Submitted:
02 May 2023
Posted:
03 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Soil-borne oomycete community diversity and compositional structure
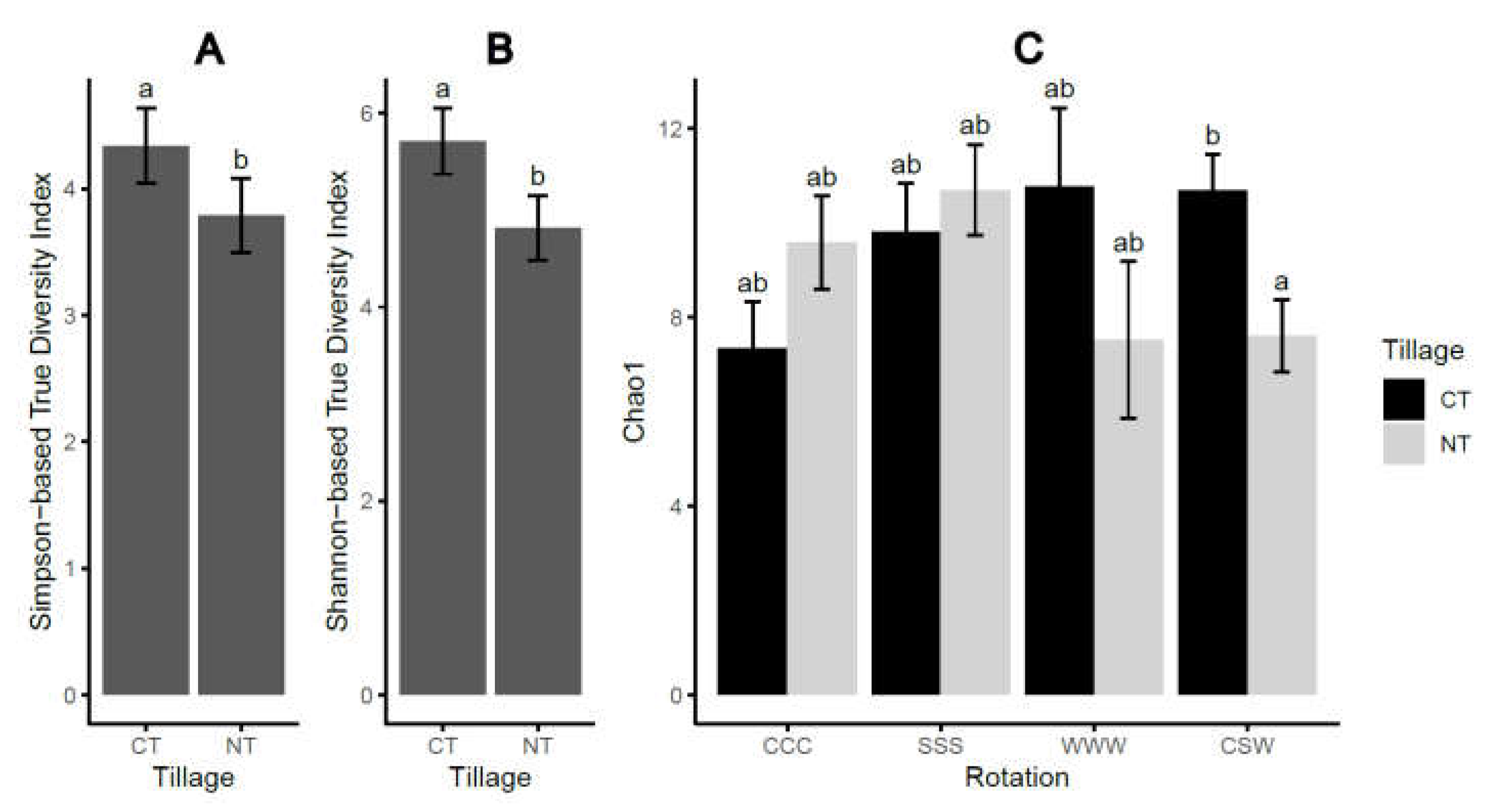
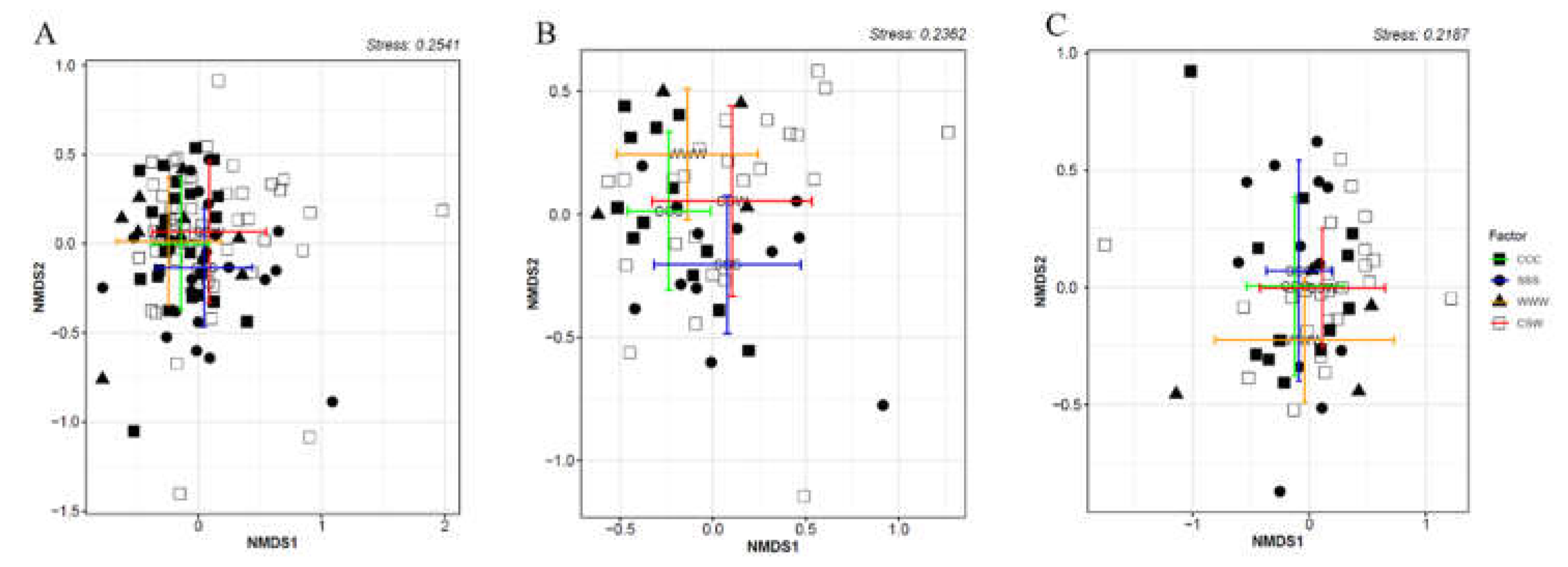
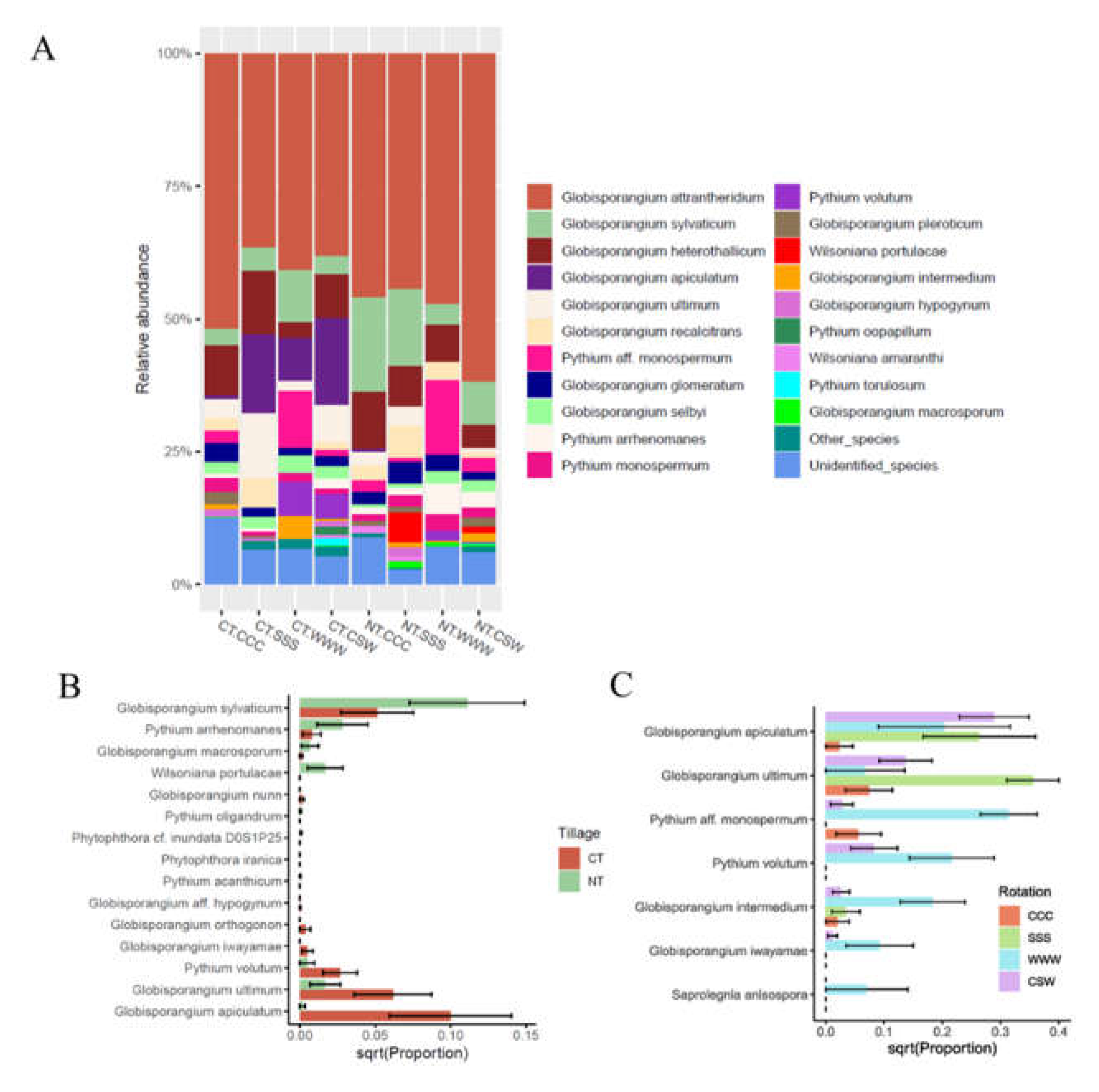
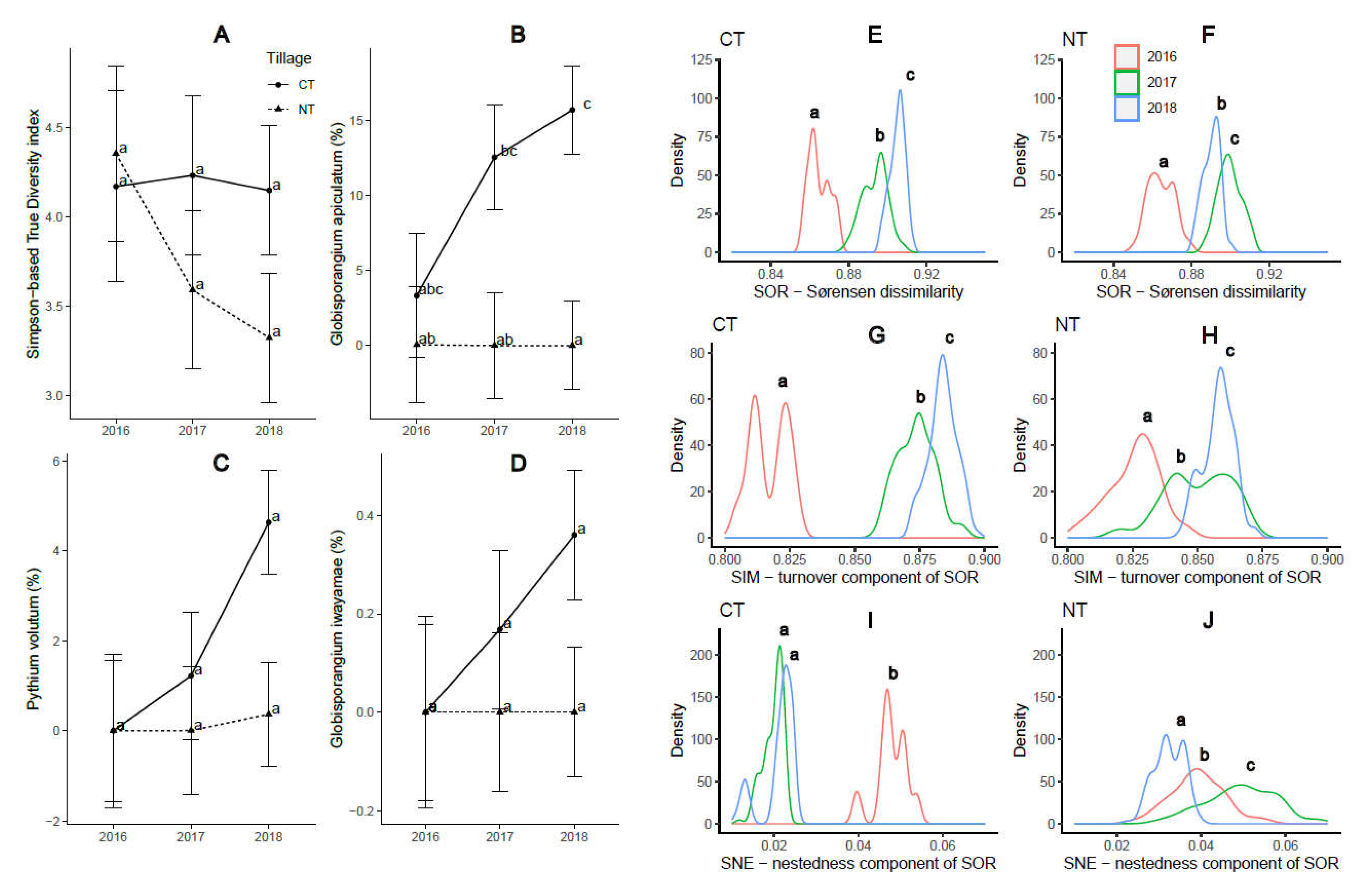
2.2. Crop yield and soybean seedling emergence
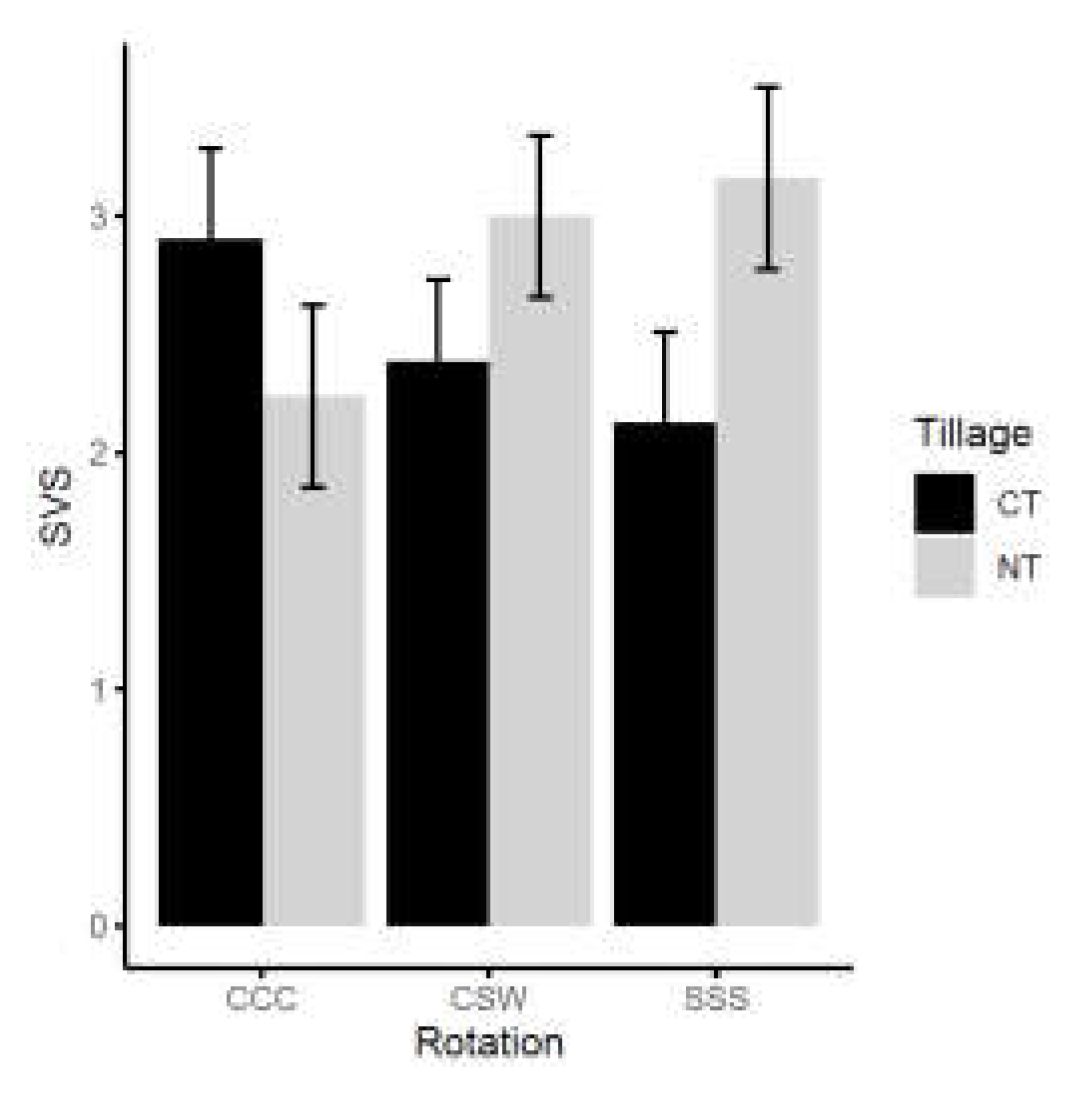
3. Discussion
4. Materials and Methods
4.1. Study site and experimental design
4.2. Soil sampling
4.3. DNA extraction
4.4. Sequencing library preparation and Illumina MiSeq sequencing
4.5. Metabarcoding data processing and analysis
4.6. Soybean seedling vitality experiment
4.7. Statistical analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Crown Copyright statement
References
- Thines, M.; Voglmayr, H.; Göker, M. Taxonomy and Phylogeny of the Downy Mildews (Peronosporaceae). In Oomycete Genetics and Genomics; 2009; pp. 47–75. [Google Scholar]
- Fry, W. Phytophthora infestans: the plant (and R gene) destroyer. Molecular Plant Pathology 2008, 9, 385–402. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.; Anderson, V.L.; Robertson, E.J.; Secombes, C.J.; van West, P. New insights into animal pathogenic oomycetes. Trends in Microbiology 2008, 16, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Cahill, D.M.; Rookes, J.E.; Wilson, B.A.; Gibson, L.; McDougall, K.L. Phytophthora cinnamomi and Australia’s biodiversity: impacts, predictions and progress towards control. Australian Journal of Botany 2008, 56. [Google Scholar] [CrossRef]
- Cohen, Y.; Coffey, M.D. SYSTEMIC FUNGICIDES AND THE CONTROL OF OOMYCETES. Annual Reviews in Phytopathology 1986, 24, 311–338. [Google Scholar] [CrossRef]
- Dorrance, A.; Grünwald, N.J. Phytophthora sojae: Diversity among and within Populations. In Oomycete Genetics and Genomics; 2009; pp. 197–212. [Google Scholar]
- Cook, R.; Sitton, J.; Haglund, W. Influence of soil treatments on growth and yield of wheat and implications for control of Pythium root rot. Phytopathology 1987, 77, 1192–1198. [Google Scholar] [CrossRef]
- Cook, R.J.; Haglund, W.A. Wheat yield depression associated with conservation tillage caused by root pathogens in the soil not phytotoxins from the straw. Soil Biology and Biochemistry 1991, 23, 1125–1132. [Google Scholar] [CrossRef]
- Bongiorno, G.; Postma, J.; Bünemann, E.K.; Brussaard, L.; de Goede, R.G.M.; Mäder, P.; Tamm, L.; Thuerig, B. Soil suppressiveness to Pythium ultimum in ten European long-term field experiments and its relation with soil parameters. Soil Biology and Biochemistry 2019, 133, 174–187. [Google Scholar] [CrossRef]
- Peters, R.D.; Sturz, A.V.; Carter, M.R.; Sanderson, J.B. Developing disease-suppressive soils through crop rotation and tillage management practices. Soil and Tillage Research 2003, 72, 181–192. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, B.; Hong, S.; Xiong, W.; Shen, Z.; Ruan, Y.; Li, R.; Shen, Q.; Dini-Andreote, F. Promoting soil microbial-mediated suppressiveness against Fusarium wilt disease by the enrichment of specific fungal taxa via crop rotation. Biology and Fertility of Soils 2021, 57, 1137–1153. [Google Scholar] [CrossRef]
- Santín-Montanyá, M. I.; Sombrero Sacristán, A. The effects of soil tillage techniques on weed flora in high input barley systems in northern Spain. Canadian Journal of Plant Science 2020, 100, 245–252. [Google Scholar] [CrossRef]
- Bogunovic, I.; Pereira, P.; Kisic, I.; Sajko, K.; Sraka, M. Tillage management impacts on soil compaction, erosion and crop yield in Stagnosols (Croatia). CATENA 2018, 160, 376–384. [Google Scholar] [CrossRef]
- Abdollahi, L.; Schjønning, P.; Elmholt, S.; Munkholm, L.J. The effects of organic matter application and intensive tillage and traffic on soil structure formation and stability. Soil and Tillage Research 2014, 136, 28–37. [Google Scholar] [CrossRef]
- Srour, A.Y.; Ammar, H.A.; Subedi, A.; Pimentel, M.; Cook, R.L.; Bond, J.; Fakhoury, A.M. Microbial Communities Associated With Long-Term Tillage and Fertility Treatments in a Corn-Soybean Cropping System. Front Microbiol 2020, 11, 1363. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, D.; Liang, S.; Dang, P.; Qin, X.; Liao, Y.; Siddique, K.H.M. Effect of no-tillage on soil bacterial and fungal community diversity: A meta-analysis. Soil Tillage Res. 2020, 204, 104721. [Google Scholar] [CrossRef]
- Sipilä Timo, P.; Yrjälä, K.; Alakukku, L.; Palojärvi, A. Cross-Site Soil Microbial Communities under Tillage Regimes: Fungistasis and Microbial Biomarkers. Applied and Environmental Microbiology 2012, 78, 8191–8201. [Google Scholar] [CrossRef]
- Liu, B.; Gumpertz, M.L.; Hu, S.; Ristaino, J.B. Effect of prior tillage and soil fertility amendments on dispersal of Phytophthora capsici and infection of pepper. European Journal of Plant Pathology 2008, 120, 273–287. [Google Scholar] [CrossRef]
- Peralta, A.L.; Sun, Y.; McDaniel, M.D.; Lennon, J.T. Crop rotational diversity increases disease suppressive capacity of soil microbiomes. 2018, 9, e02235. [Google Scholar] [CrossRef]
- Pedersen, E.A. The Effect of Crop Rotation on Development of the Septoria Disease Complex on Spring Wheat in Saskatchewan. Canadian Journal of Plant Pathology 1992, 14, 152–158. [Google Scholar] [CrossRef]
- Larkin, R.P.; Halloran, J.M. Management Effects of Disease-Suppressive Rotation Crops on Potato Yield and Soilborne Disease and Their Economic Implications in Potato Production. American Journal of Potato Research 2014, 91, 429–439. [Google Scholar] [CrossRef]
- Lyu, J.; Jin, L.; Jin, N.; Xie, J.; Xiao, X.; Hu, L.; Tang, Z.; Wu, Y.; Niu, L.; Yu, J. Effects of Different Vegetable Rotations on Fungal Community Structure in Continuous Tomato Cropping Matrix in Greenhouse. Frontiers in Microbiology 2020, 11. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Sui, P.; Liu, X.; Zhang, T.; Li, X. Crop rotation to diversify the soil microbiome in the semi-arid area of Inner Mongolia, China. Archives of Agronomy and Soil Science 2022, 1–16. [Google Scholar] [CrossRef]
- Hwang, S.; Ahmed, H.; Gossen, B.; Kutcher, H.; Brandt, S.; Strelkov, S.; Chang, K.; Turnbull, G. Effect of crop rotation on the soil pathogen population dynamics and canola seedling establishment. Plant Pathology Journal 2009, 8, 106–112. [Google Scholar] [CrossRef]
- Radmer, L.; Anderson, G.; Malvick, D.M.; Kurle, J.E.; Rendahl, A.; Mallik, A. Pythium, Phytophthora, and Phytopythium spp. Associated with Soybean in Minnesota, Their Relative Aggressiveness on Soybean and Corn, and Their Sensitivity to Seed Treatment Fungicides. 2017, 101, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Zitnick-Anderson, K.K.; Nelson, B.D. Identification and Pathogenicity of Pythium on Soybean in North Dakota. 2015, 99, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Rojas, J.A.; Jacobs, J.L.; Napieralski, S.; Karaj, B.; Bradley, C.A.; Chase, T.; Esker, P.D.; Giesler, L.J.; Jardine, D.J.; Malvick, D.K.; et al. Oomycete Species Associated with Soybean Seedlings in North America—Part I: Identification and Pathogenicity Characterization. 2017, 107, 280–292. [Google Scholar] [CrossRef]
- Rojas, J.A.; Jacobs, J.L.; Napieralski, S.; Karaj, B.; Bradley, C.A.; Chase, T.; Esker, P.D.; Giesler, L.J.; Jardine, D.J.; Malvick, D.K.; et al. Oomycete Species Associated with Soybean Seedlings in North America—Part II: Diversity and Ecology in Relation to Environmental and Edaphic Factors. 2017, 107, 293–304. [Google Scholar] [CrossRef]
- Morrison, M.J.; Cober, E.R.; Gregorich, E.; Voldeng, H.D.; Ma, B.; Topp, G.C. Tillage and crop rotation effects on the yield of corn, soybean and wheat in eastern Canada. Canadian Journal of Plant Science 2017, 98, 183–191. [Google Scholar] [CrossRef]
- Morrison, M.J.; Cober, E.R.; Gregorich, E.G.; Voldeng, H.D.; Ma, B.; Topp, G.C. Tillage and crop rotation effects on the yield of corn, soybean, and wheat in eastern Canada. 2018, 98, 183–191. [Google Scholar] [CrossRef]
- Büchi, L.; Wendling, M.; Amossé, C.; Jeangros, B.; Sinaj, S.; Charles, R. Long and short term changes in crop yield and soil properties induced by the reduction of soil tillage in a long term experiment in Switzerland. Soil and Tillage Research 2017, 174, 201–129. [Google Scholar] [CrossRef]
- Sapp, M.; Tyborski, N.; Linstädter, A.; López Sánchez, A.; Mansfeldt, T.; Waldhoff, G.; Bareth, G.; Bonkowski, M.; Rose, L.E. Site-specific distribution of oak rhizosphere-associated oomycetes revealed by cytochrome c oxidase subunit II metabarcoding. 2019, 9, 10567–10581. [Google Scholar] [CrossRef]
- Rossmann, S.; Lysøe, E.; Skogen, M.; Talgø, V.; Brurberg, M.B. DNA Metabarcoding Reveals Broad Presence of Plant Pathogenic Oomycetes in Soil From Internationally Traded Plants. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.N.; Loper, J.E. Soilborne Plant Diseases Caused by Pythium spp.: Ecology, Epidemiology, and Prospects for Biological Control. Critical Reviews in Plant Sciences 1999, 18, 111–181. [Google Scholar] [CrossRef]
- Uzuhashi, S.; Kakishima, M.; Tojo, M. Phylogeny of the genus Pythium and description of new genera. Mycoscience 2010, 51, 337–365. [Google Scholar] [CrossRef]
- Gan, H.; Chai, Z.; Lou, B.; Li, J. Pythium heterothallicum new to China and its pathogenicity. Mycosystem 2010, 29, 494–501. [Google Scholar]
- Coffua, L.S.; Veterano, S.T.; Clipman, S.J.; Mena-Ali, J.I.; Blair, J.E. Characterization of Pythium spp. Associated with Asymptomatic Soybean in Southeastern Pennsylvania. 2016, 100, 1870–1879. [Google Scholar] [CrossRef]
- Broders, K.D.; Lipps, P.E.; Paul, P.A.; Dorrance, A.E. Characterization of Pythium spp. Associated with Corn and Soybean Seed and Seedling Disease in Ohio. Plant Disease 2007, 91, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.G.; Moorman, T.B.; Kaspar, T.C.; Manter, D.K. Isolation of Cultivation-Resistant Oomycetes, First Detected as Amplicon Sequences, from Roots of Herbicide-Terminated Winter Rye. 2017, 1, 24–35. [Google Scholar] [CrossRef]
- Reyes-Tena, A.; Vallejo-González, R.; Santillán-Mendoza, R.; Rodríguez-Alvarado, G.; Larsen, J.; Fernández-Pavía, S.P. Pythium arrhenomanes causal agent of root rot on yellow maize in Mexico. Australasian Plant Disease Notes 2018, 13, 6. [Google Scholar] [CrossRef]
- Chamswarng, C.; Cook, R. Identification and comparative pathogenicity of pythium species from wheat roots and wheat-field soils in the Pacific Northwest. Phytopathology 1985, 75, 821–827. [Google Scholar] [CrossRef]
- Paulitz, T.C. Low Input No-till Cereal Production in the Pacific Northwest of the U.S.: The Challenges of Root Diseases. European Journal of Plant Pathology 2006, 115, 271–281. [Google Scholar] [CrossRef]
- Waller, J.M. Observations on Pythium root rot of wheat and barley. 1979, 28, 17–24. [Google Scholar] [CrossRef]
- Yin, C.; McLaughlin, K.; Paulitz, T.C.; Kroese, D.R.; Hagerty, C.H. Population Dynamics of Wheat Root Pathogens Under Different Tillage Systems in Northeast Oregon. 2020, 104, 2649–2657. [Google Scholar] [CrossRef] [PubMed]
- Shittu, K.A.; Oyedele, D.J.; Babatunde, K.M. The effects of moisture content at tillage on soil strength in maize production. Egyptian Journal of Basic and Applied Sciences 2017, 4, 139–142. [Google Scholar] [CrossRef]
- Yang, T.; Siddique, K.H.M.; Liu, K. Cropping systems in agriculture and their impact on soil health-A review. Global Ecology and Conservation 2020, 23. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends in Plant Science 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Hoitink, H.; Boehm, M. Biocontrol within the context of soil microbial communities: a substrate-dependent phenomenon. Annual review of phytopathology 1999, 37, 427. [Google Scholar] [CrossRef]
- Cook, R.; Sitton, J.; Waldher, J. Evidence for Pythium as a pathogen of direct-drilled wheat in the Pacific Northwest. Plant Disease 1980, 64, 61–61. [Google Scholar] [CrossRef]
- Pankhurst, C.E.; McDonald, H.J.; Hawke, B.G. Influence of tillage and crop rotation on the epidemiology of Pythium infections of wheat in a red-brown earth of South Australia. Soil Biology and Biochemistry 1995, 27, 1065–1073. [Google Scholar] [CrossRef]
- Paul, B. A new species of Pythium isolated from a vineyard in France. FEMS Microbiology Letters 2006, 263, 194–199. [Google Scholar] [CrossRef]
- Jiang, Y.N.; Haudenshield, J.S.; Hartman, G.L. Characterization of Pythium spp. from soil samples in Illinois. Canadian Journal of Plant Pathology 2012, 34, 448–454. [Google Scholar] [CrossRef]
- Wei, L.; Xue, A.; Cober, E.; Babcock, C.; Zhang, J.; Zhang, S.; Li, W.; Wu, J.; Liu, L.J.P. Pathogenicity of Pythium species causing seed rot and damping-off in soybean under controlled conditions. 2010, 91, 3–10. [Google Scholar] [CrossRef]
- Broders, K.D.; Wallhead, M.W.; Austin, G.D.; Lipps, P.E.; Paul, P.A.; Mullen, R.W.; Dorrance, A.E. Association of Soil Chemical and Physical Properties with Pythium Species Diversity, Community Composition, and Disease Incidence. 2009, 99, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Hancock, J. Association of chemical and biological factors in soils suppressive to Pythium ultimum. Phytopathology 1986, 76, 1221–1231. [Google Scholar] [CrossRef]
- Rojas, J.A.; Witte, A.; Noel, Z.A.; Jacobs, J.L.; Chilvers, M.I. Diversity and Characterization of Oomycetes Associated with Corn Seedlings in Michigan. 2019, 3, 224–234. [Google Scholar] [CrossRef]
- Bockus, W.; Shroyer, J. The impact of reduced tillage on soilborne plant pathogens. Annual review of phytopathology 1998, 36, 485–500. [Google Scholar] [CrossRef] [PubMed]
- Asad, N.I.; Wang, X.-B.; Dozois, J.; Azarbad, H.; Constant, P.; Yergeau, E. Early season soil microbiome best predicts wheat grain quality. FEMS Microbiology Ecology 2023, 99, fiac144. [Google Scholar] [CrossRef]
- Sapkota, R.; Nicolaisen, M. Cropping history shapes fungal, oomycete and nematode communities in arable soils and affects cavity spot in carrot. Agric. Ecosyst. Environ. 2018, 257, 120–131. [Google Scholar] [CrossRef]
- EMMOND, G.S.; LEDINGHAM, R.J. EFFECTS OF CROP ROTATION ON SOME SOIL-BORNE PATHOGENS OF POTATO. 1972, 52, 605–611. [Google Scholar] [CrossRef]
- Frank, J.A.; Murphy, H. The effect of crop rotations on Rhizoctonia disease of potatoes. American Potato Journal 1977, 54, 315–322. [Google Scholar] [CrossRef]
- Scholte, K.H. The effect of crop rotation and granular nematicides on the incidence ofRhizoctonia solani in potato. J Potato Research 1987, 30, 187–199. [Google Scholar] [CrossRef]
- Bargués-Ribera, M.; Gokhale, C.S. Eco-evolutionary agriculture: Host-pathogen dynamics in crop rotations. PLoS computational biology 2020, 16, e1007546. [Google Scholar] [CrossRef]
- Lipps, P.; Bruehl, G. Snow rot of winter wheat in Washington. Phytopathology 1978, 68, 120–121. [Google Scholar] [CrossRef]
- Spies, C.F.J.; Mazzola, M.; McLeod, A. Characterisation and detection of Pythium and Phytophthora species associated with grapevines in South Africa. European journal of plant pathology 2011, 131, 103–119. [Google Scholar] [CrossRef]
- Masigol, H.; Khodaparast, S.A.; Mostowfizadeh-Ghalamfarsa, R.; Rojas-Jimenez, K.; Woodhouse, J.N.; Neubauer, D.; Grossart, H.-P. Taxonomical and functional diversity of Saprolegniales in Anzali lagoon, Iran. Aquatic Ecology 2020, 54, 323–336. [Google Scholar] [CrossRef]
- Rizvi, S.; Yang, X. Fungi associated with soybean seedling disease in Iowa. Plant disease 1996, 80, 57–60. [Google Scholar] [CrossRef]
- Morrison, M.J.; Cober, E.R.; Gregorich, E.G.; Voldeng, H.D.; Ma, B.; Topp, G.C. Tillage and crop rotation effects on the yield of corn, soybean, and wheat in eastern Canada. Canadian Journal of Plant Science 2017, 98, 183–191. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EBMnet.journal 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; de Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler2, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6. [Google Scholar] [CrossRef]
- Zahariev, M.A.-O.; Chen, W. Cluster oligonucleotide signatures for rapid identification by sequencing. BMC Bioinformatics 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Radford, D.; Hambleton, S. Towards improved detection and identification of rust fungal pathogens in environmental samples using a metabarcoding approach. Phytopathology 2021, 112, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 2010, 5, e9490. [Google Scholar] [CrossRef]
- Team, R.C. R: A language and environment for statistical computing. 2021.
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O'Hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. vegan: Community Ecology Package. R package version 2.6-2. https://CRAN.R-project.org/package=vegan 2022, 2. [Google Scholar]
- Kindt, R. BiodiversityR: Package for community ecology suitability analysis. 2020. Available online: https://github.com/cran/BiodiversityR.
- Jost, L. Entropy and diversity. Oikos 2006, 113, 363–375. [Google Scholar] [CrossRef]
- Pinheiro, J.; Bates, D. nlme: Linear and Nonlinear Mixed Effects Models. 2023. Available online: https://CRAN.R-project.org/package=nlme.
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA). In Wiley StatsRef: Statistics Reference Online; 2014; pp. 1–15. [Google Scholar]
- Hervé, M. RVAideMemoire: Testing and plotting procedures for biostatistics. R package version 0.9-81-2 2022, https://CRAN.R-project.org/package=RVAideMemoire, doi:https://CRAN.Rproject. org/package=RVAideMemoire.
- Baselga, A.; Orme, C.D.L. betapart: an R package for the study of beta diversity. Methods Ecol. Evol. 2012, 3, 808–812. [Google Scholar] [CrossRef]
| DF | Simpson-TD1 | Shannon-TD2 | Chao1 | |
| Tillage (T) | 1 | 0.014 | 0.007 | 0.160 |
| Rotation (R) | 3 | 0.613 | 0.563 | 0.4292 |
| T×R | 3 | 0.217 | 0.041 | <0.001 |
| Species a | Relative abundance (%) | Analysis of variance (P value) b | Disease Note c | Known Hosts c,d | ||||||||||||||
| CT | NT | T | R | T*R | R CT | R NT | ||||||||||||
| No. ASV | CCC | SSS | WWW | CSW | CCC | SSS | WWW | CSW | ||||||||||
| Globisporangium aff. hypogynum | 1 | 0.000 | 0.000 | 0.000 | 0.153 | 0.000 | 0.000 | 0.000 | 0.000 | 0.320 | 0.78 | 0.772 | 0.7817 | N/A | ||||
| Globisporangium apiculatum | 20 | 0.664 | 16.294 | 7.970 | 16.522 | 0.000 | 0.000 | 0.000 | 0.002 | <0.001 | 0.036 | 0.031 | 0.041 | 0.761 | grape* | |||
| Globisporangium attrantheridium | 49 | 57.290 | 33.493 | 40.079 | 41.743 | 38.209 | 46.590 | 47.258 | 59.105 | 0.230 | 0.448 | 0.021 | 0.066 | 0.138 | Cavity spot lesions | Daucus carota, Prunus, soybean | ||
| Globisporangium glomeratum | 11 | 2.273 | 2.341 | 1.381 | 2.454 | 1.770 | 2.658 | 3.176 | 2.208 | 0.96 | 0.833 | 0.936 | 0.988 | 0.734 | soybean | |||
| Globisporangium heterothallicum | 27 | 10.438 | 11.844 | 2.942 | 6.979 | 13.878 | 7.603 | 7.139 | 3.802 | 0.539 | 0.039 | 0.558 | N/A | 0.076 | Damping off | Pepper, soybean, corn, spinach, wheat, lentils | ||
| Globisporangium hypogynum | 3 | 0.000 | 0.930 | 0.000 | 0.000 | 1.115 | 2.277 | 0.000 | 0.728 | 0.055 | 0.075 | 0.902 | 0.061 | 0.381 | Root rots | soybean | ||
| Globisporangium intermedium | 3 | 1.000 | 0.000 | 4.290 | 0.388 | 0.000 | 0.995 | 0.281 | 1.700 | 0.755 | 0.074 | 0.001 | 0.004 | 0.407 | Damping-off, rots | abutilon, antirrhinum, arabis, beet, begonia, carrot, cauliflower, chamaecyparis, cherry laurel, chrysanthemum, cotoneaster, cucumber, erica, ferns, godetia, hazel, hop, hyacinth, lettuce, leyland cypress, lupin, nasturtium, pea, pear, pelargonium, pepper, saintpaulia, strawberry, fragaria vesca, tomato, violet, yew, soybean, corn | ||
| Globisporangium irregulare | 2 | 0.000 | 0.000 | 0.000 | 0.247 | 0.074 | 0.000 | 0.000 | 0.000 | 0.559 | 0.867 | 0.563 | 0.782 | 0.347 | Blight, damping off, root and other rots, etc | soybean, wheat, corn | ||
| Globisporangium iwayamae | 2 | 0.000 | 0.000 | 1.860 | 0.163 | 0.000 | 0.000 | 0.000 | 0.000 | 0.024 | 0.001 | 0.0007 | 0.002 | N/A | Rots | Poaceae, wheat | ||
| Globisporangium macrosporum | 2 | 0.000 | 0.000 | 0.000 | 0.181 | 0.055 | 1.220 | 0.797 | 0.546 | 0.029 | 0.658 | 0.738 | 0.782 | 0.682 | Damping-off, root rot | iris, soybean, corn | ||
| Globisporangium nunn | 1 | 0.000 | 0.000 | 0.000 | 0.000 | 1.042 | 0.000 | 0.000 | 0.000 | 0.333 | 0.349 | 0.339 | N/A | 0.3415 | soybean | |||
| Globisporangium orthogonon | 1 | 0.000 | 1.578 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.305 | 0.323 | 0.264 | 0.3061 | N/A | corn | |||
| Globisporangium pleroticum | 5 | 2.38 | 0.3683 | 0.000 | 0.193 | 0.952 | 1.170 | 0.000 | 1.760 | 0.7545 | 0.074 | 0.009 | 0.711 | 0.841 | peas, soybean, lupins | |||
| Globisporangium recalcitrans | 14 | 1.103 | 6.190 | 0.000 | 1.321 | 3.613 | 4.969 | 3.326 | 1.693 | 0.131 | 0.194 | 0.566 | 0.219 | 0.435 | Root rot, damping off. | beet, hebe | ||
| Globisporangium rostratifingens | 5 | 0.000 | 0.000 | 0.000 | 0.329 | 0.725 | 0.233 | 0.000 | 0.000 | 0.699 | 0.671 | 0.107 | 0.529 | 0.174 | Root rot | pea, soybean, corn, wheat | ||
| Globisporangium selbyi | 8 | 2.200 | 2.223 | 3.130 | 2.350 | 0.591 | 0.774 | 2.410 | 2.290 | 0.702 | 0.859 | 0.996 | 0.943 | 0.750 | Lesions | corn, soybean | ||
| Globisporangium sylvaticum | 25 | 4.970 | 2.344 | 9.556 | 3.796 | 19.620 | 12.984 | 3.839 | 7.282 | 0.006 | 0.330 | 0.086 | 0.334 | 0.063 | Root disease, rots | apples, carrot, cherry laurel, cress, chrysanthemum, cucumber, garlic, lettuce, onion, pea, radish, rhododendron, spinach, strawberry, yew, wheat | ||
| Globisporangium ultimum | 11 | 2.346 | 14.656 | 1.834 | 6.589 | 2.490 | 2.820 | 0.000 | 1.348 | <0.001 | 0.008 | 0.011 | 0.002 | 0.991 | Blight, damping off, root rot, etc | soybean, garlic, grape, hyacinth, lettuce, lily, lupin, melon, mustard, onion, parsley, pea, pear, pelargonium, pepper, poinsettia, primula, radish, rhododendron, rhubarb, spinach, strawberry, sweet pea, tomato, tulip, wallflower, yew | ||
| Phytophthora cf. inundata D0S1P25 | 1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.237 | 0.000 | 0.000 | 0.180 | 0.093 | 0.152 | N/A | 0.092 | ||||
| Pythium acanthicum | 1 | 0.000 | 0.000 | 0.000 | 0.073 | 0.000 | 0.000 | 0.000 | 0.000 | 0.315 | 0.724 | 0.763 | 0.711 | N/A | Downy mildew. Blight, damping off, rots, etc. | soybean, corn | ||
| Pythium aff. monospermum | 3 | 1.956 | 0.000 | 10.579 | 0.949 | 2.768 | 0.774 | 14.025 | 2.828 | 0.263 | <0.001 | 0.794 | <0.001 | 0.073 | grapevine | |||
| Pythium arrhenomanes | 8 | 1.116 | 0.000 | 0.000 | 0.872 | 1.257 | 3.545 | 5.713 | 2.795 | 0.014 | 0.782 | 0.223 | 0.492 | 0.409 | Blight, root rot | corn, rice, barley, wheat | ||
| Pythium monospermum | 12 | 2.109 | 0.872 | 1.683 | 1.187 | 1.274 | 1.928 | 3.108 | 2.005 | 0.367 | 0.857 | 0.725 | 0.476 | 0.962 | Downy mildew. Root necrosis, not known as a strong pathogen | cherry, juniper, spinach | ||
| Pythium oligandrum | 1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.200 | 0.338 | 0.763 | 0.772 | N/A | 0.762 | Damping-off; root, stem, and fruit rots | soybean, wheat | ||
| Pythium oopapillum | 2 | 0.000 | 0.158 | 0.000 | 1.536 | 0.000 | 0.000 | 0.000 | 0.404 | 0.35 | 0.48 | 0.931 | 0.678 | 0.761 | Root rot | soybean | ||
| Pythium torulosum | 1 | 0.000 | 0.000 | 0.000 | 1.326 | 0.000 | 0.000 | 0.000 | 0.000 | 0.144 | 0.503 | 0.477 | 0.469 | N/A | damping-off, root rot | pea, soybean, corn | ||
| Pythium volutum | 6 | 0.000 | 0.000 | 6.287 | 4.399 | 0.000 | 0.002 | 1.895 | 0.053 | 0.020 | 0.004 | 0.143 | 0.034 | 0.014 | Root rot | melon, morning glory, wheat, barley, turfgrass | ||
| Saprolegnia anisospora | 1 | 0.000 | 0.000 | 1.997 | 0.000 | 0.000 | 0.238 | 0.000 | 0.000 | 0.704 | 0.024 | 0.01 | 0.006 | 0.379 | ||||
| Saprolegnia torulosa | 1 | 0.000 | 0.175 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 1.093 | 0.142 | 0.384 | 0.181 | 0.306 | 0.265 | ||||
| Wilsoniana amaranthi | 1 | 0.000 | 0.000 | 0.000 | 0.638 | 1.252 | 0.743 | 0.000 | 0.000 | 0.646 | 0.876 | 0.369 | 0.782 | 0.515 | White blister rust | amaranth | ||
| Wilsoniana portulacae | 6 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 5.157 | 0.000 | 1.132 | 0.031 | 0.19 | 0.211 | N/A | N/A | White blister rust | portulacaceae | ||
| Corn yield | Soybean yield | Wheat yield | |
| Tillage | |||
| CT | 9177b1 | 2257 | 1854 |
| NT | 10806a | 2652 | 1795 |
| Rotation | |||
| Monoculture | 9735 | 2404 | 1298b |
| Rotation | 10248 | 2446 | 2305a |
| Analysis of variance (P-values) | |||
| Tillage (T) | 0.018 | 0.226 | 0.548 |
| Rotation (R) | 0.442 | 0.577 | <0.001 |
| T×R | 0.163 | 0.343 | 0.652 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





