Submitted:
29 May 2023
Posted:
30 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Histone Modifiers on p53
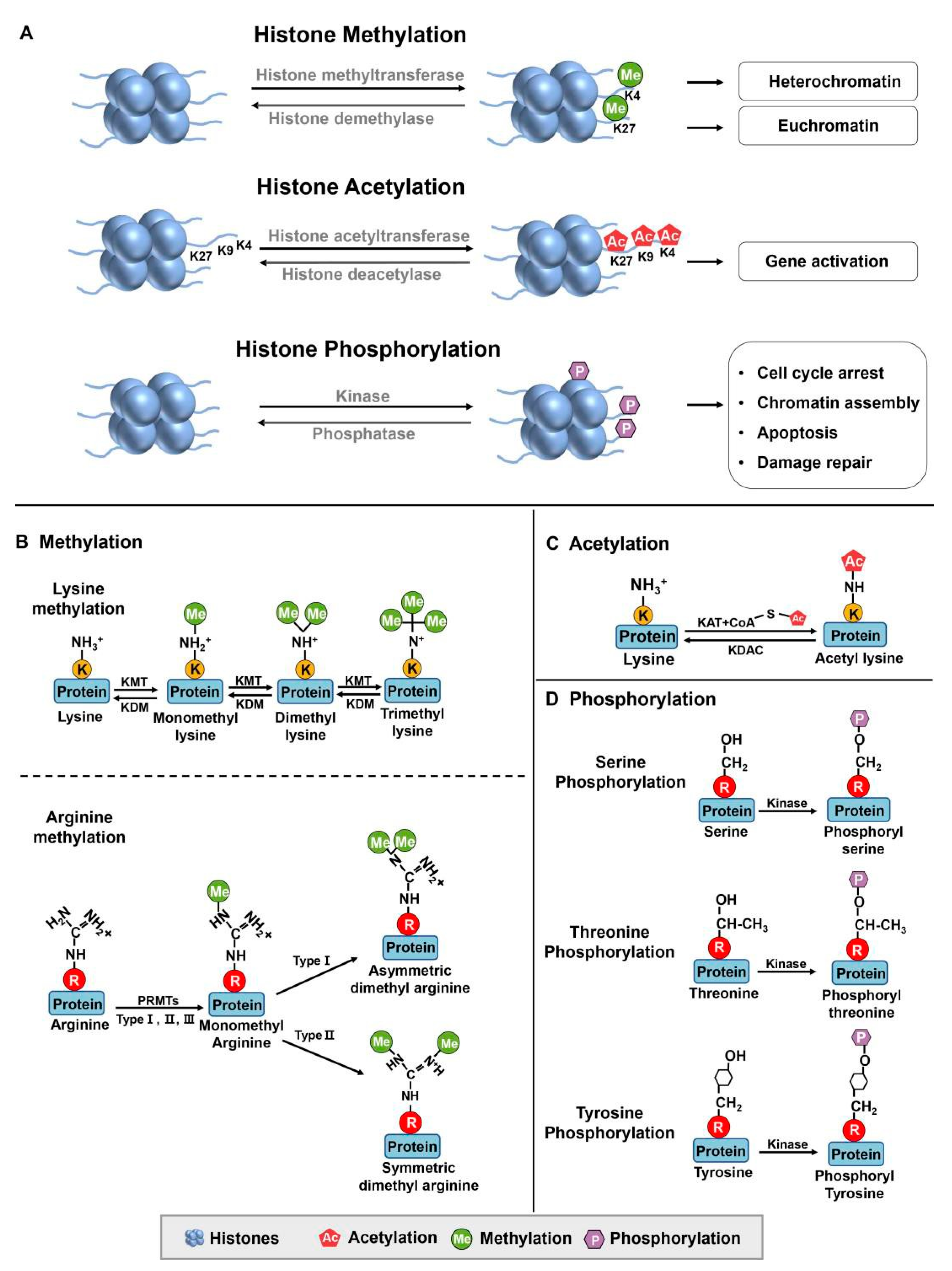
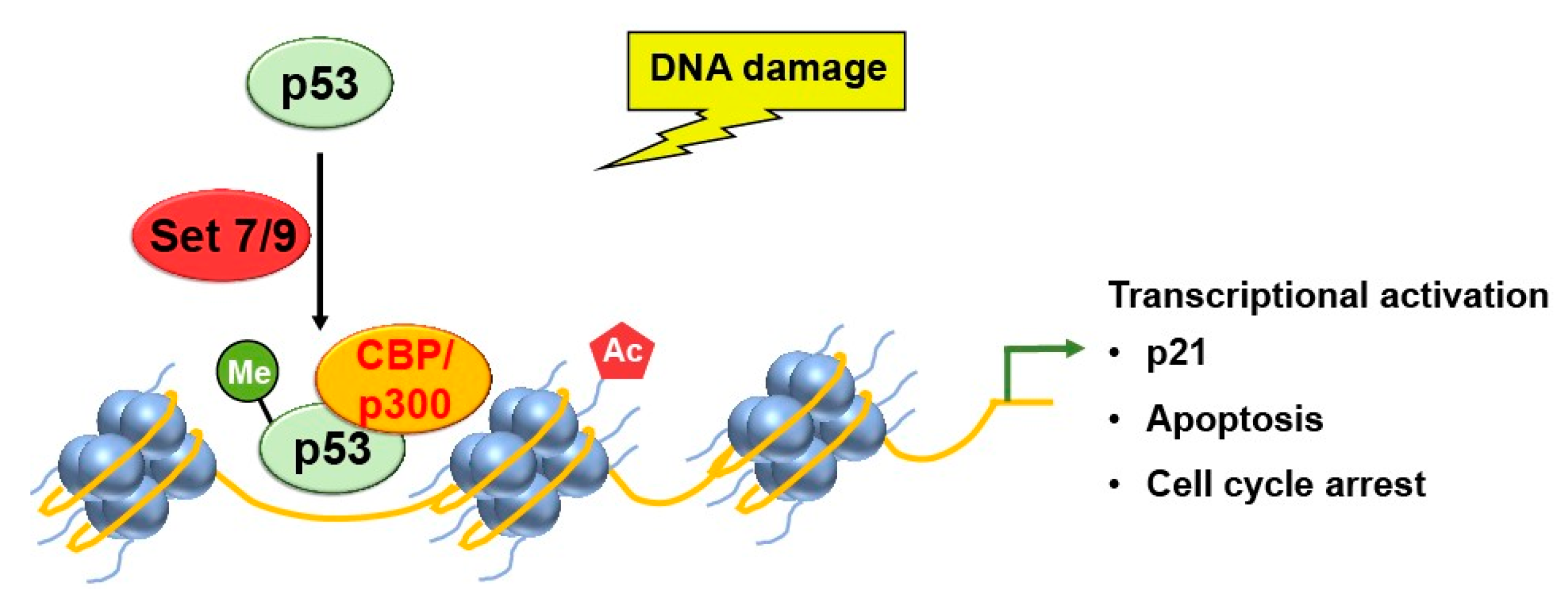
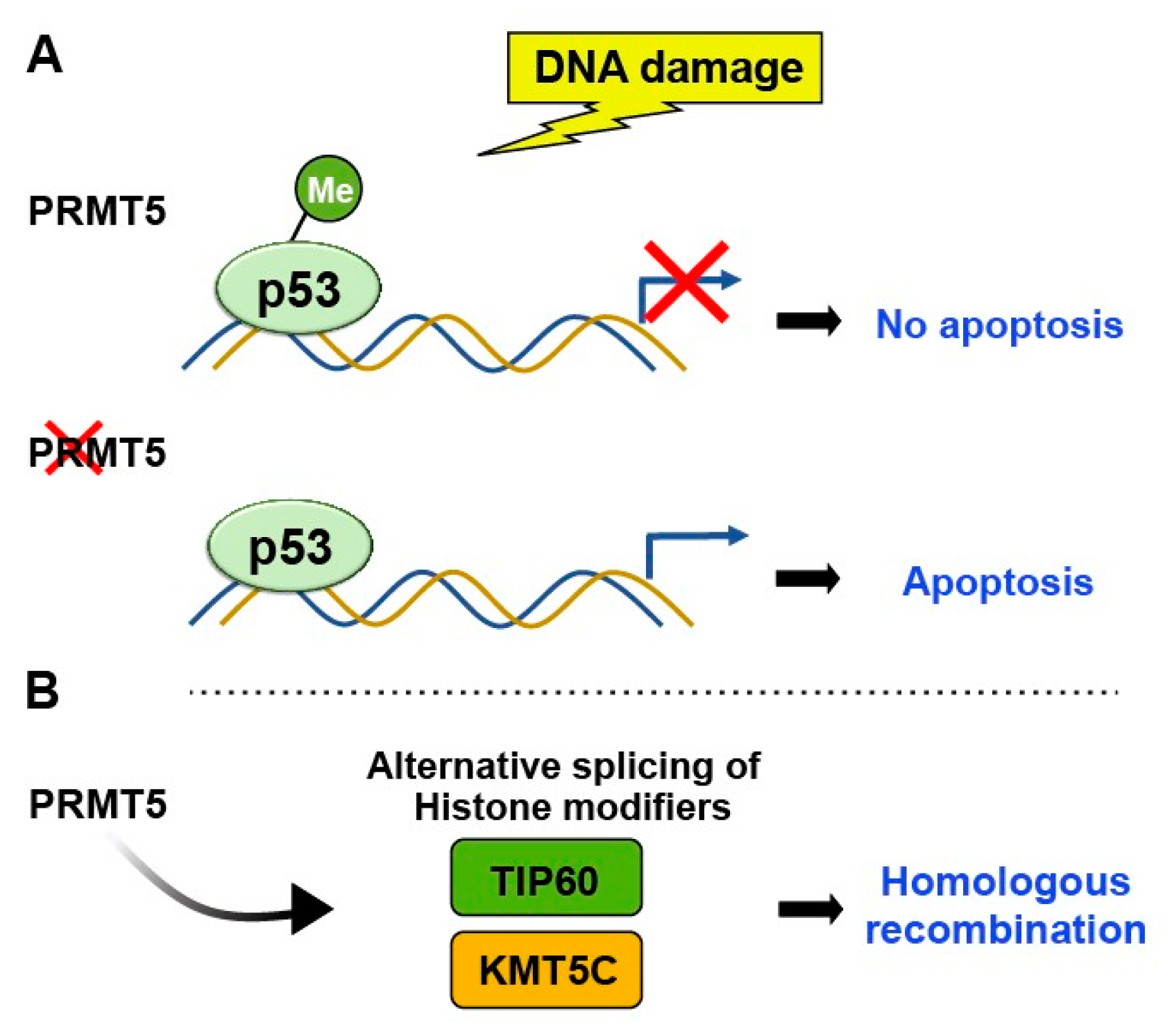
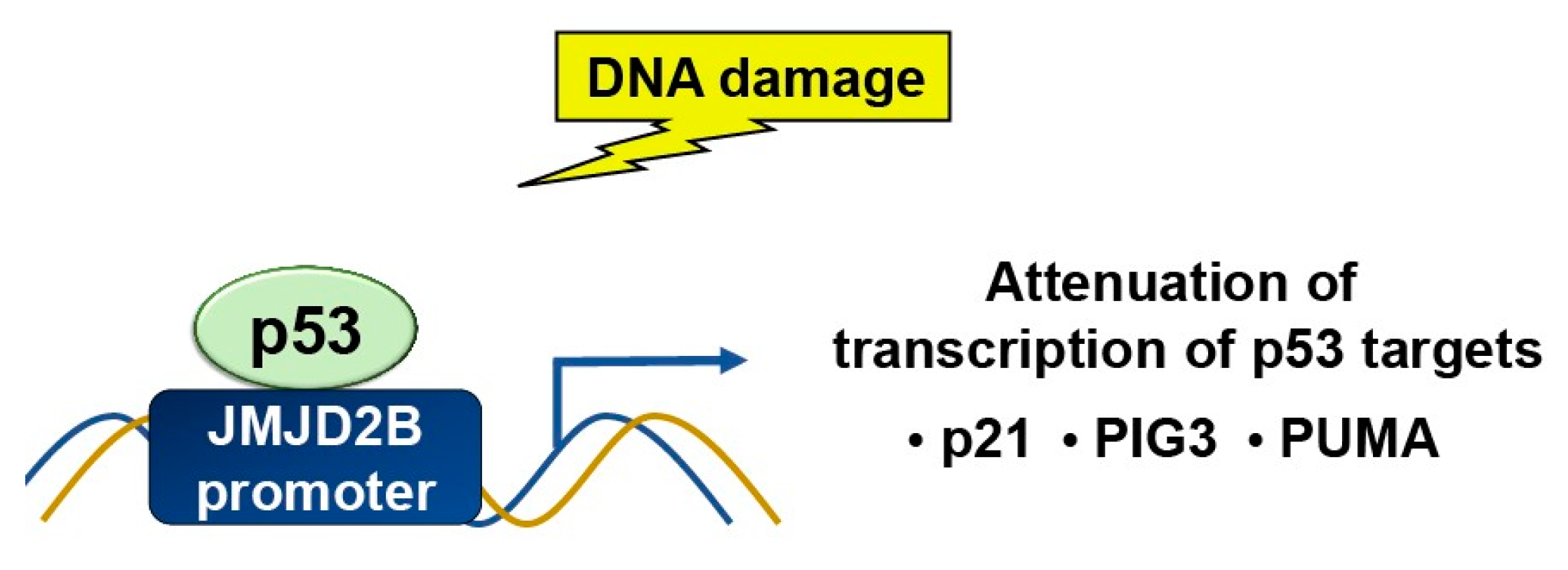
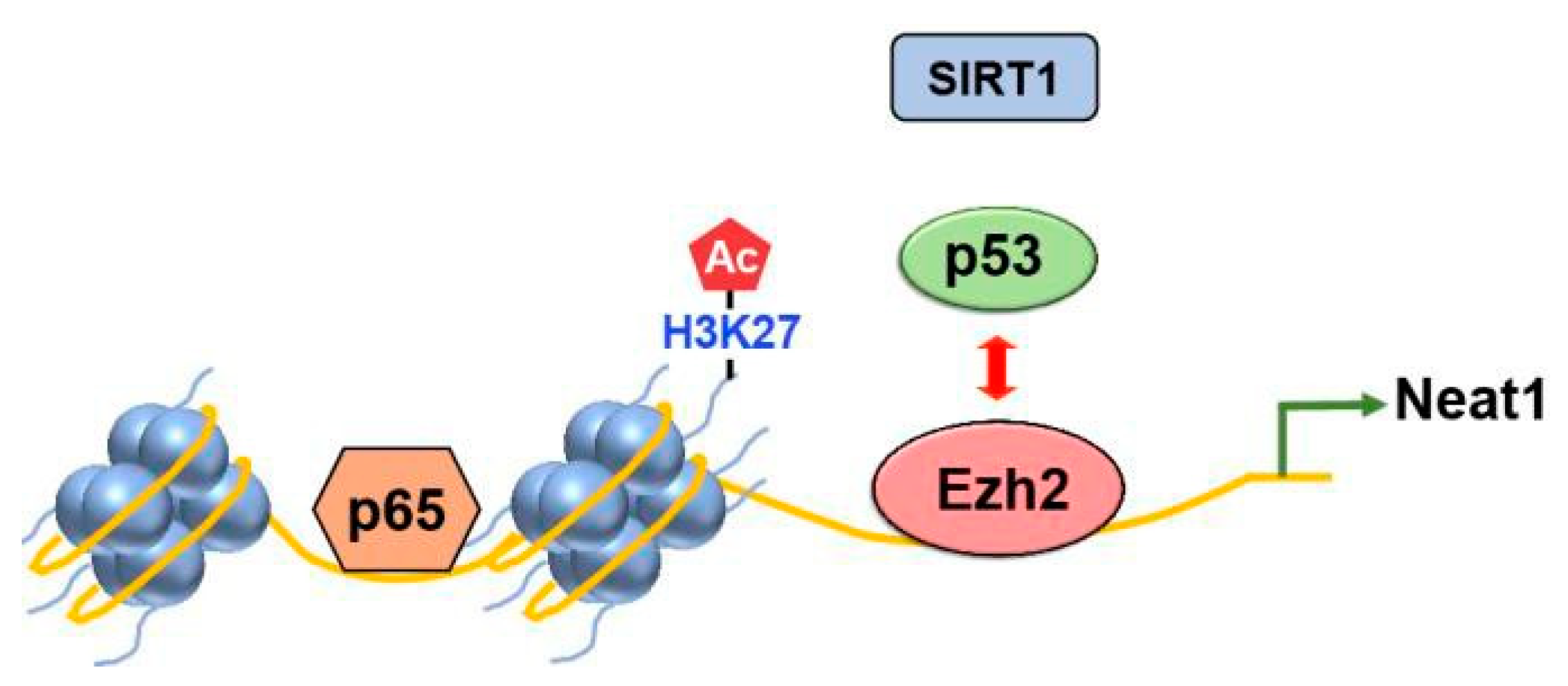
2.1. Methylation
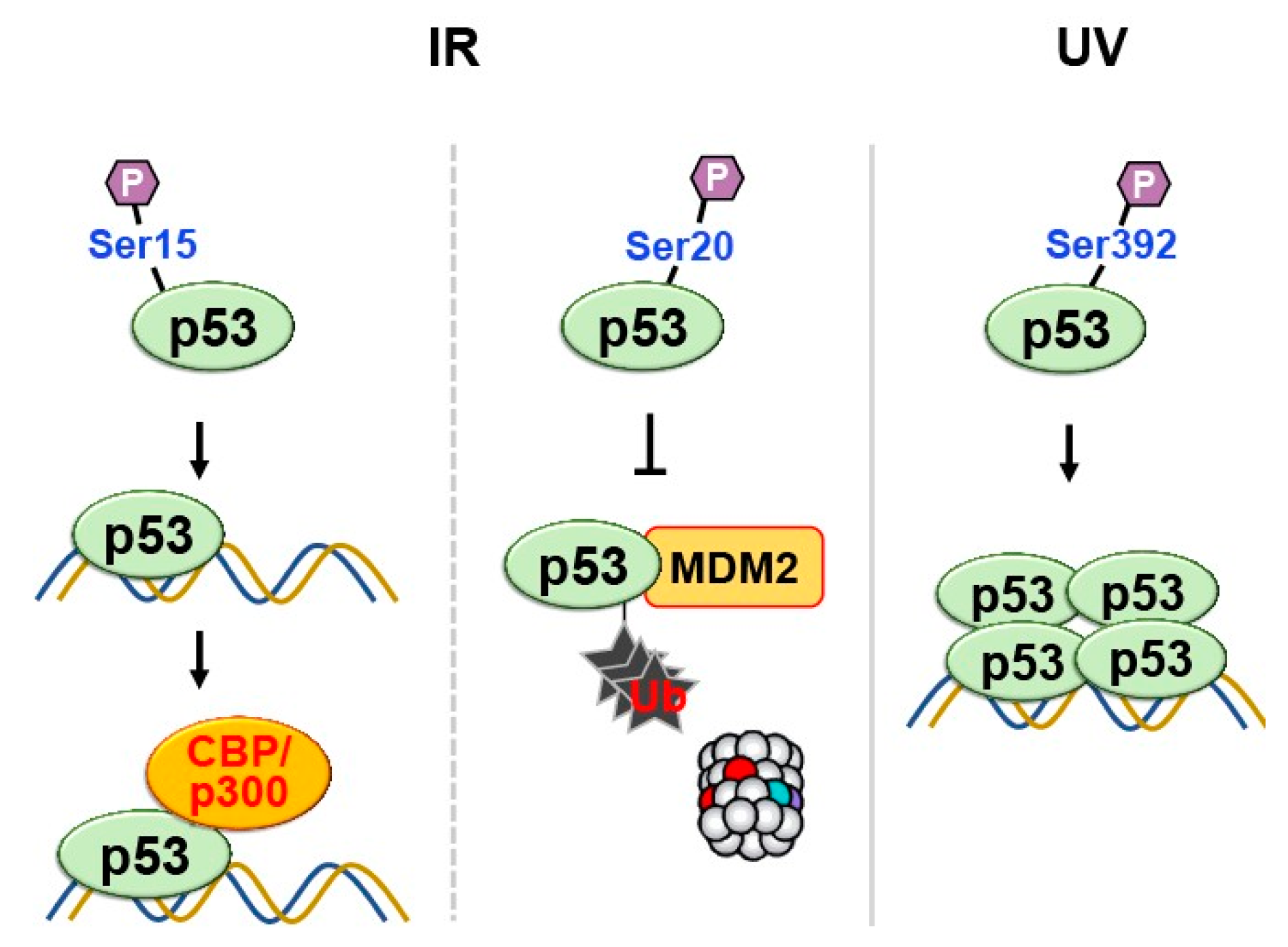
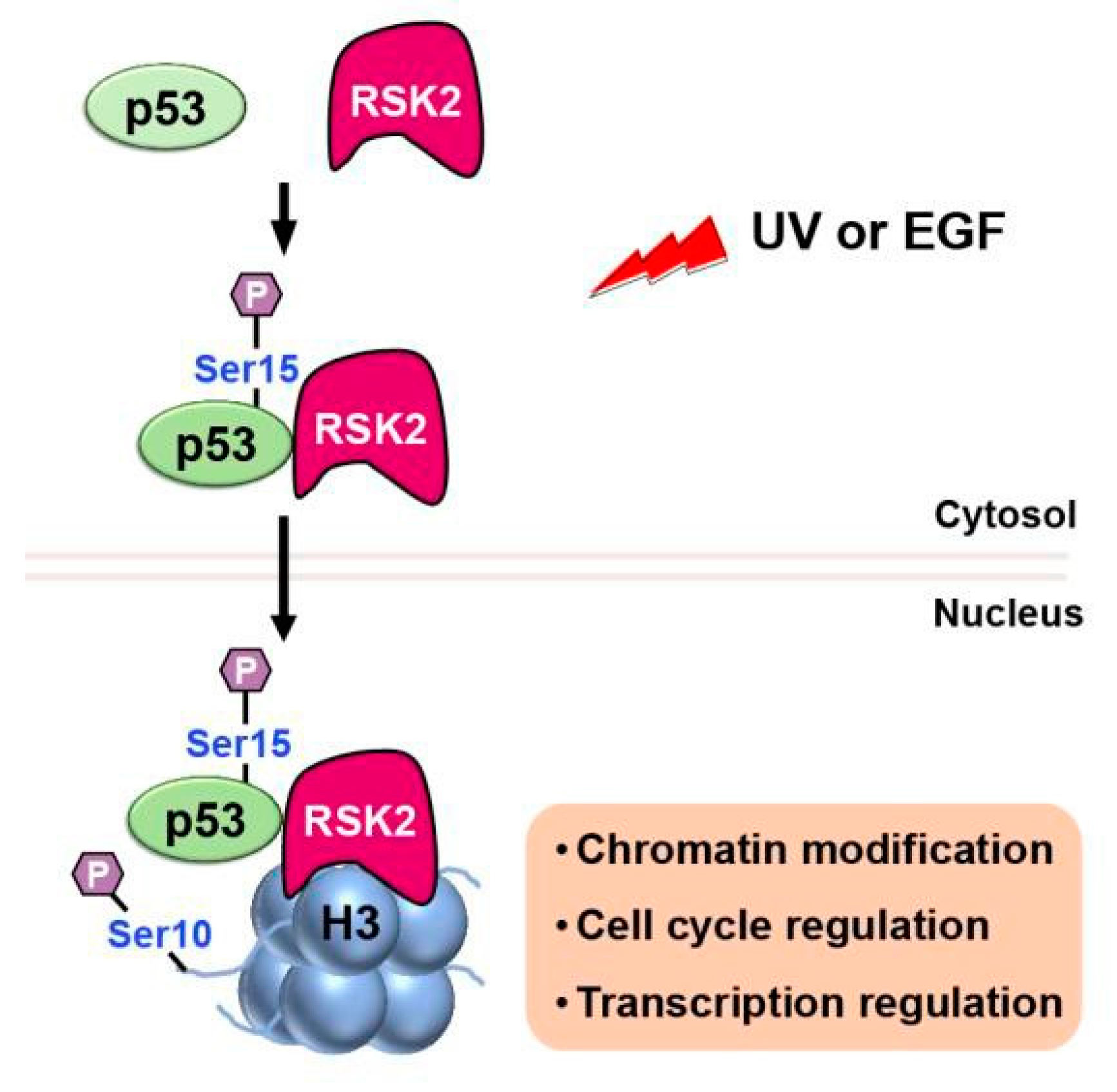
2.2. Phosphorylation
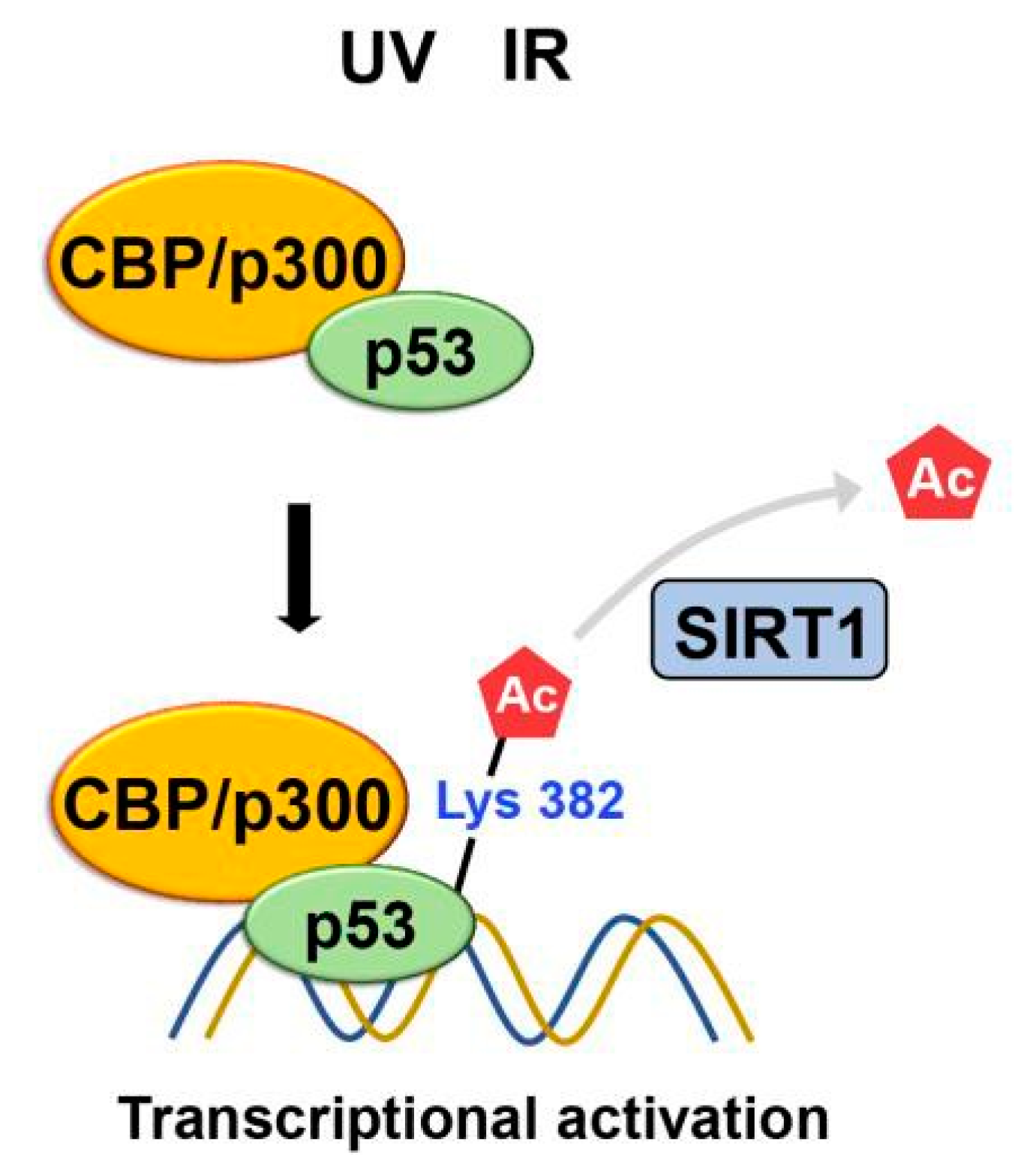
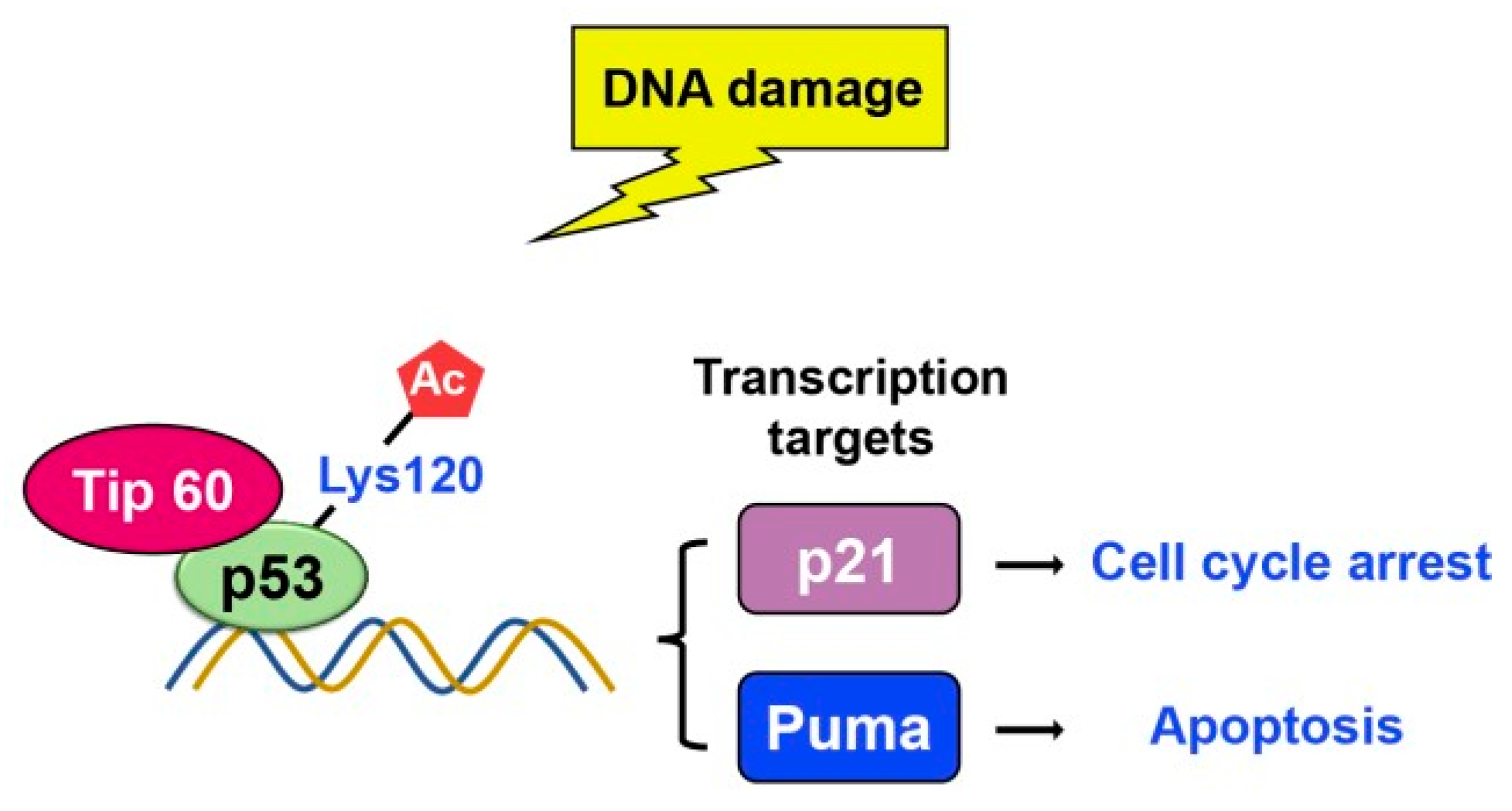
2.3. Acetylation
3. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berger, S.L. Histone modifications in transcriptional regulation. Curr Opin Genet Dev 2002, 12, 142–148. [Google Scholar] [CrossRef]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Bode, A.M.; Dong, Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer 2004, 4, 793–805. [Google Scholar] [CrossRef]
- Lee, H.T.; et al. The Key Role of DNA Methylation and Histone Acetylation in Epigenetics of Atherosclerosis. J Lipid Atheroscler 2020, 9, 419–434. [Google Scholar] [CrossRef]
- Keck, F.; et al. Phosphorylation of Single Stranded RNA Virus Proteins and Potential for Novel Therapeutic Strategies. Viruses 2015, 7, 5257–5273. [Google Scholar] [CrossRef] [PubMed]
- Blanc, R.S.; Richard, S. Arginine Methylation: The Coming of Age. Mol Cell 2017, 65, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.E. ; Epigenetic Therapies for Cancer. N Engl J Med 2020, 383, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Chuikov, S.; et al. Regulation of p53 activity through lysine methylation. Nature 2004, 432, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, G.S.; et al. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol 2007, 27, 6756–6769. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, M.; et al. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem 2001, 276, 25309–25317. [Google Scholar] [CrossRef]
- Oh, S.T.; et al. H3K9 histone methyltransferase G9a-mediated transcriptional activation of p21. FEBS Lett 2014, 588, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Rada, M.; et al. Human EHMT2/G9a activates p53 through methylation-independent mechanism. Oncogene 2017, 36, 922–932. [Google Scholar] [CrossRef]
- Bedford, M.T.; Clarke, S.G. Protein arginine methylation in mammals: who, what, and why. Mol Cell 2009, 33, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Karkhanis, V.; et al. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem Sci 2011, 36, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Jansson, M.; et al. Arginine methylation regulates the p53 response. Nat Cell Biol 2008, 10, 1431–1439. [Google Scholar] [CrossRef]
- Li, Y.; Diehl, J.A. PRMT5-dependent p53 escape in tumorigenesis. Oncoscience 2015, 2, 700–702. [Google Scholar] [CrossRef] [PubMed]
- Custodio, G.; et al. Impact of neonatal screening and surveillance for the TP53 R337H mutation on early detection of childhood adrenocortical tumors. J Clin Oncol 2013, 31, 2619–2626. [Google Scholar] [CrossRef]
- McBride, K.A.; et al. Li-Fraumeni syndrome: cancer risk assessment and clinical management. Nat Rev Clin Oncol 2014, 11, 260–271. [Google Scholar] [CrossRef]
- Hamard, P.J.; et al. PRMT5 Regulates DNA Repair by Controlling the Alternative Splicing of Histone-Modifying Enzymes. Cell Rep 2018, 24, 2643–2657. [Google Scholar] [CrossRef]
- Castellini, L.; et al. KDM4B/JMJD2B is a p53 target gene that modulates the amplitude of p53 response after DNA damage. Nucleic Acids Res 2017, 45, 3674–3692. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; et al. The histone demethylase JMJD2B is critical for p53-mediated autophagy and survival in Nutlin-treated cancer cells. J Biol Chem 2019, 294, 9186–9197. [Google Scholar] [CrossRef] [PubMed]
- Vire, E.; et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006, 439, 871–874. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; et al. Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene 2004, 23, 5759–5769. [Google Scholar] [CrossRef]
- Yuan, J.; et al. Ezh2 competes with p53 to license lncRNA Neat1 transcription for inflammasome activation. Cell Death Differ 2022, 29, 2009–2023. [Google Scholar] [CrossRef]
- Lambert, P.F.; et al. Phosphorylation of p53 serine 15 increases interaction with CBP. J Biol Chem 1998, 273, 33048–33053. [Google Scholar] [CrossRef]
- Chehab, N.H.; et al. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci U S A 1999, 96, 13777–13782. [Google Scholar] [CrossRef]
- Shieh, S.Y.; Taya, Y.; Prives, C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J 1999, 18, 1815–1823. [Google Scholar] [CrossRef]
- Unger, T.; et al. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J 1999, 18, 1805–1814. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Lozano, G. Functional activation of p53 via phosphorylation following DNA damage by UV but not gamma radiation. Proc Natl Acad Sci U S A 1998, 95, 2834–2837. [Google Scholar] [CrossRef]
- Lu, H.; et al. Ultraviolet radiation, but not gamma radiation or etoposide-induced DNA damage, results in the phosphorylation of the murine p53 protein at serine-389. Proc Natl Acad Sci U S A 1998, 95, 6399–6402. [Google Scholar] [CrossRef]
- Hupp, T.R.; et al. Regulation of the specific DNA binding function of p53. Cell 1992, 71, 875–886. [Google Scholar] [CrossRef]
- Sakaguchi, K.; et al. Phosphorylation of serine 392 stabilizes the tetramer formation of tumor suppressor protein p53. Biochemistry 1997, 36, 10117–10124. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, N.; Premkumar Reddy, E. Signaling by dual specificity kinases. Oncogene 1998, 17, 1447–55. [Google Scholar] [CrossRef] [PubMed]
- She, Q.B.; Chen, N.; Dong, Z. ERKs and p38 kinase phosphorylate p53 protein at serine 15 in response to UV radiation. J Biol Chem 2000, 275, 20444–20449. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.L.; Meek, D.W. Phosphorylation of serine 392 in p53 is a common and integral event during p53 induction by diverse stimuli. Cell Signal 2010, 22, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Shieh, S.Y.; et al. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997, 91, 325–334. [Google Scholar] [CrossRef]
- Sattarifard, H.; et al. Mitogen- and stress-activated protein kinase (MSK1/2) regulated gene expression in normal and disease states. Biochem Cell Biol 2023. [Google Scholar] [CrossRef] [PubMed]
- Crosio, C.; et al. Chromatin remodeling and neuronal response: multiple signaling pathways induce specific histone H3 modifications and early gene expression in hippocampal neurons. J Cell Sci 2003, 116 Pt 24, 4905–4914. [Google Scholar] [CrossRef]
- Clayton, A.L.; et al. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J 2000, 19, 3714–3726. [Google Scholar] [CrossRef]
- Ahn, J.; et al. MSK1 functions as a transcriptional coactivator of p53 in the regulation of p21 gene expression. Exp Mol Med 2018, 50, 1–12. [Google Scholar] [CrossRef]
- Xing, J.; Ginty, D.D.; Greenberg, M.E. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor- regulated CREB kinase. Science 1996, 273, 959–963. [Google Scholar] [CrossRef]
- Cho, Y.Y.; et al. The p53 protein is a novel substrate of ribosomal S6 kinase 2 and a critical intermediary for ribosomal S6 kinase 2 and histone H3 interaction. Cancer Res 2005, 65, 3596–3603. [Google Scholar] [CrossRef]
- Gu, W.; Roeder, R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997, 90, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Lill, N.L.; et al. Binding and modulation of p53 by p300/CBP coactivators. Nature 1997, 387, 823–827. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, K.; et al. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev 1998, 12, 2831–2841. [Google Scholar] [CrossRef] [PubMed]
- Barlev, N.A.; et al. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell 2001, 8, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, H.; et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 2001, 107, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Ikura, T.; et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 2000, 102, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; et al. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 2006, 24, 827–839. [Google Scholar] [CrossRef]
- Sykes, S.M.; et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 2006, 24, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Murr, R.; et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol 2006, 8, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Robert, F.; et al. The transcriptional histone acetyltransferase cofactor TRRAP associates with the MRN repair complex and plays a role in DNA double-strand break repair. Mol Cell Biol 2006, 26, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Tyteca, S.; et al. Tip60 and p400 are both required for UV-induced apoptosis but play antagonistic roles in cell cycle progression. EMBO J 2006, 25, 1680–1689. [Google Scholar] [CrossRef]
- Li, T.; et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 2012, 149, 1269–1283. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; et al. Acetylation is indispensable for p53 activation. Cell 2008, 133, 612–626. [Google Scholar] [CrossRef]
- Gao, Y.; et al. Dual inhibitors of histone deacetylases and other cancer-related targets: A pharmacological perspective. Biochem Pharmacol 2020, 182, 114224. [Google Scholar] [CrossRef]
- Seo, Y.H. Dual Inhibitors Against Topoisomerases and Histone Deacetylases. J Cancer Prev 2015, 20, 85–91. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




