Submitted:
11 May 2023
Posted:
12 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
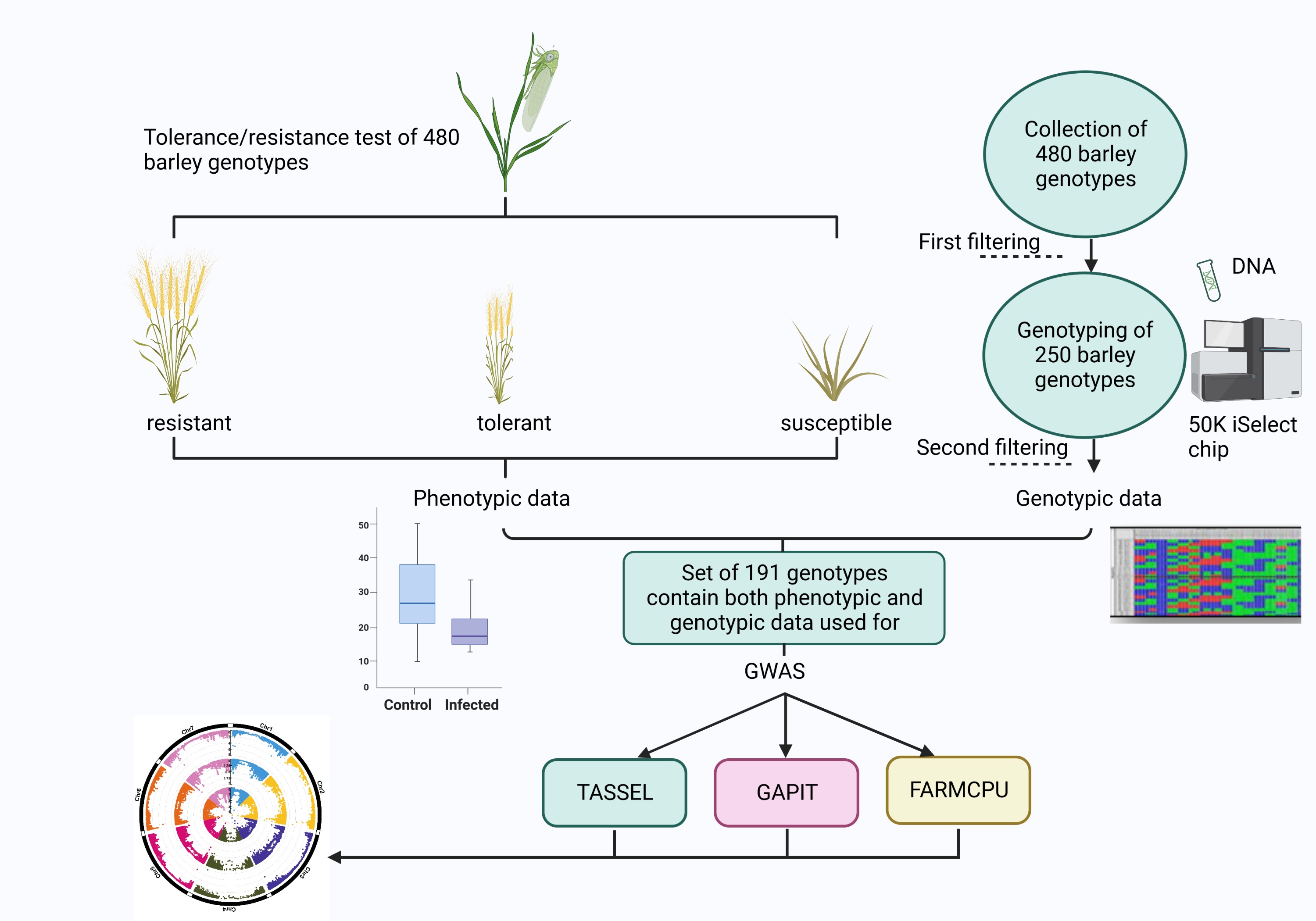
2. Materials and Methods
2.1. Plant Material
2.1.1. Phenotyping
2.2. Statistical Analysis
2.3. Genome Wide Association Study (GWAS)
3. Results
3.1. Phenotypic Data
3.2. Genotypic Data
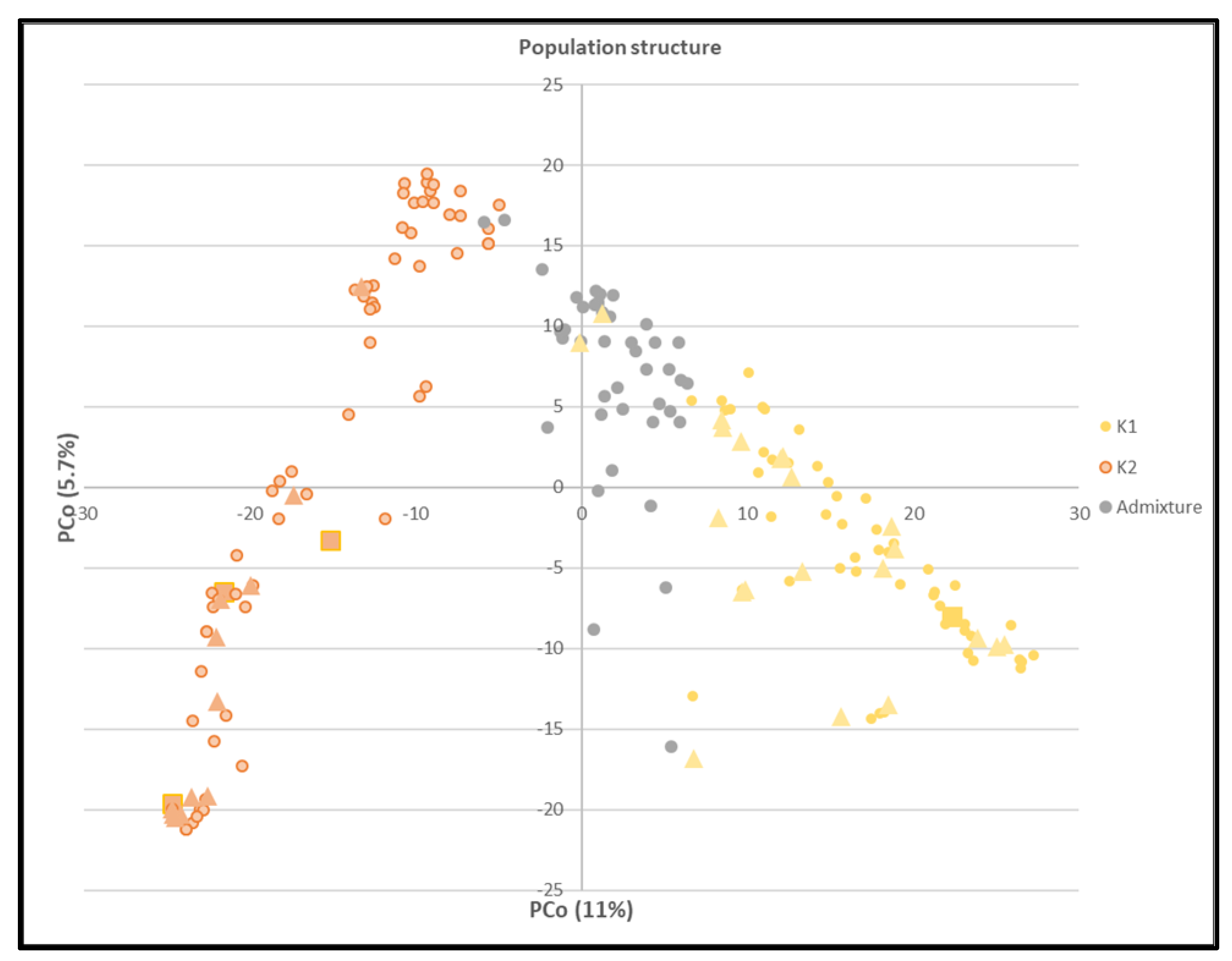
3.2.1. Population Structure
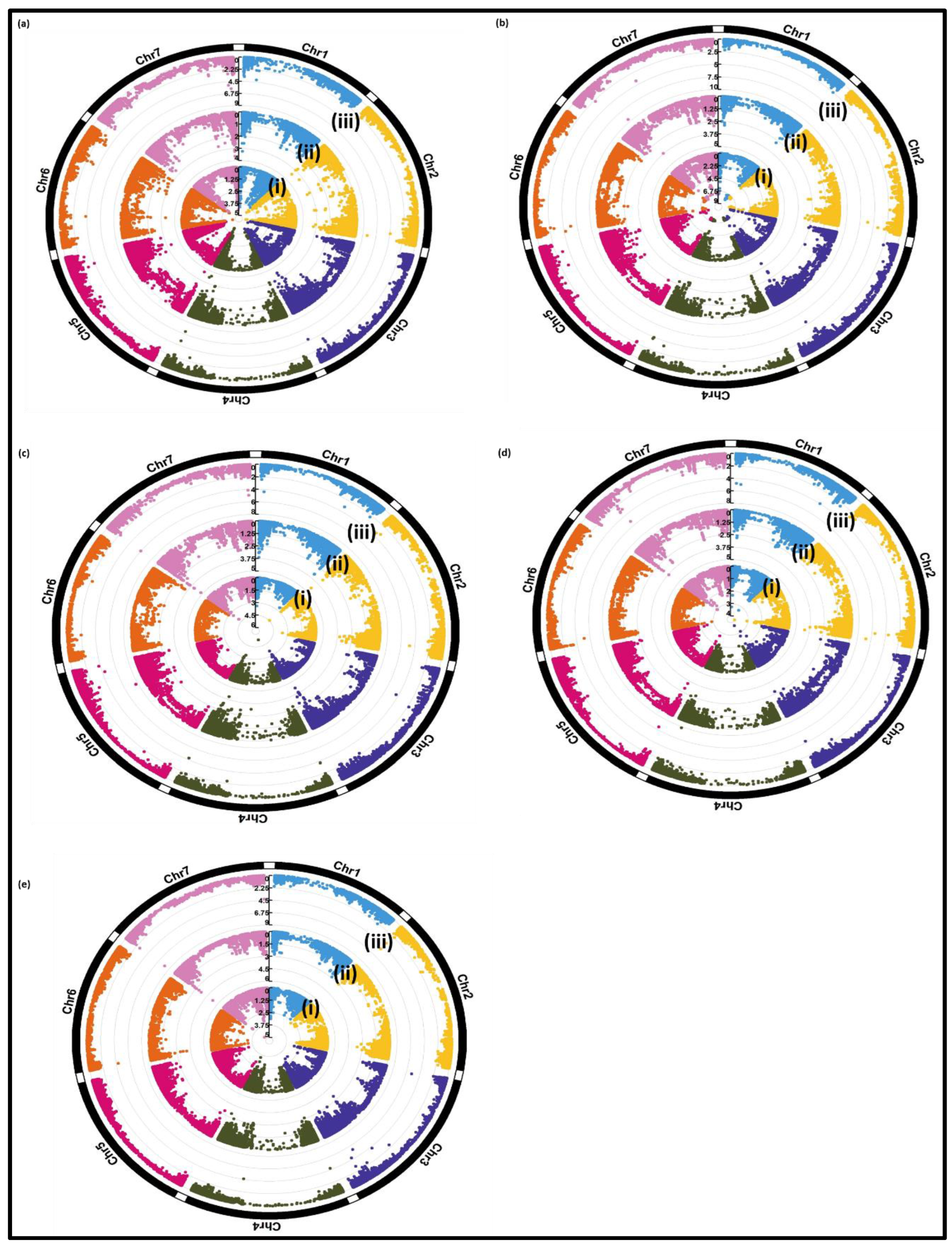
3.2.2. Genome-Wide Association Study (GWAS):
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mayer, K.F.X. A physical, genetic and functional sequence assembly of the barley genome. Nature 2012, 491, 711. [Google Scholar] [PubMed]
- Habekuß, A. Breeding for resistance to insect-transmitted viruses in barley-an emerging challenge due to global warming. J. Für Kult. 2009, 6, 53–61. [Google Scholar]
- Nygren, J. Variation in Susceptibility to Wheat dwarf virus among Wild and Domesticated Wheat. PLoS ONE 2015, 10, e0121580. [Google Scholar] [CrossRef] [PubMed]
- Cejnar, C.; et al. Two mutations in the truncated Rep gene RBR domain delayed the Wheat dwarf virus infection in transgenic barley plants. J. Integr. Agric. 2018, 17, 2492–2500. [Google Scholar] [CrossRef]
- Vacke, J. Host plants range and symptoms of wheat dwarf virus. ěVěd Pr Výz Ústavú Rostl Výroby Praha-Ruzyn 1972, 17, 151–162. [Google Scholar]
- Vacke, J.; Cibulka, R. Reactions of registered winter barley varieties to wheat dwarf virus infection. Czech J. Genet. Plant Breed. -UZPI 2001, 37, 50–52. [Google Scholar]
- Vacke, J. Wheat dwarf virus. Biol. Plant. 1961, 3, 228–233. [Google Scholar] [CrossRef]
- Koklu, G.; Ramsell, J.N.E.; Kvarnheden, A. The complete genome sequence for a Turkish isolate of Wheat dwarf virus (WDV) from barley confirms the presence of two distinct WDV strains. Virus Genes 2007, 34, 359–366. [Google Scholar] [CrossRef]
- Kanzi, A.M.; et al. Next Generation Sequencing and Bioinformatics Analysis of Family Genetic Inheritance. Front. Genet. 2020, 11, 544162. [Google Scholar] [CrossRef]
- Wang, S.C.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef]
- Bradbury, P.J.; et al. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; et al. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Liu, X.; et al. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS ONE 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.Y. Genomic prediction and GWAS of yield, quality and disease-related traits in spring barley and winter wheat. Sci. Rep. 2020, 10, 3347. [Google Scholar] [CrossRef] [PubMed]
- Gyawali, S.; et al. Genome wide association studies (GWAS) of spot blotch resistance at the seedling and the adult plant stages in a collection of spring barley. Mol. Breed. 2018, 38, 62. [Google Scholar] [CrossRef]
- Novakazi, F.; et al. Genome-wide association studies in a barley (Hordeum vulgare) diversity set reveal a limited number of loci for resistance to spot blotch (Bipolaris sorokiniana). Plant Breed. 2020, 139, 521–535. [Google Scholar] [CrossRef]
- Wehner, G.G.; et al. Identification of genomic regions involved in tolerance to drought stress and drought stress induced leaf senescence in juvenile barley. BMC Plant Biology 2015, 15, 125. [Google Scholar] [CrossRef]
- Jabbari, M.; et al. GWAS analysis in spring barley (Hordeum vulgare L.) for morphological traits exposed to drought. PLoS ONE 2018, 13, e0204952. [Google Scholar] [CrossRef]
- Rode, J.; et al. Identification of marker-trait associations in the German winter barley breeding gene pool (Hordeum vulgare L.). Mol. Breed. 2012, 30, 831–843. [Google Scholar] [CrossRef]
- Pfrieme, A.K. Identification and validation of Quantitative Trait Loci for Wheat dwarf virus resistance in wheat (Triticum spp.). Front. Plant Sci. 2022; 13. [Google Scholar]
- Bayer, M.M.; et al. Development and Evaluation of a Barley 50k iSelect SNP Array. Front. Plant Sci. 2017, 8, 1792. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.F.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Mascher, M. Pseudomolecules and annotation of the second version of the reference genome sequence assembly of barley cv. Morex [Morex V2]. 2019.
- Browning, B.L.; Browning, S.R. A Unified Approach to Genotype Imputation and Haplotype-Phase Inference for Large Data Sets of Trios and Unrelated Individuals. Am. J. Hum. Genet. 2009, 84, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Browning, S.R.; Browning, B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007, 81, 1084–1097. [Google Scholar] [CrossRef] [PubMed]
- Reif, J.C.; Melchinger, A.E.; Frisch, M. Genetical and mathematical properties of similarity and dissimilarity coefficients applied in plant breeding and seed bank management. Crop Sci. 2005, 45, 1–7. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Perrier, X.; Jacquemoud-Collet, J. DARwin software: Dissimilarity analysis and representation for windows. 2006. Available online: http://darwin. cirad. fr/darwin (accessed on 1 March 2013).
- Earl, D.A.; Vonholdt, B.M. , STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: http://www.R-project.org/. 2014.
- Shin, J.H.; et al. LDheatmap: An R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Softw. 2006, 16, 1–9. [Google Scholar] [CrossRef]
- Warnes, G. et al. Genetics: Population Genetics. R package version 1.3.8.1. 2013. Available online: http://CRAN.R-project.org/package=genetics.
- Voss-Fels, K.; et al. Subgenomic Diversity Patterns Caused by Directional Selection in Bread Wheat Gene Pools. Plant Genome 2015, 8, 13. [Google Scholar] [CrossRef]
- Sannemann, W.; et al. Multi-parent advanced generation inter-cross in barley: High-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol. Breed. 2015, 35, 86. [Google Scholar] [CrossRef]
- Lehnert, H.; et al. Genome-Wide Association Studies Reveal Genomic Regions Associated With the Response of Wheat (Triticum aestivum L.) to Mycorrhizae Under Drought Stress Conditions. Front. Plant Sci. 2018, 9, 1728. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.; et al. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J Genet Eng Biotechnol 2021, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Buerstmayr, M.; Buerstmayr, H. Two major quantitative trait loci control wheat dwarf virus resistance in four related winter wheat populations. Theor. Appl. Genet. 2023, 136, 103. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, E.; et al. Virus epidemics, plant-controlled population bottlenecks and the durability of plant resistance. Philos. Trans. R. Soc. B-Biol. Sci. 2019, 374. [Google Scholar] [CrossRef] [PubMed]
- Kaler, A.S.; et al. Comparing different statistical models and multiple testing corrections for association mapping in soybean and maize. Front. Plant Sci. 2020, 1794. [Google Scholar] [CrossRef]
- He, T., T. T.; Li, C. Pleiotropy structures plant height and seed weight scaling in barley despite long history of domestication and breeding selection. Plant Phenomics 2020, 5, 0015. [Google Scholar] [CrossRef]
- Rao, K.N.; Venkatachalam, S.R. Inhibition of dihydrofolate reductase and cell growth activity by the phenanthroindolizidine alkaloids pergularinine and tylophorinidine: The in vitro cytotoxicity of these plant alkaloids and their potential as antimicrobial and anticancer agents. Toxicol. Vitr. 2000, 14, 53–59. [Google Scholar] [CrossRef]
- Gorelova, V.; et al. Dihydrofolate Reductase/Thymidylate Synthase Fine-Tunes the Folate Status and Controls Redox Homeostasis in Plants. Plant Cell 2017, 29, 2831–2853. [Google Scholar] [CrossRef]
- Maule, A.J.; Caranta, C.; Boulton, M.I. Sources of natural resistance to plant viruses: Status and prospects. Mol. Plant Pathol. 2007, 8, 223–231. [Google Scholar] [CrossRef]
- Kristiansen, K.N.; Rohde, W. Structure of the Hordeum vulgare gene encoding dihydroflavonol-4-reductase and molecular analysis of ant 18 mutants blocked in flavonoid synthesis. Mol. Gen. Genet. MGG 1991, 230, 49–59. [Google Scholar] [CrossRef]
- Badshah, S.L.; et al. Antiviral activities of flavonoids. Biomed. Pharmacother. 2021, 140, 111596. [Google Scholar] [CrossRef] [PubMed]
- Masoud, S.A.; et al. Expression of a Cysteine Proteinase-Inhibitor (Oryzacystatin-I) in Transgenic Tobacco Plants. Plant Mol. Biol. 1993, 21, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Campos, R.; et al. The use of cysteine proteinase inhibitors to engineer resistance against potyviruses in transgenic tobacco plants. Nat. Biotechnol. 1999, 17, 1223–1226. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, E.; Michaud, D.; Cloutier, C. Molecular interactions between an insect predator and its herbivore prey on transgenic potato expressing a cysteine proteinase inhibitor from rice. Mol. Ecol. 2003, 12, 2429–2437. [Google Scholar] [CrossRef]
- Carrillo, L.; et al. A barley cysteine-proteinase inhibitor reduces the performance of two aphid species in artificial diets and transgenic Arabidopsis plants. Transgenic Res. 2011, 20, 305–319. [Google Scholar] [CrossRef]
| Trait | Abbreviation | Method of measurement | Unit |
|---|---|---|---|
| ELISA-60 | double antibody sandwich enzyme-linked immunosorbent assay | ||
| Total grain weight | ToGW | Weight all harvested seeds per plant | g |
| Plant height | HEI | Measure plant length from basis to leaf tip | cm |
| Number of ears per plant | NEP | Count number of ears after harvesting | |
| Thousand grain weight | TGW | Weigh 1000 grain after threshing | g |
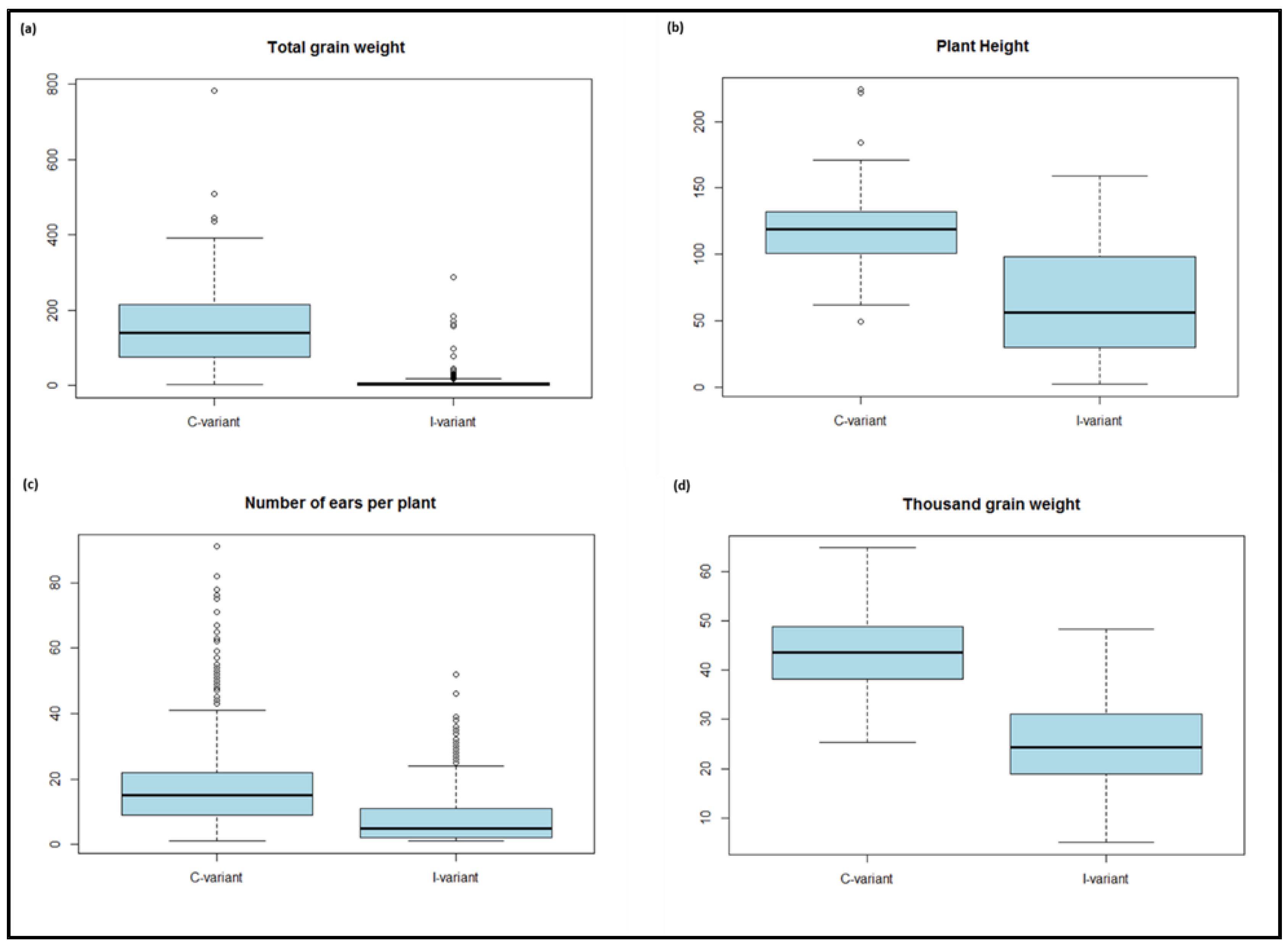
| Trait | Treatmenta | Nb | Meanc | Minimumd | Maximumd | Sde | CVf |
|---|---|---|---|---|---|---|---|
| ELISA-60 | I-variant | 1866.0 | 0.3 | -0.03 | 1.84 | 0.6 | 179.7 |
| Total grain weight | I-variant | 209 | 11.3 | 0 | 287.4 | 31.7 | 280.7 |
| C-variant | 229 | 156.3 | 2.2 | 781.7 | 102.6 | 65.7 | |
| Plant height | I-variant | 520 | 62.9 | 1 | 159 | 40.4 | 64.2 |
| C-variant | 1163 | 117.5 | 49 | 224 | 20.3 | 17.2 | |
| Number of ears per plant | I-variant | 524 | 6.9 | 0 | 52 | 8.2 | 118.3 |
| C-variant | 1160 | 18.4 | 1 | 117 | 12.1 | 65.8 | |
| Thousand grain weight | I-variant | 208 | 25.5 | 5 | 48.3 | 8.7 | 34.1 |
| C-variant | 229 | 43.5 | 25.37 | 64.8 | 7.8 | 17.9 |
| Trait | Effect | Degrees of freedom | F Value | Pr > F |
|---|---|---|---|---|
| ELISA-60 | Genotype (G) | 190 | 2.26 | <.0001 |
| Total grain weight | Genotype (G) | 190 | 1.05 | 0.49 |
| Treatment (T) | 1 | 364.08 | <.0001 | |
| G*T | 184 | 0.84 | 0.74 | |
| Plant height | Genotype (G) | 190 | 5.43 | <.0001 |
| Treatment (T) | 1 | 3651.82 | <.0001 | |
| G*T | 186 | 9.26 | <.0001 | |
| Number of ears per plant | Genotype (G) | 190 | 1.04 | 0.50 |
| Treatment (T) | 1 | 443.86 | <.0001 | |
| G*T | 186 | 1.83 | <.0001 | |
| Thousand grain weight | Genotype (G) | 190 | 3.05 | <.0001 |
| Treatment (T) | 1 | 1233.26 | <.0001 | |
| G*T | 184 | 2.19 | <.0001 |
| Trait | Marker name | Chra | Posb | P value | Identified genes in QTL region | ||
|---|---|---|---|---|---|---|---|
| Gapitc | Tasselc | FarmCPU | |||||
| ELISA-60 | JHI-Hv50k-2016-202912 | 3H | 562758917 | 3.3E-04 | 2.6E-04 | 2.2E-04 | |
| Relative total grain weight | JHI-Hv50k-2016-196649 | 3H | 534052013 | 6.8E-05 | 7.7E-04 | 2.8E-06 | |
| Relative total grain weight | BOPA1_2955-452 | 4H | 552300974 | 9.5E-04 | 2.4E-05 | 9.4E-05 | Cysteine proteinase inhibitor |
| Relative total grain weight | BOPA2_12_10333 | 5H | 554416618 | 3.4E-04 | 1.1E-04 | 1.7E-04 | |
| Relative plant height | BOPA2_12_21049 | 2H | 31329721 | 3.3E-05 | 3.5E-05 | 6.6E-04 | Dihydrofolate reductase |
| Relative plant height | JHI-Hv50k-2016-435708 | 7H | 1402273 | 1.5E-05 | 7.4E-05 | 1.1E-06 | |
| Relative number of ears per plant | JHI-Hv50k-2016-123144 | 2H | 631278948 | 1.8E-04 | 8E-04 | 2.7E-06 | NBS-LRR disease resistance protein |
| Relative number of ears per plant | JHI-Hv50k-2016-142550 | 2H | 666139797 | 6.4E-05 | 1.0E-04 | 1.7E-07 | Dihydroflavonol 4-reductase |
| Relative thousand grain weight | JHI-Hv50k-2016-435708 | 7H | 1402273 | 4.8E-06 | 1.7E-05 | 8.7E-09 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





