Submitted:
02 June 2023
Posted:
05 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Animals
2.3. Isolation of the virus.
2.4. Sequencing and phylogenetic analysis.
2.5. Mean death time (MDT) assays
2.6. Analysis of the pathogenicity and contagiousness of the virus in chicken.
2.7. Analysis of the pathogenicity of the virus in mice.
2.8. Ethics Statement
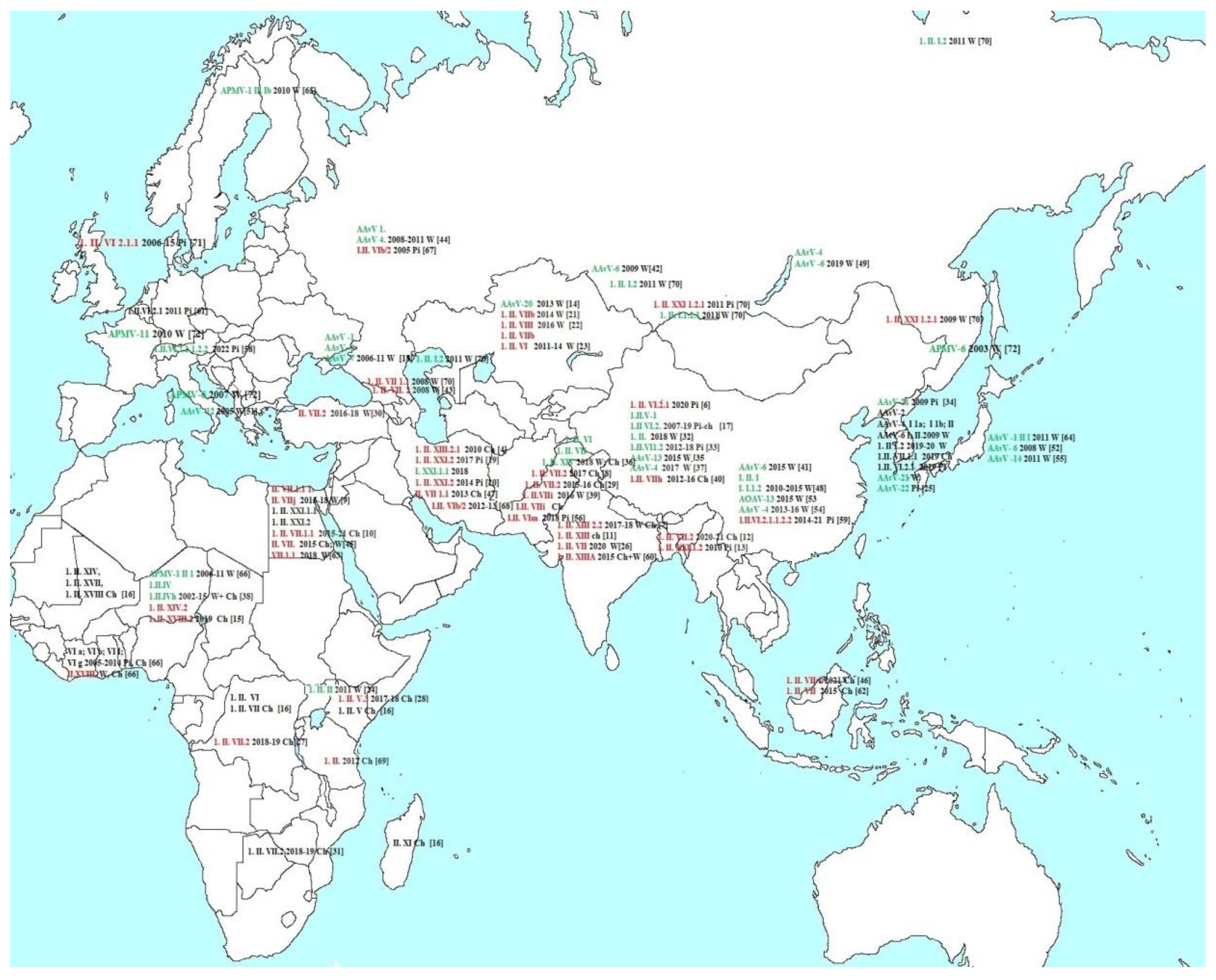
3. Results
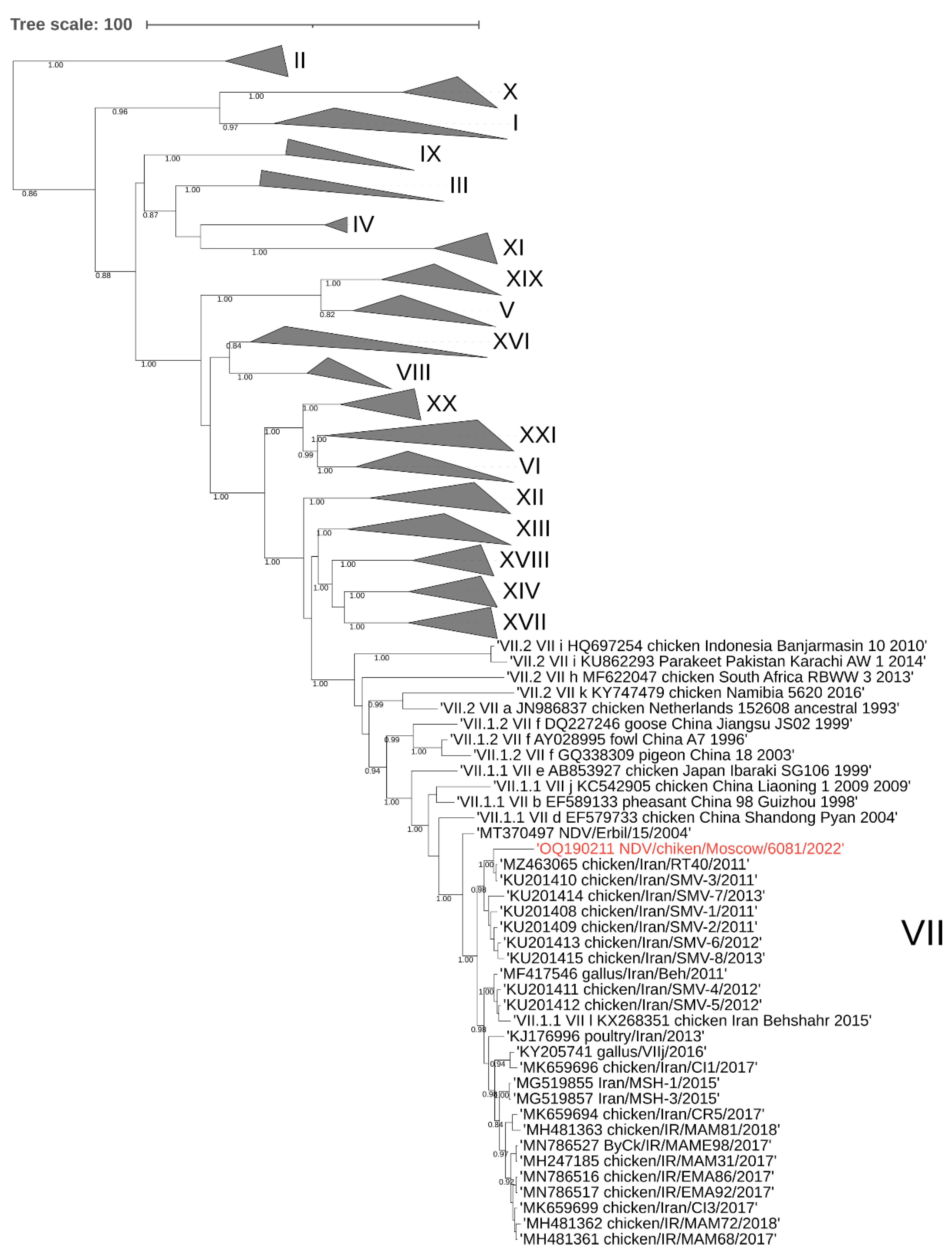
3.1. Phylogenetic analysis of the NDV/ Chicken/Moscow/6081/2022.
3.2. Pathogenicity and contagiousness of the NDV/Moscow/6081/2022 for chickens.
3.3. Pathogenicity of the NDV/Moscow/6081/2022 for mice.
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rima, B.; Balkema-Buschmann, A.; Dundon, W.G.; Duprex, P.; Easton, A.; Fouchier, R.; Kurath, G.; Lamb, R.; Lee, B.; Rota, P.; et al. ICTV Virus Taxonomy Profile: Paramyxoviridae. J. Gen. Virol. 2019, 100, 1593–1594. [Google Scholar] [CrossRef]
- Ross, C.S.; Mahmood, S.; Skinner, P.; Mayers, J.; Reid, S.M.; Hansen, R.D.E.; Banyard, A.C. JMM Profile: Avian paramyxovirus type-1 and Newcastle disease: a highly infectious vaccine-preventable viral disease of poultry with low zoonotic potential. J. Med Microbiol. 2022, 71, 001489. [Google Scholar] [CrossRef]
- K. Ganar, M. Das, S. Sinha, S. Kumar Newcastle disease virus: current status and our understanding Virus Res., 184 (2014), pp. 71–81.
- Hejazi, Z.; Tabatabaeizadeh, S.; Toroghi, R.; Farzin, H.; Saffarian, P. First detection and characterisation of sub-genotype XIII.2.1 Newcastle disease virus isolated from backyard chickens in Iran. Veter- Med. Sci. 2022, 8, 2521–2531. [Google Scholar] [CrossRef]
- Eid, A.A.M.; Hussein, A.; Hassanin, O.; Elbakrey, R.M.; Daines, R.; Sadeyen, J.-R.; Abdien, H.M.F.; Chrzastek, K.; Iqbal, M. Newcastle Disease Genotype VII Prevalence in Poultry and Wild Birds in Egypt. Viruses 2022, 14, 2244. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, W.; Li, Y.; Liu, J.; Wang, W.; Bai, J.; Yang, Z.; Liu, H.; Xiao, S. A pigeon paramyxovirus type 1 isolated from racing pigeon as an inactivated vaccine candidate provides effective protection. Poult. Sci. 2022, 101, 102097. [Google Scholar] [CrossRef]
- Balam, D.; Doddamane, R.; Rayudu, R.P.; Isloor, S.; Belamaranahally, V.; Maddireddy, H.; Metta, M. Simultaneous detection of velogenic Newcastle disease virus of genotype XIII 2.2 from spot-billed pelican and backyard chicken: implications to the viral maintenance and spread. Acta Virol. 2022, 66, 238–248. [Google Scholar] [CrossRef]
- Awais, M.; Wajid, A.; Goraichuk, I.V.; Batool, A.; Rahim, A.; Anif, A.; Ahmed, N.; Yin, R. Surveillance and Assessment of Risk Factors for Newcastle Disease Virus from Live Bird Retail Stalls in Lahore District of Pakistan. Avian Dis. 2022, 66, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sultan, S.; Eldamarany, N.M.I.; Abdelazeem, M.W.; Fahmy, H.A. Active Surveillance and Genetic Characterization of Prevalent Velogenic Newcastle Disease and Highly Pathogenic Avian Influenza H5N8 Viruses Among Migratory Wild Birds in Southern Egypt During 2015–2018. Food Environ. Virol. 2022, 14, 280–294. [Google Scholar] [CrossRef]
- Abozaid, K.G.A.; Abdel-Moneim, A.S. Epidemiological surveillance of Newcastle disease virus in Egypt — a 6-year cohort study. Trop. Anim. Heal. Prod. 2022, 54, 1–9. [Google Scholar] [CrossRef]
- Deka, P.; Nath, M.K.; Das, S.; Das, B.C.; Phukan, A.; Lahkar, D.; Bora, B.; Shokeen, K.; Kumar, A.; Deka, P. A study of risk factors associated with Newcastle disease and molecular characterization of genotype XIII Newcastle disease virus in backyard and commercial poultry in Assam, India. Res. Veter- Sci. 2022, 150, 122–130. [Google Scholar] [CrossRef]
- Nooruzzaman, M.; Hossain, I.; Begum, J.A.; Moula, M.; Khaled, S.A.; Parvin, R.; Chowdhury, E.H.; Islam, M.R.; Diel, D.G.; Dimitrov, K.M. The First Report of a Virulent Newcastle Disease Virus of Genotype VII.2 Causing Outbreaks in Chickens in Bangladesh. Viruses 2022, 14, 2627. [Google Scholar] [CrossRef]
- Nooruzzaman, M.; Barman, L.R.; Mumu, T.T.; Chowdhury, E.H.; Dimitrov, K.M.; Islam, M.R. A Pigeon-Derived Sub-Genotype XXI.1.2 Newcastle Disease Virus from Bangladesh Induces High Mortality in Chickens. Viruses 2021, 13, 1520. [Google Scholar] [CrossRef]
- Karamendin, K.O.; Sayatov, M.K.; Kydyrmanov, A.I.; Kasymbekov, E.T.; Asanova, S.E.; Daulbayeva, K.D.; Khan, E.Y. Molecular-genetic characterization of Avian avulavirus 20 strains isolated from wild birds. Probl. Virol. 2019, 64, 185–192. [Google Scholar] [CrossRef]
- Souley, M.M.; Ibrahim, A.I.; Kouato, B.S.; Abdou, A.; Issa, R.; Yaou, B.; Amadou, H.; Hama, H.; Adakal, H.; Abdou, N.; et al. Co-circulation of genotypes XIV.2 and XVIII.2 of avian paramyxovirus-1 (Newcastle disease virus) in backyard poultry in Niger. Virus Genes 2020, 57, 100–105. [Google Scholar] [CrossRef]
- Mngumi, E.B.; Mpenda, F.N.; Buza, J. Epidemiology of Newcastle disease in poultry in Africa: systematic review and meta-analysis. Trop. Anim. Heal. Prod. 2022, 54, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Lu, X.; He, D.; Gao, X.; Chen, Y.; Hu, Z.; Wang, X.; Hu, S.; Liu, X. Phylogenetic analysis and pathogenicity assessment of pigeon paramyxovirus type 1 circulating in China during 2007–2019. Transbound. Emerg. Dis. 2021, 69, 2076–2088. [Google Scholar] [CrossRef]
- Muzyka, D.; Pantin-Jackwood, M.; Stegniy, B.; Rula, O.; Bolotin, V.; Stegniy, A.; Gerilovych, A.; Shutchenko, P.; Stegniy, M.; Koshelev, V.; et al. Wild Bird Surveillance for Avian Paramyxoviruses in the Azov-Black Sea Region of Ukraine (2006 to 2011) Reveals Epidemiological Connections with Europe and Africa. Appl. Environ. Microbiol. 2014, 80, 5427–5438. [Google Scholar] [CrossRef]
- Esmaeelzadeh-Dizaji, R.; Molouki, A.; Hosseini, H.; Fallah-Mehrabadi, M.H.; Ziafati-Kafi, Z.; Takalou, A.; Eram, N.; Kumar, N.; Ashuri, A.; Sadri, N.; et al. Molecular characterization of a pigeon paramyxovirus type 1 virus isolated from Eurasian collared doves in Iran, 2017. J. Veter- Sci. 2022, 23, e29. [Google Scholar] [CrossRef]
- Dizaji, R.E.; Ghalyanchilangeroudi, A.; Marandi, M.V.; Hosseini, H.; Karimi, V.; Ziafatikafi, Z.; Molouki, A.; Mehrabadi, M.H.F. Complete genome sequence of a subgenotype XXI.1.1 pigeon paramyxovirus type 1 virus (PPMV-1) isolated from Iran in 2018 and phylogenetic analysis of a possible novel, but unassigned, PPMV-1 group isolated in 2014. Comp. Immunol. Microbiol. Infect. Dis. 2020, 73, 101565. [Google Scholar] [CrossRef]
- Karamendin, K.; Kydyrmanov, A.; Kasymbekov, Y.; Daulbayeva, K.; Khan, E.; Seidalina, A.; Sayatov, M.; Gavrilov, A.; Fereidouni, S. Cormorants as Potential Victims and Reservoirs of Velogenic Newcastle Disease Virus (Orthoavulavirus-1) in Central Asia. Avian Dis. 2019, 63, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Karamendin, K.; Kydyrmanov, A. Cormorants as a Potentially Important Reservoir and Carrier of Newcastle Disease Virus on the Asian Continent. Front. Veter- Sci. 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Orynbayev, M.B.; Fereidouni, S.; Sansyzbai, A.R.; Seidakhmetova, B.A.; Strochkov, V.M.; Nametov, A.M.; Sadikaliyeva, S.O.; Nurgazieva, A.; Tabynov, K.K.; Rametov, N.M.; et al. Genetic diversity of avian avulavirus 1 (Newcastle disease virus genotypes VIg and VIIb) circulating in wild birds in Kazakhstan. Arch. Virol. 2018, 163, 1949–1954. [Google Scholar] [CrossRef] [PubMed]
- Wanyana, A.; Mugimba, K.K.; Bosco, O.J.; Kirunda, H.; Nakavuma, J.L.; Teillaud, A.; Ducatez, M.F.; Byarugaba, D.K. Genotypic characterisation of Avian paramyxovirus type-1 viruses isolated from aquatic birds in Uganda. Onderstepoort J. Veter- Res. 2018, 85, 7–e7. [Google Scholar] [CrossRef]
- Liu, Y.-P.; Lee, F.; Cheng, M.-C.; Chang, C.-Y.; Chiou, C.-J.; Tsai, H.-J. Genetic diversity of avian paramyxoviruses isolated from wild birds and domestic poultry in Taiwan between 2009 and 2020. J. Veter- Med Sci. 2022, 84, 378–389. [Google Scholar] [CrossRef]
- Gaurav, S.; Deka, P.; Das, S.; Deka, P.; Hazarika, R.; Kakati, P.; Kumar, A.; Kumar, S. Isolation of genotype VII avian orthoavulavirus serotype 1 from barn owl from Northeast India. Avian Pathol. 2021, 51, 45–50. [Google Scholar] [CrossRef]
- Twabela, A.T.; Nguyen, L.T.; Masumu, J.; Mpoyo, P.; Mpiana, S.; Sumbu, J.; Okamatsu, M.; Matsuno, K.; Isoda, N.; Zecchin, B.; et al. A New Variant among Newcastle Disease Viruses Isolated in the Democratic Republic of the Congo in 2018 and 2019. Viruses 2021, 13, 151. [Google Scholar] [CrossRef]
- Kariithi, H.M.; Ferreira, H.L.; Welch, C.N.; Ateya, L.O.; Apopo, A.A.; Zoller, R.; Volkening, J.D.; Williams-Coplin, D.; Parris, D.J.; Olivier, T.L.; et al. Surveillance and Genetic Characterization of Virulent Newcastle Disease Virus Subgenotype V.3 in Indigenous Chickens from Backyard Poultry Farms and Live Bird Markets in Kenya. Viruses 2021, 13, 103. [Google Scholar] [CrossRef]
- Wajid, A.; Mayahi, V.; Yin, R.; Ain, Q.; Mohiuddin, A.; Khalid, F.; Rehim, A.; Manan, A.; Baksh, M. Genomic and biological characteristics of Avian Orthoavulavirus-1 strains isolated from multiple wild birds and backyard chickens in Pakistan. Trop. Anim. Heal. Prod. 2021, 53, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Turan, N.; Ozsemir, C.; Yilmaz, A.; Cizmecigil, U.Y.; Aydin, O.; Bamac, O.E.; Gurel, A.; Kutukcu, A.; Ozsemir, K.; Tali, H.E.; et al. Identification of Newcastle disease virus subgenotype VII.2 in wild birds in Turkey. BMC Veter- Res. 2020, 16, 1–8. [Google Scholar] [CrossRef]
- Kgotlele, T.; Modise, B.; Nyange, J.F.; Thanda, C.; Cattoli, G.; Dundon, W.G. First molecular characterization of avian paramyxovirus-1 (Newcastle disease virus) in Botswana. Virus Genes 2020, 56, 646–650. [Google Scholar] [CrossRef]
- Li, J.; Ling, M.; Sun, Y.; Di, H.; Cong, Y.; Yu, H.; Cong, Y. Characterization of Newcastle disease virus obtained from toco toucan. J. Veter- Sci. 2020, 21, e19. [Google Scholar] [CrossRef]
- He, Y.; Lu, B.; Dimitrov, K.M.; Liang, J.; Chen, Z.; Zhao, W.; Qin, Y.; Duan, Q.; Zhou, Y.; Liu, L.; et al. Complete Genome Sequencing, Molecular Epidemiological, and Pathogenicity Analysis of Pigeon Paramyxoviruses Type 1 Isolated in Guangxi, China during 2012–2018. Viruses 2020, 12, 366. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-P.; Kuo, S.-T.; Chiou, C.-J.; Terregino, C.; Tsai, H.-J. Novel avian metaavulavirus isolated from birds of the family Columbidae in Taiwan. Veter- Microbiol. 2019, 236, 108377. [Google Scholar] [CrossRef] [PubMed]
- Fei, Y.; Liu, X.; Mu, J.; Li, J.; Yu, X.; Chang, J.; Bi, Y.; Stoeger, T.; Wajid, A.; Muzyka, D.; et al. The Emergence of Avian Orthoavulavirus 13 in Wild Migratory Waterfowl in China Revealed the Existence of Diversified Trailer Region Sequences and HN Gene Lengths within this Serotype. Viruses 2019, 11, 646. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.-U.; Munir, M.; Shabbir, M.Z. A comparative genomic and evolutionary analysis of circulating strains of Avian avulavirus 1 in Pakistan. Mol. Genet. Genom. 2019, 294, 1289–1309. [Google Scholar] [CrossRef]
- Yuan, X.; Meng, K.; Zhang, Y.; Yu, Z.; Wang, Y.; Ai, W. Genetic characterization of one duck-origin paramyxovirus type 4 strain in China. Transbound. Emerg. Dis. 2019, 66, 2171–2174. [Google Scholar] [CrossRef] [PubMed]
- Welch, C.N.; Shittu, I.; Abolnik, C.; Solomon, P.; Dimitrov, K.M.; Taylor, T.L.; Williams-Coplin, D.; Goraichuk, I.V.; Meseko, C.A.; Ibu, J.O.; et al. Genomic comparison of Newcastle disease viruses isolated in Nigeria between 2002 and 2015 reveals circulation of highly diverse genotypes and spillover into wild birds. Arch. Virol. 2019, 164, 2031–2047. [Google Scholar] [CrossRef] [PubMed]
- Yaqub, T.; Imran, M.; Habib, M.; Sohail, T.; Shahid, M.F.; Munir, M.; Shabbir, M.Z. Phylogenomics and Infectious Potential of Avian Avulaviruses Species-Type 1 Isolated from Healthy Green-Winged Teal (Anas carolinensis) from a Wetland Sanctuary of Indus River. Avian Dis. 2018, 62, 404–415. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; Ge, S.; Lv, Y.; Li, Y.; Zheng, D.; Zhao, Y.; Castellan, D.; Wang, Z. Molecular characterization of new emerging sub-genotype VIIh Newcastle disease viruses in China. Virus Genes 2019, 55, 314–321. [Google Scholar] [CrossRef]
- Chen, Y.; Ding, Z.; Liu, X.; Chen, J.; Li, J.; Fei, Y.; Liu, Z.; Stoeger, T.; Bi, Y.; Yin, R. Biological and phylogenetic characterization of a novel hemagglutination-negative avian avulavirus 6 isolated from wild waterfowl in China. Transbound. Emerg. Dis. 2018, 65, 1421–1428. [Google Scholar] [CrossRef] [PubMed]
- Sobolev, I.A.; Sharshov, K.; Yurchenko, K.; Korneev, D.; Glushchenko, A.; Alikina, T.; Kabilov, M.; Bi, Y.; Liu, W.; Gubanova, N.; et al. Characterization of avian paramyxovirus type 6 isolated from a Eurasian teal in the intersection of migratory flyways in Russia. Arch. Virol. 2016, 161, 3275–3279. [Google Scholar] [CrossRef]
- Silko, N.I.; Glushchenko, A.V.; Shestopalova, L.V.; Iurchenko, K.S.; Korchagina, K.V.; Iushkov, I.G.; Shchelkanov, M.I.; Shestopalov, A.M. [Biological properties of velogenic strains of the Newcastle disease virus isolated in the Northern Caucasian region]. . 2013, 58, 45–48. [Google Scholar] [PubMed]
- Treshchalina, A.; Postnikova, Y.; Gambaryan, A.; Ishmukhametov, A.; Prilipov, A.; Sadykova, G.; Lomakina, N.; Boravleva, E. Monitoring of Avian Influenza Viruses and Paramyxoviruses in Ponds of Moscow and the Moscow Region. Viruses 2022, 14, 2624. [Google Scholar] [CrossRef] [PubMed]
- El Naggar, R.F.; Rohaim, M.A.; Bazid, A.H.; Ahmed, K.A.; Hussein, H.A.; Munir, M. Biological characterization of wild-bird-origin avian avulavirus 1 and efficacy of currently applied vaccines against potential infection in commercial poultry. Arch. Virol. 2018, 163, 2743–2755. [Google Scholar] [CrossRef]
- S. , S.A. Identification of Newcastle Disease Virus sub-genotype VII 1.1 isolated from chickens in Sabah, Malaysia. Trop. Biomed. 2022, 39, 579–586. [Google Scholar] [CrossRef]
- Sabouri, F.; Marandi, M.V.; Bashashati, M. Characterization of a novel VIIl sub-genotype of Newcastle disease virus circulating in Iran. Avian Pathol. 2017, 47, 90–99. [Google Scholar] [CrossRef]
- Jia, L.; Liang, B.; Wu, K.; Wang, R.; Liu, H.; Liu, D.; Chen, Q. Circulation, genomic characteristics, and evolutionary dynamics of class I Newcastle disease virus in China. Virulence 2022, 13, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Gulyaeva, M. , Badmaeva, E., Yurchenko, K., Sharshov,I., Sobolev, I., Bi,Y., Chen,J.,,, Shi,W., Diulin,I., Dorzhev,T., Shestopalov,A. Monitoring of Potentially Emerging Pathogens in Wild Birds at Baikal Lake Basin in 2019. EcoHealth 19, 335–341 (2022). [CrossRef]
- Choi, K.-S.; Kim, J.-Y.; Lee, H.-J.; Jang, M.-J.; Kwon, H.-M.; Sung, H.-W. Genetic Diversity of Avian Paramyxovirus Type 6 Isolated from Wild Ducks in the Republic of Korea. J. Wildl. Dis. 2018, 54, 558–563. [Google Scholar] [CrossRef]
- Terregino, C.; Aldous, E.W.; Heidari, A.; Fuller, C.M.; De Nardi, R.; Manvell, R.J.; Beato, M.S.; Shell, W.M.; Monne, I.; Brown, I.H.; et al. Antigenic and genetic analyses of isolate APMV/wigeon/Italy/3920-1/2005 indicate that it represents a new avian paramyxovirus (APMV-12). Arch. Virol. 2013, 158, 2233–2243. [Google Scholar] [CrossRef] [PubMed]
- Bui, V.N.; Mizutani, T.; Nguyen, T.H.; Trinh, D.Q.; Awad, S.S.A.; Minoungou, G.L.; Yamamoto, Y.; Nakamura, K.; Saito, K.; Watanabe, Y.; et al. Characterization of a genetic and antigenic variant of avian paramyxovirus 6 isolated from a migratory wild bird, the red-necked stint (Calidris ruficollis). Arch. Virol. 2014, 159, 3101–3105. [Google Scholar] [CrossRef]
- Fei, Y.; Liu, X.; Mu, J.; Li, J.; Yu, X.; Chang, J.; Bi, Y.; Stoeger, T.; Wajid, A.; Muzyka, D.; et al. The Emergence of Avian Orthoavulavirus 13 in Wild Migratory Waterfowl in China Revealed the Existence of Diversified Trailer Region Sequences and HN Gene Lengths within this Serotype. Viruses 2019, 11, 646. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Zhang, P.; Liu, X.; Chen, Y.; Tao, Z.; Ai, L.; Li, J.; Yang, Y.; Li, M.; Xue, C.; et al. Dispersal and Transmission of Avian Paramyxovirus Serotype 4 among Wild Birds and Domestic Poultry. Front. Cell. Infect. Microbiol. 2017, 7, 212. [Google Scholar] [CrossRef]
- Thampaisarn, R.; Bui, V.N.; Trinh, D.Q.; Nagai, M.; Mizutani, T.; Omatsu, T.; Katayama, Y.; Gronsang, D.; Le, D.H.; Ogawa, H.; et al. Characterization of avian paramyxovirus serotype 14, a novel serotype, isolated from a duck fecal sample in Japan. Virus Res. 2016, 228, 46–57. [Google Scholar] [CrossRef]
- Hussain, A.; Wajid, A.; Ather, S.; Alyas, K.; Awais, M.; Khan, M.R.; Hussain, T.; Babar, M.E. Isolation and genetic characterization of virulent strains of avian paramyxovirus-1 from multiple avian species in Azad Jammu and Kashmir 2017–2018. Braz. J. Microbiol. 2019, 51, 385–394. [Google Scholar] [CrossRef]
- Sun, J.; Ai, H.; Chen, L.; Li, L.; Shi, Q.; Liu, T.; Zhao, R.; Zhang, C.; Han, Z.; Liu, S. Surveillance of Class I Newcastle Disease Virus at Live Bird Markets in China and Identification of Variants with Increased Virulence and Replication Capacity. J. Virol. 2022, 96, e0024122. [Google Scholar] [CrossRef]
- Annaheim, D.; Vogler, B.R.; Sigrist, B.; Vögtlin, A.; Hüssy, D.; Breitler, C.; Hartnack, S.; Grund, C.; King, J.; Wolfrum, N.; et al. Screening of Healthy Feral Pigeons (Columba livia domestica) in the City of Zurich Reveals Continuous Circulation of Pigeon Paramyxovirus-1 and a Serious Threat of Transmission to Domestic Poultry. Microorganisms 2022, 10, 1656. [Google Scholar] [CrossRef]
- Yu, X.; Luo, Y.; Wang, J.; Shu, B.; Jiang, W.; Liu, S.; Li, Y.; Li, J.; Hou, G.; Peng, C.; et al. A molecular, epidemiological and pathogenicity analysis of pigeon paramyxovirus type 1 viruses isolated from live bird markets in China in 2014–2021. Virus Res. 2022, 318, 198846. [Google Scholar] [CrossRef] [PubMed]
- Mariappan, A.K.; Munusamy, P.; Kumar, D.; Latheef, S.K.; Singh, S.D.; Singh, R.; Dhama, K. Pathological and molecular investigation of velogenic viscerotropic Newcastle disease outbreak in a vaccinated chicken flocks. VirusDisease 2018, 29, 180–191. [Google Scholar] [CrossRef]
- Van Borm, S.; Rosseel, T.; Steensels, M.; Berg, T.v.D.; Lambrecht, B. What's in a strain? Viral metagenomics identifies genetic variation and contaminating circoviruses in laboratory isolates of pigeon paramyxovirus type 1. Virus Res. 2013, 171, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Roohani, K.; Tan, S.W.; Yeap, S.K.; Ideris, A.; Bejo, M.H.; Omar, A.R. Characterisation of genotype VII Newcastle disease virus (NDV) isolated from NDV vaccinated chickens, and the efficacy of LaSota and recombinant genotype VII vaccines against challenge with velogenic NDV. J. Veter- Sci. 2015, 16, 447–457. [Google Scholar] [CrossRef]
- Elfatah, K.S.A.; Elabasy, M.A.; El-Khyate, F.; Elmahallawy, E.K.; Mosad, S.M.; El-Gohary, F.A.; Abdo, W.; Al-Brakati, A.; Seadawy, M.G.; Tahoon, A.E.; et al. Molecular Characterization of Velogenic Newcastle Disease Virus (Sub-Genotype VII.1.1) from Wild Birds, with Assessment of Its Pathogenicity in Susceptible Chickens. Animals 2021, 11, 505. [Google Scholar] [CrossRef] [PubMed]
- Wille, M.; Avril, A.; Tolf, C.; Schager, A.; Larsson, S.; Borg, O.; Olsen, B.; Waldenström, J. Temporal dynamics, diversity, and interplay in three components of the virodiversity of a Mallard population: Influenza A virus, avian paramyxovirus and avian coronavirus. Infect. Genet. Evol. 2014, 29, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Tolf, C.; Wille, M.; Haidar, A.-K.; Avril, A.; Zohari, S.; Waldenström, J. Prevalence of avian paramyxovirus type 1 in Mallards during autumn migration in the western Baltic Sea region. Virol. J. 2013, 10, 285–285. [Google Scholar] [CrossRef]
- Snoeck, C.J.; Adeyanju, A.T.; Owoade, A.A.; Couacy-Hymann, E.; Alkali, B.R.; Ottosson, U.; Muller, C.P. Genetic Diversity of Newcastle Disease Virus in Wild Birds and Pigeons in West Africa. Appl. Environ. Microbiol. 2013, 79, 7867–7874. [Google Scholar] [CrossRef]
- Pchelkina, I.P.; Manin, T.B.; Kolosov, S.N.; Starov, S.K.; Andriyasov, A.V.; Chvala, I.A.; Drygin, V.V.; Yu, Q.; Miller, P.J.; Suarez, D.L. Characteristics of Pigeon Paramyxovirus Serotype-1 Isolates (PPMV-1) from the Russian Federation from 2001 to 2009. Avian Dis. 2013, 57, 2–7. [Google Scholar] [CrossRef]
- Far, A.R.; Peighambari, S.M.; Pourbakhsh, S.A.; Ashtari, A.; Soltani, M. Co-circulation of genetically distinct groups of avian paramyxovirus type 1 in pigeon Newcastle disease in Iran. Avian Pathol. 2016, 46, 36–43. [Google Scholar] [CrossRef]
- Msoffe, P.L.M.; Chiwanga, G.H.; Cardona, C.J.; Miller, P.J.; Suarez, D.L. Isolation and Characterization of Newcastle Disease Virus from Live Bird Markets in Tanzania. Avian Dis. 2019, 63, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Glushchenko, A.V.; Yurchenko, K.S.; Yurlov, A.K.; Yushkov, Y.G.; Shchelkanov, M.Y.; Шестoпалoв, A. . THE ROLE OF WILD BIRDS IN PRESERVATION AND PREVALENCE OF AVIAN PARAMYXOVIRUS SEROTYPE 1 (NEWCASTLE DISEASE VIRUSES) IN SIBERIA AND THE FAR EAST, RUSSIA. South Russ. Ecol. Dev. 2016, 11, 50–58. [Google Scholar] [CrossRef]
- Ross, C.S.; Sutton, D.; Skinner, P.; Mahmood, S.; Wynne, F.; Londt, B.; Fuller, C.M.; Mayers, J.; Nunez, A.; Hicks, D.J.; et al. Comparative pathogenesis of two genotype VI.2 avian paramyxovirus type-1 viruses (APMV-1) in pheasants, partridges and chickens. Avian Pathol. 2022, 52, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Gogoi, P.; Ganar, K.; Kumar, S. Avian Paramyxovirus: A Brief Review. Transbound. Emerg. Dis. 2015, 64, 53–67. [Google Scholar] [CrossRef] [PubMed]
- OIE. Newcastle Disease Manual of Diagnostic Tests and Vaccines for Terrestrial Animals: Mammals, Birds and Bees. 1, Part 2. 8th ed Paris, France: Office International des Epizooties; 2014.
- Tan, S.W.; Ideris, A.; Omar, A.R.; Yusoff, K.; Hair-Bejo, M. Detection and differentiation of velogenic and lentogenic Newcastle disease viruses using SYBR Green I real-time PCR with nucleocapsid gene-specific primers. J. Virol. Methods 2009, 160, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Alkie, T.N.; Cox, S.; Embury-Hyatt, C.; Stevens, B.; Pople, N.; Pybus, M.J.; Xu, W.; Hisanaga, T.; Suderman, M.; Koziuk, J.; et al. Characterization of neurotropic HPAI H5N1 viruses with novel genome constellations and mammalian adaptive mutations in free-living mesocarnivores in Canada. Emerg. Microbes Infect. 2023, 12, 2186608. [Google Scholar] [CrossRef] [PubMed]
- Frolov, S.V.; Moroz, N.V.; Chvala, I.A.; Irza, V.N. Effectiveness of vaccines produced by the Federal State-Financed Institution “ARRIAH” against topical genotype VII Newcastle disease viruses. Veter- Sci. Today 2021, 1, 44–51. [Google Scholar] [CrossRef]
- Volkova, M.A.; Chvala, I.A.; Yaroslavtseva, P.S.; Sosipatorova, V.Y. Serological monitoring of Newcastle disease in Russia in 2017. Vet. Sci. 2019, 26–30. [Google Scholar] [CrossRef]
- Volkova, M.A.; Chvala, I.A.; Osipova, O.S.; Kulagina, M.A.; Andreychuk, D.B. Serological monitoring of avian influenza and Newcastle disease in the Russian Federation in 2019. Veter- Sci. Today 2020, 76-82. [CrossRef]
| Group | Mortality* | Mdd** |
|---|---|---|
| infected | 5/5 | 5 |
| direct contact | 5/5 | 6 |
| remote contact | 5/5 | 8.6 |
| Infection dose: EID50 per mouse | |||||
|---|---|---|---|---|---|
| 0 | 103 | 104 | 105 | 106 | |
| Mortality* | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 |
| Weight** | 100% | 100% | 97% | 96% | 94% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




