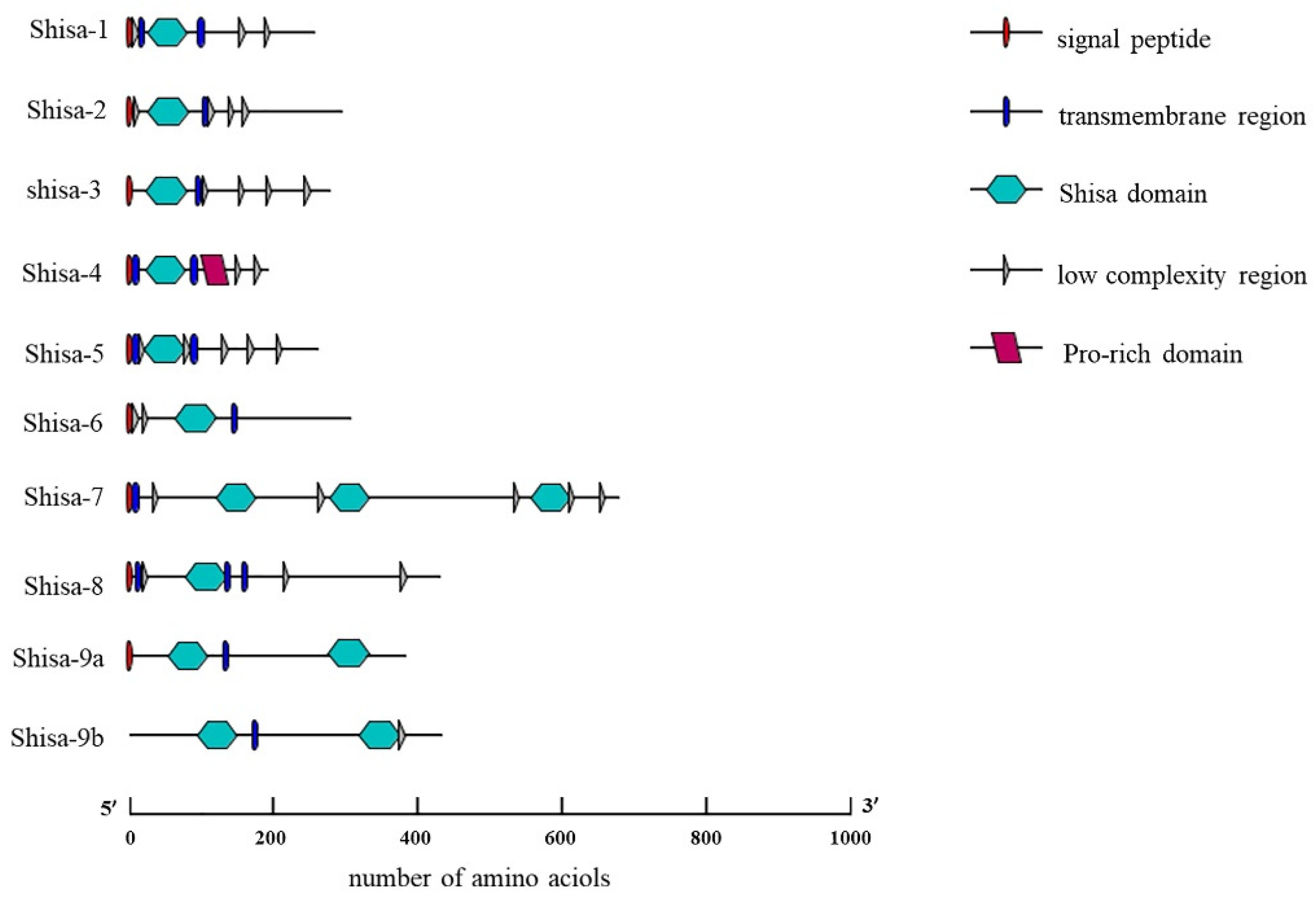
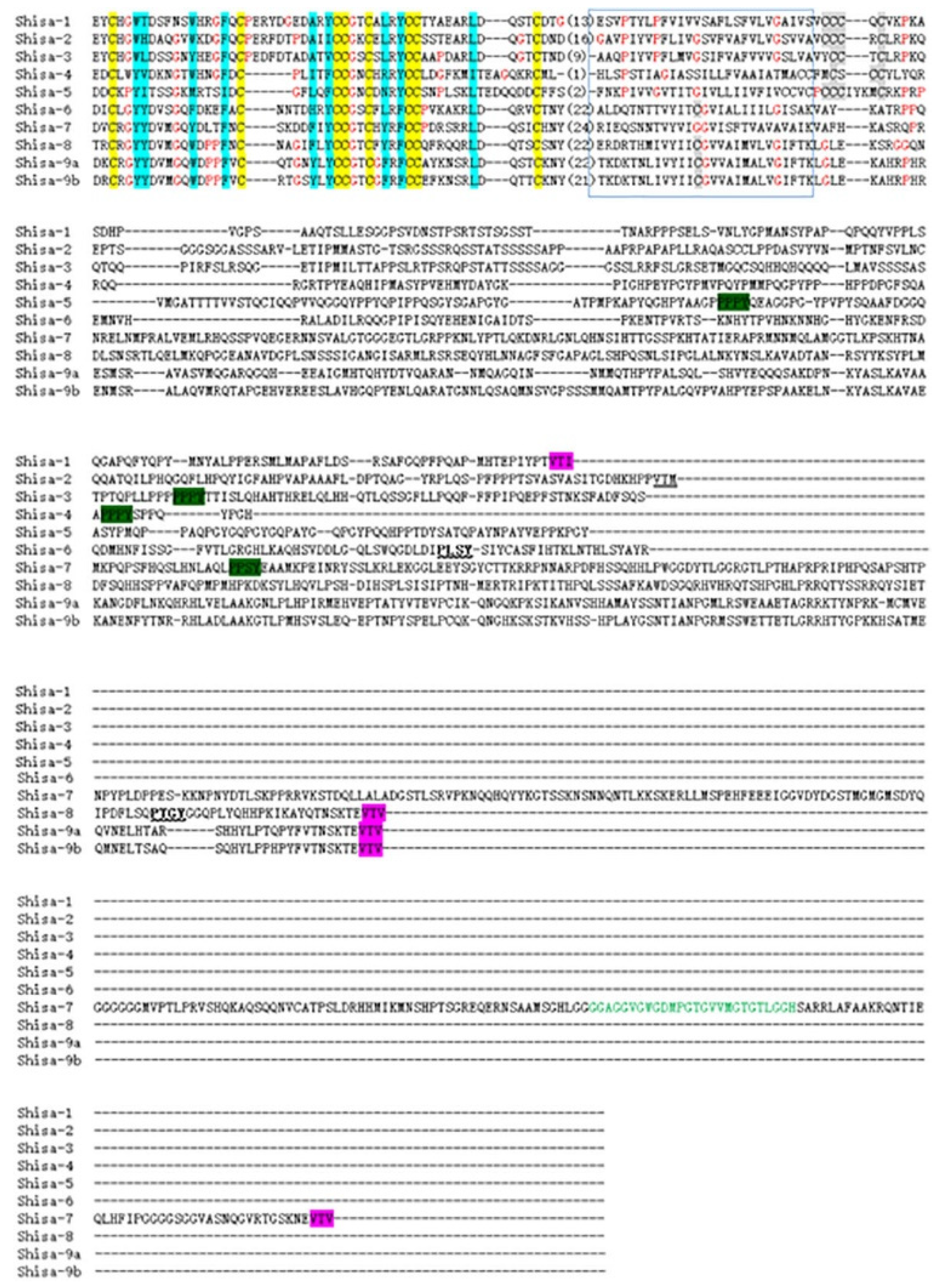
2.2. Conserved Domain Among Shisa Proteins
 ’; C-terminal PDZ-binding like motif, ‘
’; C-terminal PDZ-binding like motif, ‘ ’ .The GRID domain [12] is shown in green letters.
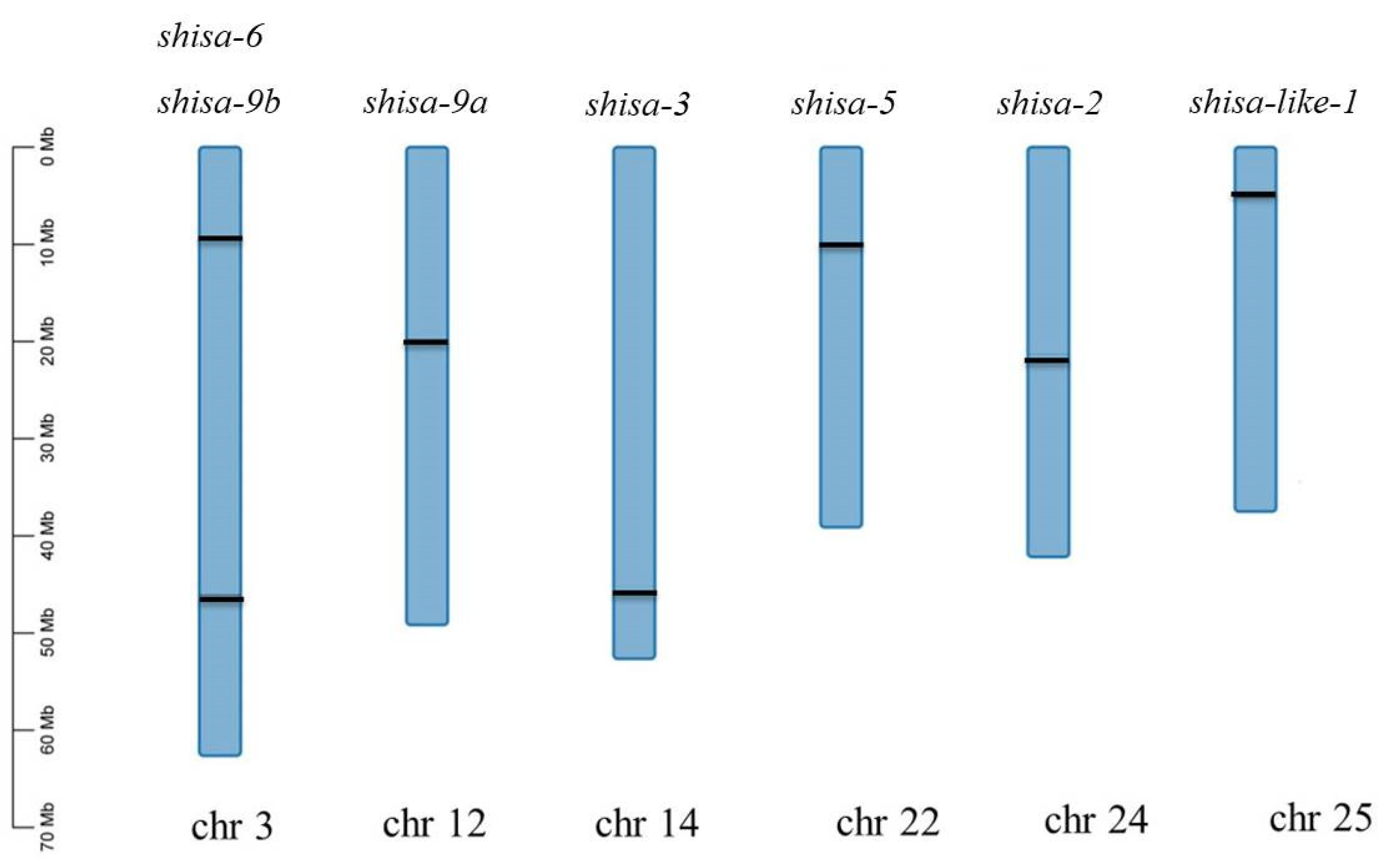
’ .The GRID domain [12] is shown in green letters.2.3. Shisa Genes Localization on Chromosomes
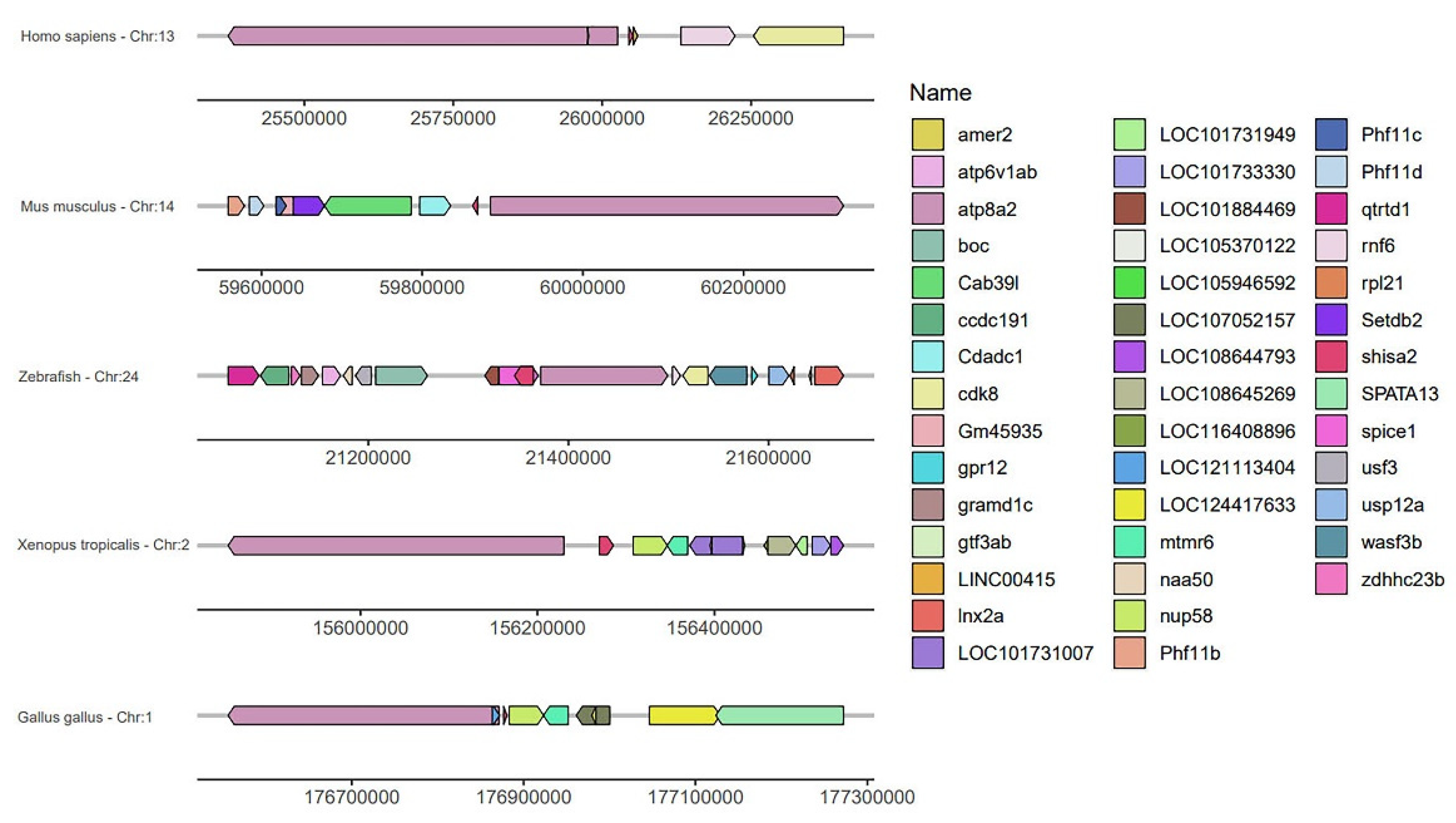
2.4. Conserved Syntenic Block Containing shisa-2
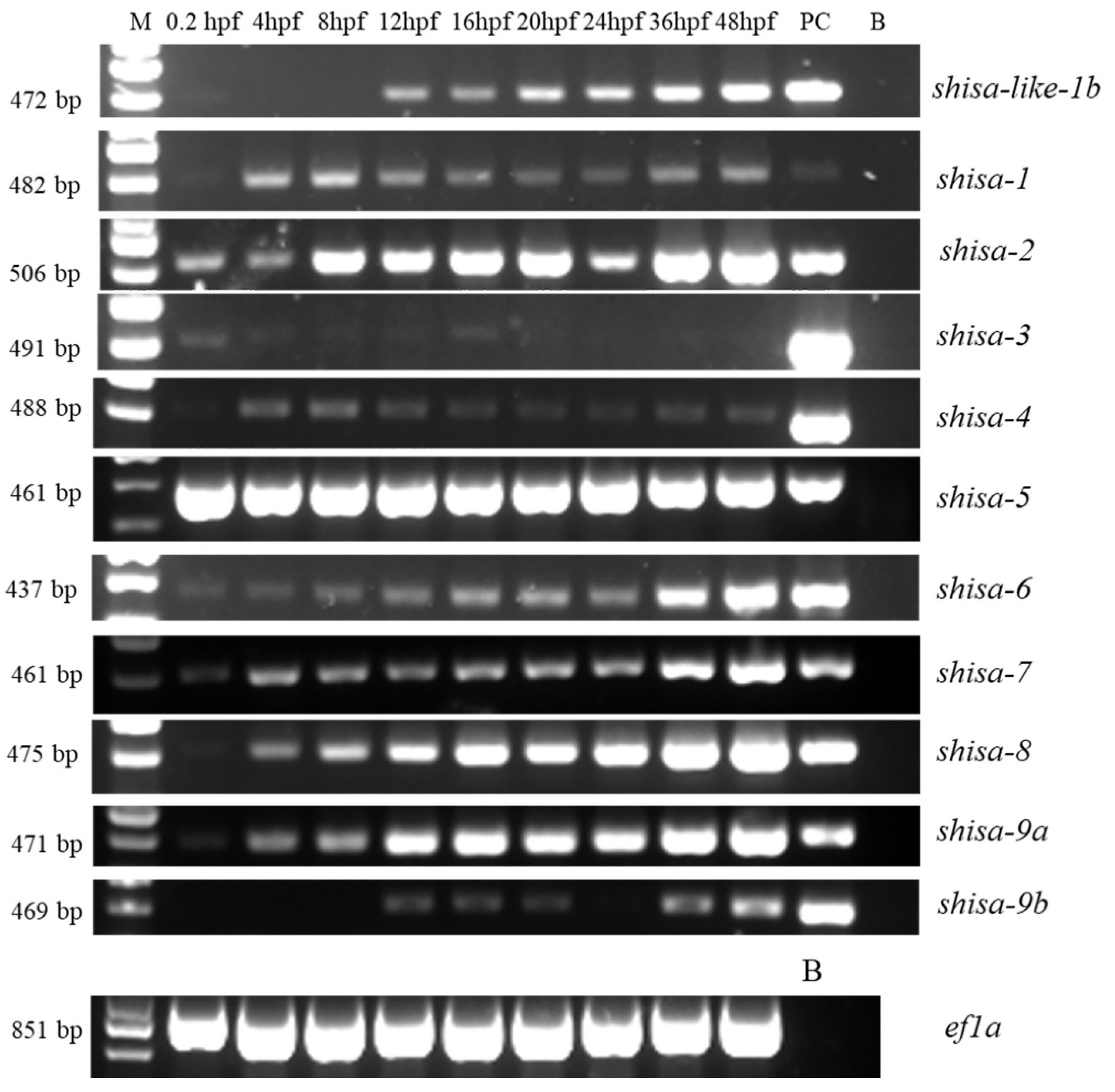
2.5. Different Expression Patterns of Shisa Family Genes During Embryonic Development
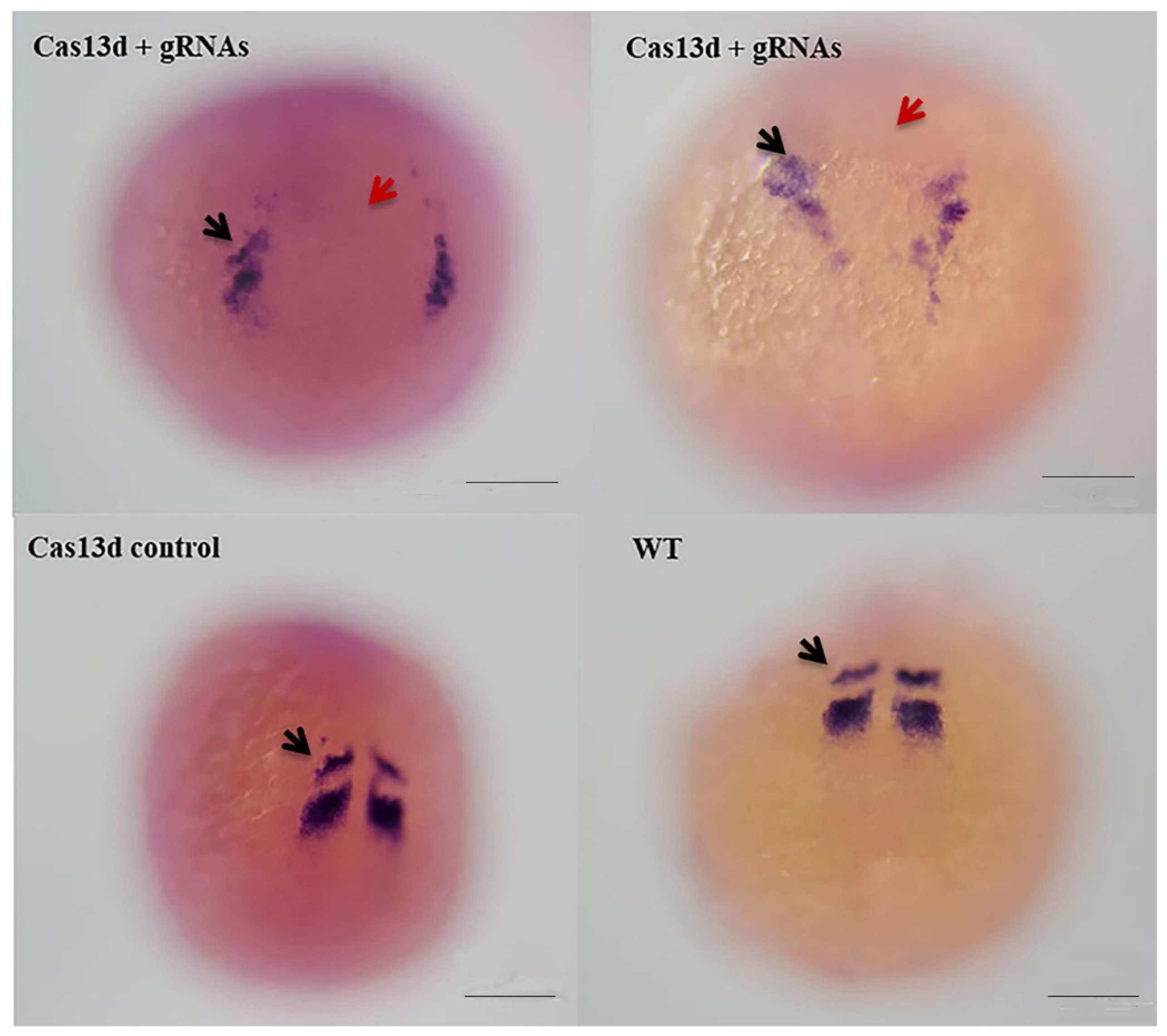
2.6. Abnormal Expression of mesp-ab by shisa-2 Knockdown
3. Discussion
3.1. Different Evolutionary Processes for Shisa Subfamily Genes
3.2. Conserved Domains and Divergent C-terminal Regions of Shisa Proteins
3.3. Extensive Participation of Shisa Family Genes During Zebrafish Embryonic Development
3.4. Shisa-2 Regulating the Convergent Extension Cell Movement of the Somatic Precursors in Zebrafish
3.5. Limitations
4. Materials and Methods
4.1. Sample Collection
4.2. Total RNA Isolation and cDNA Synthesis
4.3. Cloning of Zebrafish Shisa Family Genes
4.4. Bioinformatics Analysis
4.5. Knockdown of shisa 2 in Zebrafish Embryos Using Cas13d mRNA and gRNAs
4.6. Rescue Using Flounder shisa-2 mRNA
4.7. Semi-quantitative PCR
4.8. Whole Mount in situ Hybridization.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Pan, C.Q.; Sudol, M.; Sheetz, M.; Low, B.C. Modularity and functional plasticity of scaffold proteins as p(l)acemakers in cell signaling. Cell Signal 2012, 24, 2143–2165. [Google Scholar] [CrossRef] [PubMed]
- Zeke, A.; Lukacs, M.; Lim, W.A.; Remenyi, A. Scaffolds: interaction platforms for cellular signalling circuits. International Journal of Developmental Biology 2009, 19, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Langeberg, L.K.; Scott, J.D. Signalling scaffolds and local organization of cellular behavior. Nature Reviews Molecular Cell Biology 2015, 16, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Grishin, N.V. Unexpected diversity in Shisa-like proteins suggests the importance of their roles as transmembrane adaptors. Cellular Signalling 2012, 24, 758–769. [Google Scholar] [CrossRef]
- Yamamoto, A.; Nagano, T.; Takehara, S.; Hibi, M.; Aizawa, S. Shisa promotes head formation through the inhibition of receptor protein maturation for the caudalizing factors, Wnt and FGF. Cell 2005, 120, 223–235. [Google Scholar] [CrossRef]
- Nagano, T.; Takehara, S.; Takahashi, M.; Aizawa, S.; Yamamoto, A. Shisa2 promotes the maturation of somitic precursors and transition to the segmental fate in Xenopus embryos. Development 2006, 133, 4643–4654. [Google Scholar] [CrossRef]
- Onishi, K.; Zou, Y. Sonic Hedgehog switches on Wnt/planar cell polarity signaling in commissural axon growth cones by reducing levels of Shisa2. eLife 2017, 6, e25269. [Google Scholar] [CrossRef]
- Liu, Z.J.; Wang, C.; Liu, X.Q.; Kuang, S. Shisa2 regulates the fusion of muscle progenitors. Stem Cell Research 2018, 31, 31–41. [Google Scholar] [CrossRef]
- Chen, C.C.; Chen, H.Y.; Su, K.Y.; Hong, Q.S.; Yan, B.S.; Chen, C.H.; Pan, S.H.; Chang, Y.L.; Wang, C.J.; Hung, P.F.; Yuan, S.; Chang, G.C.; Chen, J.J.W.; Yang, P.C.; Yang, Y.C.; Yu, S.L. Shisa3 is associated with prolonged survival through promoting beta-catenin degradation in lung cancer. American Journal of Respiratory and Critical Care Medicine 2014, 190, 433–444. [Google Scholar] [CrossRef]
- von Engelhardt, J.; Mack, V.; Sprengel, R.; Kavenstock, N.; Li, K.W.; Stern-Bach, Y.; Smit, A.B.; Seeburg, P.H.; Monyer, H. CKAMP44: a brain-specific protein attenuating short-term synaptic plasticity in the dentate gyrus. Science 2010, 327, 1518–1522. [Google Scholar] [CrossRef]
- Gupta, R.K.; Tripathi, R.; Naidu, B.J.; Srinivas, U.; Shashidhara, L. Cell cycle regulation by the pro-apoptotic gene Scotin. Cell Cycle 2014, 7, 2401–2408. [Google Scholar] [CrossRef]
- Han, W.; Li, J.; Pelkey, K.A.; Pandey, S.; Chen, X.; Wang, Y.X.; Wu, K.W.; Ge, L.; Li, T.M.; Castellano, D. Shisa7 is a GABA(A) receptor auxiliary subunit controlling benzodiazepine actions. Science 2019, 366, 246–250. [Google Scholar] [CrossRef]
- Abdollahi Nejat, M.; Klaassen, R.V.; Spijker, S.; Smit, A.B. Auxiliary subunits of the AMPA receptor: The Shisa family of proteins. Current Opinion in Pharmacology 2021, 58, 52–61. [Google Scholar] [CrossRef]
- Hedge, T.A.; Mason, I. Expression of Shisa2, a modulator of both Wnt and Fgf signaling, in the chick embryo. The International Journal of Developmental Biology 2008, 52, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Bourdon, J.C.; Renzing, J.; Robertson, P.L.; Fernandes, K.N.; Lane, D.P. Scotin, a novel p53-inducible proapoptotic protein located in the ER and the nuclear membrane. Journal of Cell Biology 2002, 158, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kim, M.J.; Sung, P.S.; Bae, Y.C.; Shin, E.C.; Yoo, J.Y. Interferon-inducible protein SCOTIN interferes with HCV replication through the autolysosomal degradation of NS5A. Nature Communication 2016, 7, 10631. [Google Scholar] [CrossRef] [PubMed]
- Furushima, K.; Yamamoto, A.; Nagano, T.; Shibata, M.; Miyachi, H.; Abe, T.; Ohshima, N.; Kiyonari, H.; Aizawa, S. Mouse homologues of Shisa antagonistic to Wnt and Fgf signalings. Developmental Biology 2007, 306, 480–492. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7. 0 for Bigger Datasets. Molecular Biology and Evolution 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Shams, F.; Dyer, F.; Thompson, R.; Duncan, R.P.; Thiem, J.D.; Kilian, A.; Ezaz, T. Application of DArT seq derived SNP tags for comparative genome analysis in fishes; An alternative pipeline using sequence data from a non-traditional model species, Macquaria ambigua. PLoS One 2019, 14(12), e0226365. [Google Scholar] [CrossRef]
- Minchin, J.E.; Hughes, S.M. Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Developmental Biology 2008, 319, 530. [Google Scholar] [CrossRef]
- Minchin, J.E.N.; Williams, V.C.; Hinits, Y.; Low, S.; Tandon, P.; Fan, C.M.; Rawls, J.F.; Hughes, S.M. Oesophageal and sternohyal muscle fibres are novel Pax3-dependent migratory somite derivatives essential for ingestion (vol 140, pg 2972, 2013). Development 2013, 140, 4296. [Google Scholar] [CrossRef]
- Jiao, S.; Tan, X.G.; Wang, Q.; Li, M.J.; Du, S.J. The olive flounder (Paralichthys olivaceus) Pax3 homologues are highly conserved, encode multiple isoforms and show unique expression patterns. Comparative Biochemistry and Physiology B-Biochemistry & Molecular Miology 2015, 180, 7–15. [Google Scholar]
- Jiao, S.; Tan, X.G.; Li, M.J.; Sui, Y.L.; Du, S.J.; You, F. The duplicated paired box protein 7 (pax7) genes differentially transcribed during Japanese flounder (Paralichthys olivaceus) embryogenesis. Comparative Biochemistry and Physiology B-Biochemistry & Molecular Biology 2015, 189, 62–68. [Google Scholar]
- Venkatesh, B. Evolution and diversity of fish genomes. Current Opinion in Genetics & Development 2003, 13, 588–592. [Google Scholar]
- Linder, M.E.; Deschenes, R.J. Palmitoylation: policing protein stability and traffic. Nature Reviews Molecular Cell Biology 2007, 8, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Amacher, J.F.; Brooks, L.; Hampton, T.H.; Madden, D.R. Specificity in PDZ-peptide interaction networks: Computational analysis and review. J Struct Biol X 2020, 4, 100022. [Google Scholar] [CrossRef]
- Murciano-Calles, J. The Conformational Plasticity Vista of PDZ Domains. Life (Basel) 2020, 10(8), 123. [Google Scholar] [CrossRef]
- Kimmel, C,B; Ballard, W.W; Kimmel, S. R.; Ullmann, B.; Schilling,, T. F. Stages of embryonic development of the zebrafish. Dev Dyn 1995, 203(3), 253–310. [Google Scholar]
- Abdollahi Nejat, M.; Klaassen, R.V.; Spijker, S.; Smit, A.B. Auxiliary subunits of the AMPA receptor: The Shisa family of proteins. Current Opinion in Pharmacology. 2021, 58, 52–61. [Google Scholar] [CrossRef]
- Silva, A.C.; Filipe, M.; Vitorino, M.; Steinbeisser, H.; Belo, J.A. Developmental expression of Shisa-2 in Xenopus laevis. Int J Dev Biol 2006, 50(6), 575–579. [Google Scholar] [CrossRef]
- Cutty, S.; Fior, R.; Henriques, P.M.; Saude, L.; Wardle, F.C. Identification and expression analysis of two novel members of the Mesp family in zebrafish. International Journal of Developmental Biology 2012, 56, 285–294. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit De-veloped for Interactive Analyses of Big Biological Data. Molecular Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Springer 2016.
- Kushawah, G.; Hernandez-Huertas, L.; del Prado, J.A.N.; Martinez-Morales, J.R.; DeVore, M.L.; Hassan, H.; Moreno-Sanchez, I.; Tomas-Gallardo, L.; Diaz-Moscoso, A.; Monges, D.E.; Guelfo, J.R.; Theune, W.C.; Brannan, E.O.; Wang, W.; Corbin, T.J.; Moran, A.M.; Alvarado, A.S.; Malaga-Trillo, E.; Takacs, C.M.; Bazzini, A.A.; Moreno-Mateos, M.A. CRISPR-Cas13d Induces Efficient mRNA Knockdown in Animal Embryos. Developmental Cell 2020, 54, 805. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Rahman, J.A.; Wessels, H.H.; Méndez-Mancilla, A.; Haro, D.; Chen, X.; Sanjana, N.E. Transcriptome-wide Cas13 guide RNA design for model organisms and viral RNA pathogens. Cell Genom 2021, 1(1), 100001. [Google Scholar] [CrossRef] [PubMed]
- Wessels, H.H.; Méndez-Mancilla, A.; Guo, X.; Legut, M.; Daniloski, Z.; Sanjana, N.E. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat Biotechnol 2020, 38, 722–727. [Google Scholar] [CrossRef] [PubMed]
- Du, S.J.; Dienhart, M. The zebrafish tiggy-winkle hedgehog promoter directs notochord and floor plate GFP expression in transgenic zebrafish embryos. Dev Dyn 2001, 222, 655–666. [Google Scholar] [CrossRef]



| Gene Name (NCBI name) | Species | GenBank accession number | |
|---|---|---|---|
| shisa-like 1b | Danio rerio | XM_002667571.6 | |
| shisa-1 ( shisa 2a) | Danio rerio | NM_001003631.1 | |
| shisa-2 | Danio rerio | XM_003201303.5 | |
| shisa-3 | Danio rerio | NM_001080662.2 | |
| shisa-4 | Danio rerio | NM_001017869.1 | |
| shisa-5 | Danio rerio | NM_001044870.1 | |
| shisa -6 | Danio rerio | XM_002667291.5 | |
| shisa-7( shisa like 7) | Danio rerio | XM_021472968.1 | |
| shisa-8 | Danio rerio | XM_021480293.1 | |
| shisa-9a | Danio rerio | NM_001013509.1 | |
| shisa-9b | Danio rerio | NM_001135975.2 | |
| shisa-like 1 | Gallus gallus | XM_015291144.4 | |
| Shisa-2 | Gallus gallus | NM_204501.2 | |
| Shisa-3 | Gallus gallus | XM_040700477.2 | |
| Shisa-4 | Gallus gallus | XM_015298798.4 | |
| Shisa-5 | Gallus gallus | NM_001030591.3 | |
| Shisa-6 | Gallus gallus | XM_025141681.2 | |
| Shisa-8 | Gallus gallus | XM_003640380.6 | |
| Shisa-9 | Gallus gallus | XM_040647667.2 | |
| Shisa-2 | Homo sapiens | NM_001007538.2 | |
| Shisa-3 | Homo sapiens | NM_001080505.3 | |
| Shisa-4 | Homo sapiens | NM_198149.3 | |
| Shisa-5 | Homo sapiens | NM_001272065.3 | |
| Shisa-6 | Homo sapiens | NM_001173461.2 | |
| Shisa-7 | Homo sapiens | NM_001145176.2 | |
| Shisa-8 | Homo sapiens | NM_001207020.3 | |
| Shisa-9 | Homo sapiens | NM_001145204.3 | |
| Shisa-like 1 | Homo sapiens | NM_001099294.2 | |
| Shisa-like 2a | Homo sapiens | NM_001042693.3 | |
| Shisa-like 2b | Homo sapiens | NM_001164442.2 | |
| Shisa-2 | Mus musculus | NM_145463.5 | |
| Shisa-3 | Mus musculus | NM_001033415.3 | |
| Shisa-4 | Mus musculus | NM_175259.5 | |
| Shisa-5 | Mus musculus | NM_001284332.1 | |
| Shisa-6 | Mus musculus | NM_001034874.4 | |
| Shisa-7 | Mus musculus | NM_001290291.1 | |
| Shisa-8 | Mus musculus | NM_001207021.2 | |
| Shisa-9 | Mus musculus | NM_001174086.1 | |
| Shisa-like 1 | Mus musculus | NM_001163145.2 | |
| Shisa-like 2a | Mus musculus | NM_001099303.2 | |
| Shisa-like 2b | Mus musculus | NM_029984.1 | |
| Shisa-1 | Xenopus tropicalis | XM_004915754.4 | |
| Shisa-2 | Xenopus tropicalis | XM_002940554.3 | |
| Shisa-3 | Xenopus tropicalis | XM_002933451.5 | |
| Shisa-4 | Xenopus tropicalis | XM_018091360.2 | |
| Shisa-5 | Xenopus laevis | XM_018261282.2 | |
| Shisa-6 | Xenopus tropicalis | XM_031894479.1 | |
| Shisa-7 | Xenopus tropicalis | XM_002939964.4 | |
| Shisa-8 | Xenopus tropicalis | XM_002934724.5 | |
| Shisa-9 | Xenopus tropicalis | NM_001112925.1 | |
| Group | Phenotype (%)(n/N) | ||
|---|---|---|---|
| First | Second | Third | |
| Wildtype | 0 (0/59) |
0 (0/80) |
0 (0/102) |
| Cas13d mRNA Control | 0 (0/103) |
8.0 (2/25) |
0 (0/48) |
| gRNAs Control | 0 (0/83) |
4.4 (2/45) |
2.5 (1/40) |
| Cas13d mRNA+gRNAs | 72.9 (70/96) |
69.4 (34/49) |
90.5 (19/21) |
| Gene Name(NCBI name) | Primer Name | Sequence (5′-3′) | GenBank number |
|---|---|---|---|
| shisa- like 1 | shisa like-1-F | CTGATGGAGGACAAGAAGATG | XM_002667571.6 |
| shisa like-1-R | CTATGGTCAGTCTCAGGCT | ||
| shisa-1(Shisa 2a) | shisa-2a-F | AAGATGAAGTCATCGGCATC | NM_001003631.1 |
| shisa-2a-R | AATAATCCATGTGTAGTCC | ||
| shisa-2 | shisa-2-F | GTGGTTTGTGACACGATG | XM_003201303.5 |
| shisa-2-R | CATTGGGTTTCACATGGT | ||
| shisa-3 | shisa-3-F | AATTCAAGTTTGTCGGCGAG | NM_001080662.2 |
| shisa-3-R | GAGGTCACAGGTCAGCTCTG | ||
| shisa-4 | shisa-4-F | GATGTCCTTCTACGCTGTC | NM_001017869.1 |
| shisa-4-R | GTTATCTTCTCCTCGCAGAG | ||
| shisa-5 | shisa-5-F | GCGAGAGAGCAGCGCTATG | NM_001044870.1 |
| shisa-5-R | AAATGAACCATCCAGCTTGT | ||
| shisa-6 | shisa-6-F | GAAACACACCCTGAAGCCAT | XM_002667291.5 |
| shisa-6-R | TCCAGAGCATCCAAACAGC | ||
| shisa-7( shisa-like-7) | shisa-like-7-F | CATGTAAAGATGATGCCCACC | XM_021472968.1 |
| shisa-like-7-R | CCTCTACCATCCTCCAACTC | ||
| shisa-8b | shisa-8b-F | ATTTCTGGACAGGACCAGAG | XM_021480293.1 |
| shisa-8b-R | TGCATACAGTTATCTGAGTC | ||
| shisa-9a | shisa-9a-F | CCAGGAGACTACAGGATGA | NM_001013509.1 |
| shisa-9a-R | TCCCGCTCTCAGCTGCTTC | ||
| shisa-9b | shisa-9b-F | CCTCAAACATGAGCAGCATC | NM_001135975.2 |
| shisa-9b-R | CCACGTTCACACAGTCACC |
| Primer Name | Sequence (5′-3′) | Purpose |
|---|---|---|
| Cas13D-F-psp64-T7 | AAGCTTGGGCTGCAGGTCGACTAATACGACTCACTATAGGGAGCCACCATGAGCGAGGCCAGCATCGAAAAAAAAAAG | construction of Cas13d |
| Cas13D-R-psp64-T7 | TGGGAGCTCGCCCGGGGATCCTTAAGCGTAATCTGGAACATCGTATGGGTAAGCGGCCGCTCCGGATCCGGAATTGCCG | construction of Cas13d |
| Cas13d-Universal-F | TAATACGACTCACTATAGGAACCCCTACCAACTGGTCGGGGTTTGAAAC | synthesis of gRNA |
| Cas13D-zfshisa2-gRNA1 | ATCGTCGGCTCAGTTTTTGTGGCGTTTCAAACCCCGACCAGTTGGTAGGGGTT | synthesis of gRNA |
| Cas13D-zfshisa2-gRNA2 | TCGTCGGCTCAGTTTTTGTGGCAGTTTCAAACCCCGACCAGTTGGTAGGGGTT | synthesis of gRNA |
| Cas13D-zfshisa2-gRNA3 | CGTCGGTTCAGTTTTTGTGGCATGTTTCAAACCCCGACCAGTTGGTAGGGGTT | synthesis of gRNA |
| Cas13D-zfshisa2-gRNA4 | TGGGCTCTGTTGTTGCTGTATGCGTTTCAAACCCCGACCAGTTGGTAGGGGTT | synthesis of gRNA |
| Primer | Sequence (5′-3′) | Purpose |
|---|---|---|
| flounder- shisa-2-F | TGGTCGAGGATGTGGGGCGG | flounder shisa-2 cloning |
| flounder-shisa-2-R | GTGGCAGAGTGGACTACATG | flounder shisa-2 cloning |
| flounder-shisa2psp64-F | AAGCTTGGGCTGCAGGTCGACATGTGGGGCGGAGGTTTCCC | construction of flounder shisa-2 mRNA expression vector |
| flounder-shisa2psp64-R | TGGGAGCTCGCCCGGGGATCCGTGGCAGAGTGGACTACATG | construction of flounder shisa-2 mRNA expression vector |
| Gene Name | Primer Name | Sequence (5′-3′) |
|---|---|---|
| shisa- like 1 | shisa-like-1-RT-F | ACTCTCGGACAACAAGACGT |
| shisa-like-1-RT-R | CTATGGTCAGTCTCAGGCT | |
| shisa-1 | shisa-1-RT-F | CGGTGCGATTGTATCTGTCTG |
| shisa-1-RT-R | AATAATCCATGTGTAGTCC | |
| shisa-2 | shisa-2-RT-F | AGTACCCATCTACGTGCCCT |
| shisa-2-RT-R | GAGACTGTAACGGCCGGTAG | |
| shisa-3 | shisa-3-RT-F | CTGGACAGCAGTGGGAATTAC |
| shisa-3-RT-R | TGTGAACATTGACCCATCGT | |
| shisa-4 | shisa-4-RT-F | GATGTCCTTCTACGCTGTC |
| shisa-4-RT-R | TCATCGGATACTGAGGCACC | |
| shisa-5 | shisa-5-RT-F | GCGAGAGAGCAGCGCTATG |
| shisa-5-RT-R | TGGGCTGATATGGTGGGTAC | |
| shisa-6 | shisa-6-RT-F | GAAACACACCCTGAAGCCAT |
| shisa-6-RT-R | AGAGCAGGGTCATACGTGTC | |
| shisa-7 | shisa-7-RT-F | CATGTAAAGATGATGCCCACC |
| shisa-7-RT-R | CAGGTCCCACAGCAGTAGAT | |
| shisa-8 | shisa-8-RT-F | TGCAAACCGGAGCTACTACA |
| shisa-8-RT-R | TGCATACAGTTATCTGAGTC | |
| shisa-9a | shisa-9a-RT-F | CCAGGAGACTACAGGATGA |
| shisa-9a-RT-R | TATCCCAACCAGTGCCATGA | |
| shisa-9b | shisa-9b-RT-F | TCACCCCTATGAGCCGTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





