Submitted:
28 April 2023
Posted:
29 April 2023
You are already at the latest version
Abstract

Keywords:
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Truncation of the original aptamer
2.2.2. Secondary structures of curtailed oligonucleotides
2.2.3. Energetics and stability of curtailed oligonucleotides
2.2.4. Biolayer interferometry studies on binding affinity
2.2.5. Aptasensing of CAP
2.2.5.1. Performance of the assay
2.2.5.2. Validation in real samples
2.2.5.3. Reproducibility and specificity
3. Results and Discussion
3.1. Truncation of the original aptamer
3.2. Determination of secondary structures of curtailed oligonucleotides
3.3. Stability of curtailed oligonucleotides
3.4. Binding affinity with analyte
3.5. Aptasensing of CAP
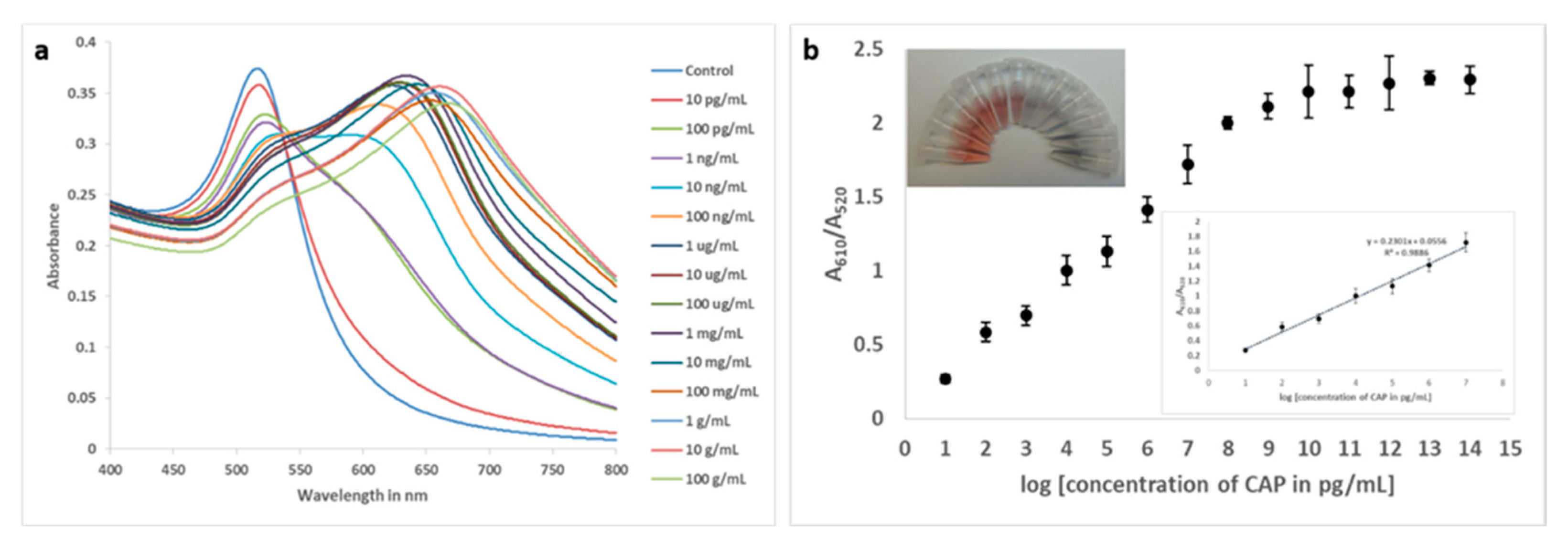
3.5.1. Performance of the assay
3.5.2. Validation in real samples
3.5.3. Specificity and reproducibility
3.5.4. Comparison with reported literature
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li J., Shao B., Shen T., Wang S. and Wu Y. Occurrence of Chloramphenicol-Resistance Genes as Environmental Pollutants from Swine Feedlots. Environ. Sci. Technol. 2013, 47(6), 2892–2897. [CrossRef] [PubMed]
- Extralabel Drug Use in Animals, Fed. Regist. 1994, 61, 57732–57745.
- Durham, M. A bitter taste of honey. The Guardian (International Edition). 21 Jul 2004. Accessed on 16th July 2018.
- Moudgil P., Bedi J. S., Aulakh R. S., Gill J. P. S., Kumar A. Validation of HPLC multi-residue method for determination of fluoroquinolones, tetracycline, sulphonamides and chloramphenicol residues in bovine milk. Food Anal. Methods. 2019, 12, 338–346. [CrossRef]
- Sharma, R. , Raghavarao KSMS. Nanoparticle-based aptasensors for food contaminant detection, in: Nanomaterials for Food Applications, A.L. Rubio et al. (eds.), Elsevier Inc. 2019; 123–145. [Google Scholar]
- Gao H., Gan N., Pan D., Chen Y., Li T., Cao Y., Fu T. A sensitive colorimetric aptasensor for chloramphenicol detection in fish and pork based on the amplification of a nano-peroxidase polymer. Anal. Methods 2015a, 7, 6528–6536. [CrossRef]
- Gao H., Pan D., Gan N., Cao J., Sun Y., Wu Z., Zeng X. An aptamer-based colorimetric assay for chloramphenicol using a polymeric HRP-antibody conjugate for signal amplification. Microchim. Acta 2015a, 182, 2551–2559. [CrossRef]
- Miao, Y. Miao Y., Gan N., Li T., Zhang H., Cao Y., Jiang Q. A colorimetric aptasensor for chloramphenicol in fish based on double-stranded DNA antibody labeled enzyme-linked polymer nanotracers for signal amplification. Sens. Actuators B Chem. 2015a, 220, 679–687. [Google Scholar] [CrossRef]
- Miao Y., Gan N., Ren H-X., Cao Y., Hu F., Yan Z., Chen Y. A triple-amplification colorimetric assay for antibiotics based on magnetic aptamer–enzyme co-immobilized platinum nanoprobes and exonuclease-assisted target recycling. Analyst. 2015b, 140, 7663–7671. [CrossRef]
- Abnous K., Danesh N. M., Ramezani M., Emrani A. S., Taghdisi S. M. A novel colorimetric sandwich aptasensor based on an indirect competitive enzyme-free method for ultrasensitive detection of chloramphenicol. Biosens. Bioelectron. 2016, 78, 80–86. [CrossRef]
- Chang CC., Wang G., Takarada T., Maeda M. Iodine-mediated etching of triangular gold nanoplates for colorimetric sensing of copper ion and aptasensing of chloramphenicol. ACS Appl. Mater. Interfaces. 2017, 9, 34518–34525. [CrossRef]
- Luan Q., Xi Y., Gan N., Cao Y., Li T., Chen Y. A facile colorimetric aptamer assay for small molecule detection infood based on a magnetic single-stranded DNA binding protein-linked composite probe. Sens. Actuators B Chem. 2017, 239, 979–987. [CrossRef]
- Javidi M., Housaindokht M. R., Verdian A., Razavizadeh B. M. Detection of chloramphenicol using a novel apta-sensing platform based on aptamer terminal-lock in milk samples. Anal. Chim. Acta 2018, 1039, 116–123. [CrossRef] [PubMed]
- Xie Y., Huang Y., Tang D., Cui H., Cao H.A. competitive colorimetric chloramphenicol assay based on the non-cross-linking deaggregation of gold nanoparticles coated with a polyadenine-modified aptamer. Mikrochim Acta 185(12), 534. [CrossRef] [PubMed]
- Yan C., Zhang J., Xue F., Lu J., Li B., Chen W. Aptamer-mediated colorimetric method for rapid and sensitive detection of chloramphenicol in food. Food Chem. 2018, 260, 208–212. [CrossRef] [PubMed]
- Huang W., Zhang H., Lai G., Liu S., Li B., Yu A. Sensitive and rapid aptasensing of chloramphenicol by colorimetric signal transduction with a DNAzyme-functionalized gold nanoprobe. Food Chem. 2019, 270, 287–292. [CrossRef]
- Li J., Yu C., Wu Y-n., Zhu Y., Xu J., Wang Y., Wang H., Guo M., Li F. Novel sensing platform based on gold nanoparticle-aptamer and Fe-metalorganic framework for multiple antibiotic detection and signal amplification. Environ. Int. 2019, 125, 135–141. [CrossRef]
- Wu Y-y., Liu, B-w., Huang, P., Wu, F-y. A novel colorimetric aptasensor for detection of chloramphenicol based on lanthanum ion–assisted gold nanoparticle aggregation and smartphone imaging. Anal Bioanal Chem. 2019, 411(28), 7511–7518. [CrossRef]
- Sharma, R., Ragavan K.V., Raghavarao K.S.M.S., Thakur M.S. Nano-aptamer based quantitative detection of chloramphenicol in: Biotechnology and Biochemical Engineering, Select Proceedings of ICACE 2015, B.D. Prasanna et al. (eds.), Springer. 2016; 187–195.
- McKeague M., DeRosa M. C. Challenges and opportunities for small molecule aptamer development. J. Nucleic Acids 2012, 748913, 20 pages.
- Duan, Y., Gao, Z., Wang, L., Wang, H., Zhang, H., and Li, H. Selection and identification of chloramphenicol-specific DNA aptamers by Mag-SELEX. Appl. Biochem. Biotechnol. 2016, 180(8), 1644–1656. [CrossRef]
- Mehta, J., Van Dorst, B., Rouah-Martin, E., Herrebout, W., Scippo, M. L., Blust, R., Robbens, J. In vitro selection and characterization of DNA aptamers recognizing chloramphenicol. J. Biotechnol. 2011, 155(4), 361–369. [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31(13), 3406–3415. [Google Scholar] [CrossRef]
- Searle, M. S., Williams, D. H.. On the stability of nucleic acid structures in solution: enthalpy-entropy compensations, internal rotations and reversibility. Nucleic Acids Res. 1993, 21(9), 2051–2056. [CrossRef] [PubMed]
- Owczarzy, R., You, Y., Groth, C. L., and Tataurov, A. V. Stability and mismatch discrimination of locked nucleic acid–DNA duplexes. Biochemistry 2011, 50(43), 9352–9367. [CrossRef] [PubMed]
- Yang Y., Yin Y., Li X., Wang S., Dong Y. Development of a chimeric aptamer and an AuNPs aptasensor for highly sensitive and specific identification of Aflatoxin B1. Sens. Actuators B Chem. 2020, 319, 128250. [CrossRef]
- Alawad A., Istamboulié G., Calas-Blanchard C., Noguer T. A reagentless aptasensor based on intrinsic aptamer redox activity for the detection of tetracycline in water. Sens. Actuators B Chem. 2019, 288, 141–146. [CrossRef]
- Macdonald, J. Houghton, P. Xiang, D. Duan, W., Shigdar, S. Truncation and mutation of a transferrin receptor aptamer enhances binding affinity. Nucleic Acid Therapeutics. 2016, 26, 6, 348–354.
- Sun, Y., Duan, N., Ma, P., Liang, Y., Zhu, X., Wang, Z. Colorimetric aptasensor based on truncated aptamer and trivalent DNAzyme for Vibrio parahemolyticus determination. J. Agric. Food Chem. 2019, 67, 8, 2313–2320.
- Sharma, R., Akshath, U.A.S., Bhatt, P., Raghavarao, KSMS. Fluorescent aptaswitch for chloramphenicol detection -quantification enabled by immobilization of aptamer. Sens. Actuators B Chem. 2019, 290, 110–117. [CrossRef]
- Abnous K., Danesh N.M., Ramezani M., Taghdisi S.M., Emrani A.S. A novel electrochemical aptasensor based on H-shape structure of aptamer-complimentary strands conjugate for ultrasensitive detection of cocaine. Sens. Actuators B Chem. 2016, 224, 351–355. [CrossRef]
- Auer S., Koho T., Uusi-Kerttula H., Vesikari T., Blazevic V., Hytönen V.P. Rapid and sensitive detection of norovirus antibodies in human serum with a biolayer interferometry biosensor. Sens. Actuators B Chem. 2015, 221, 507–514. [CrossRef]
- Gandhi, I., Narendran, K., Jackson, G.W. Rapid DNA Aptamer Binding Characterization and ELASA Development Using Biolayer Interferometry (BLI). https://www.basepairbio.com. 2011, Accessed on: 17th January 2017.
- Tao, X., He, F., Liu, X., Zhang, F., Wang, X, Peng, Y., Liu, J. Detection of chloramphenicol with an aptamer-based colorimetric assay: critical evaluation of specific and unspecific binding of analyte molecules. Microchim Acta 2020, 187, 668. [CrossRef]
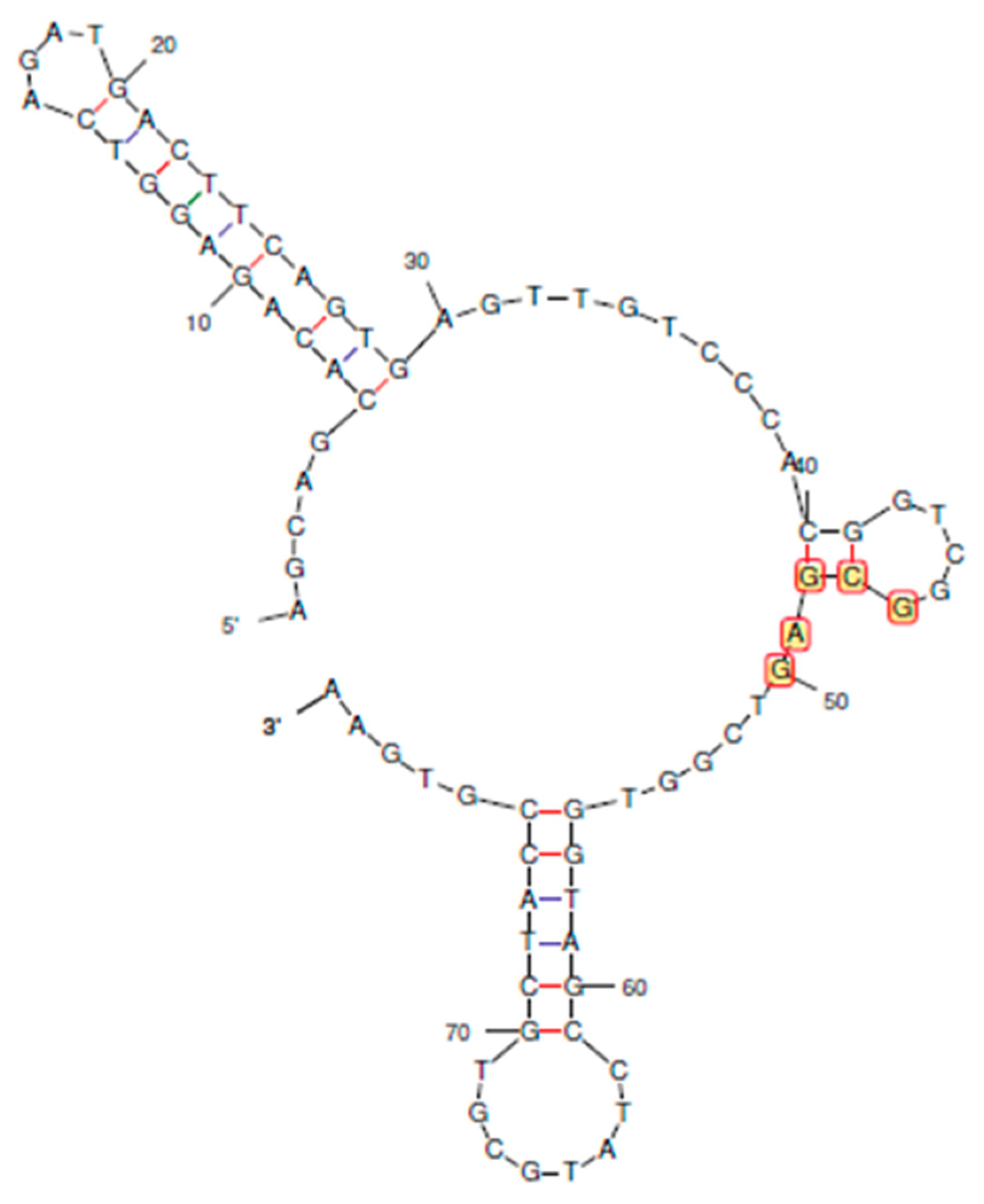
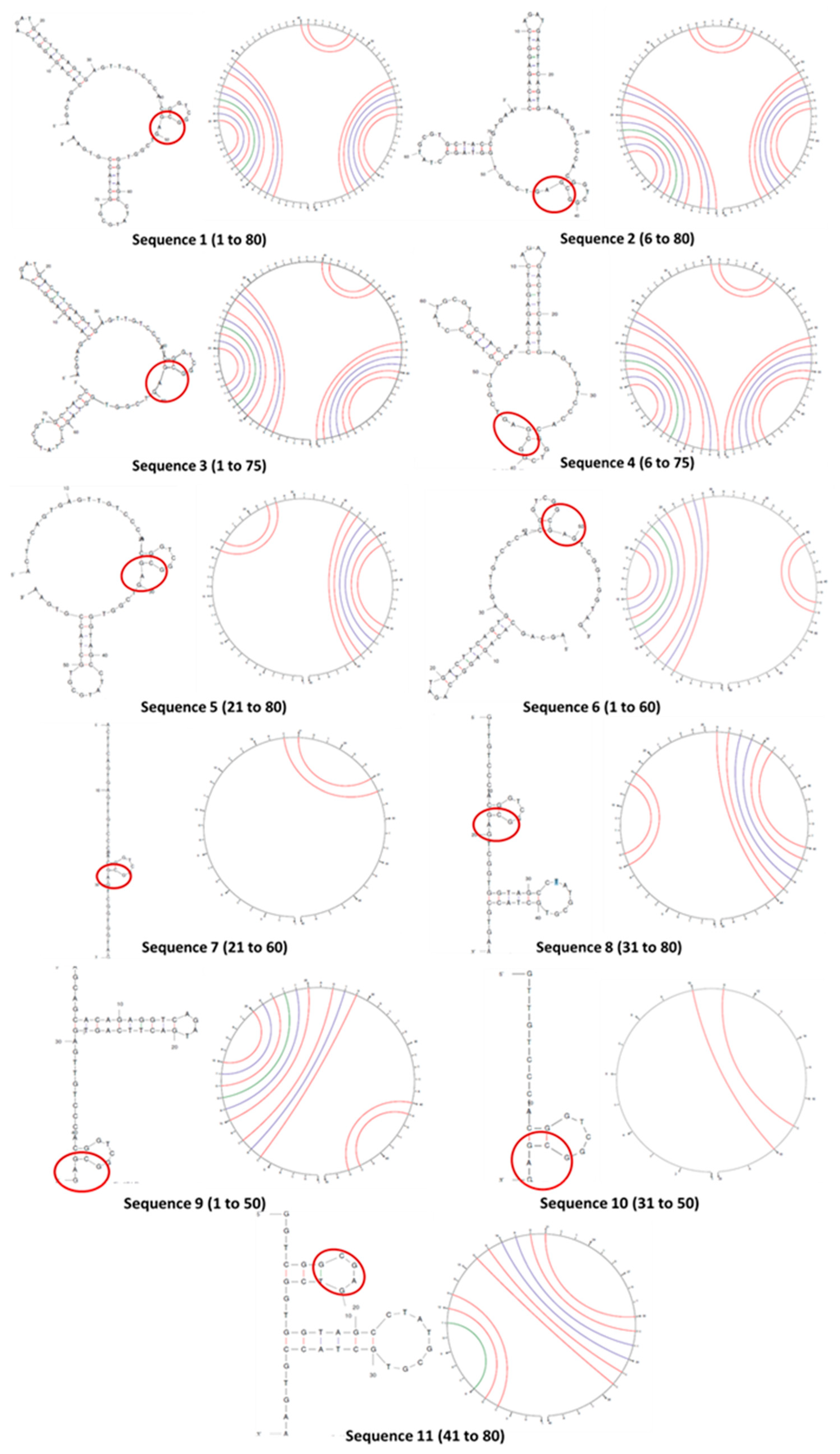
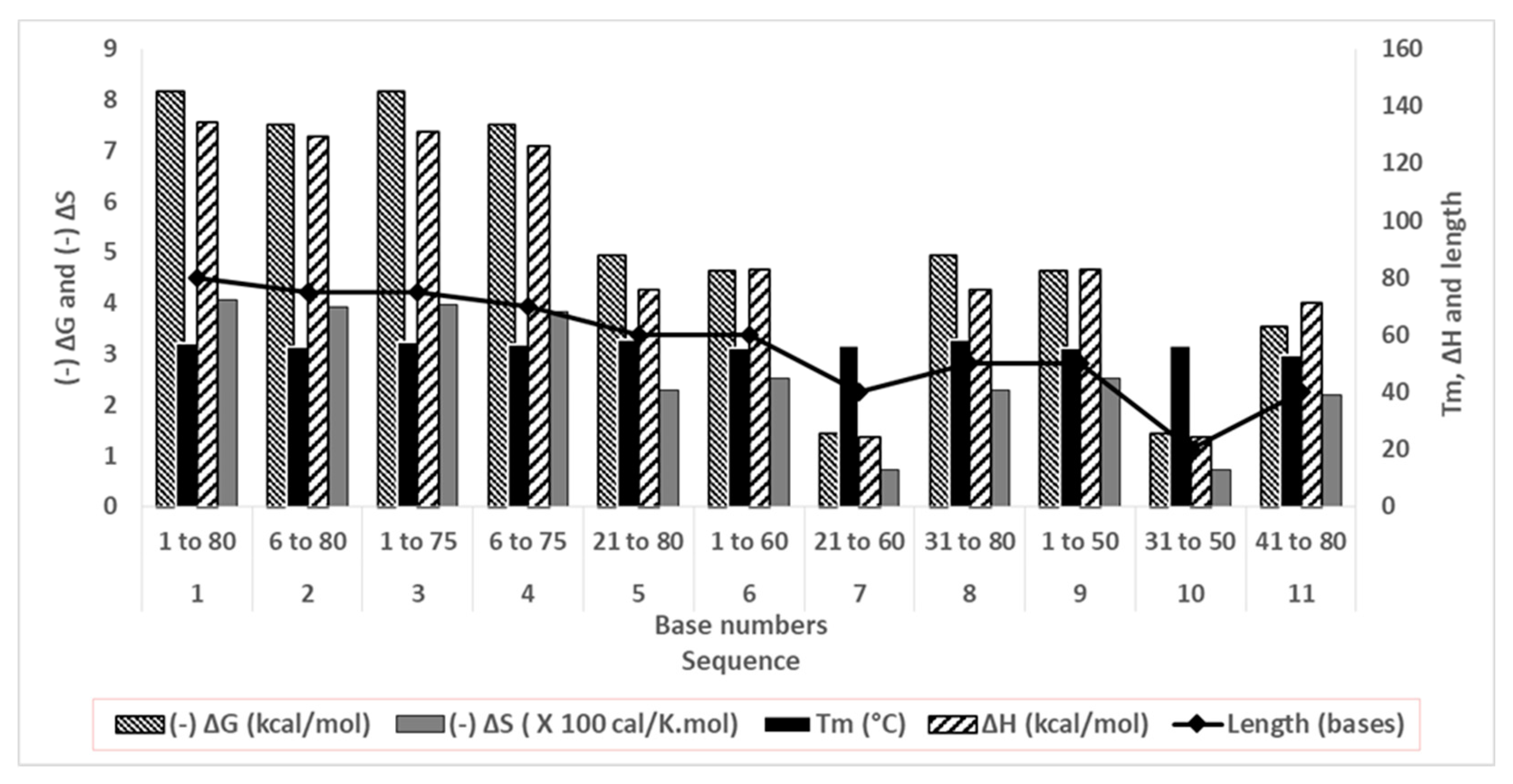
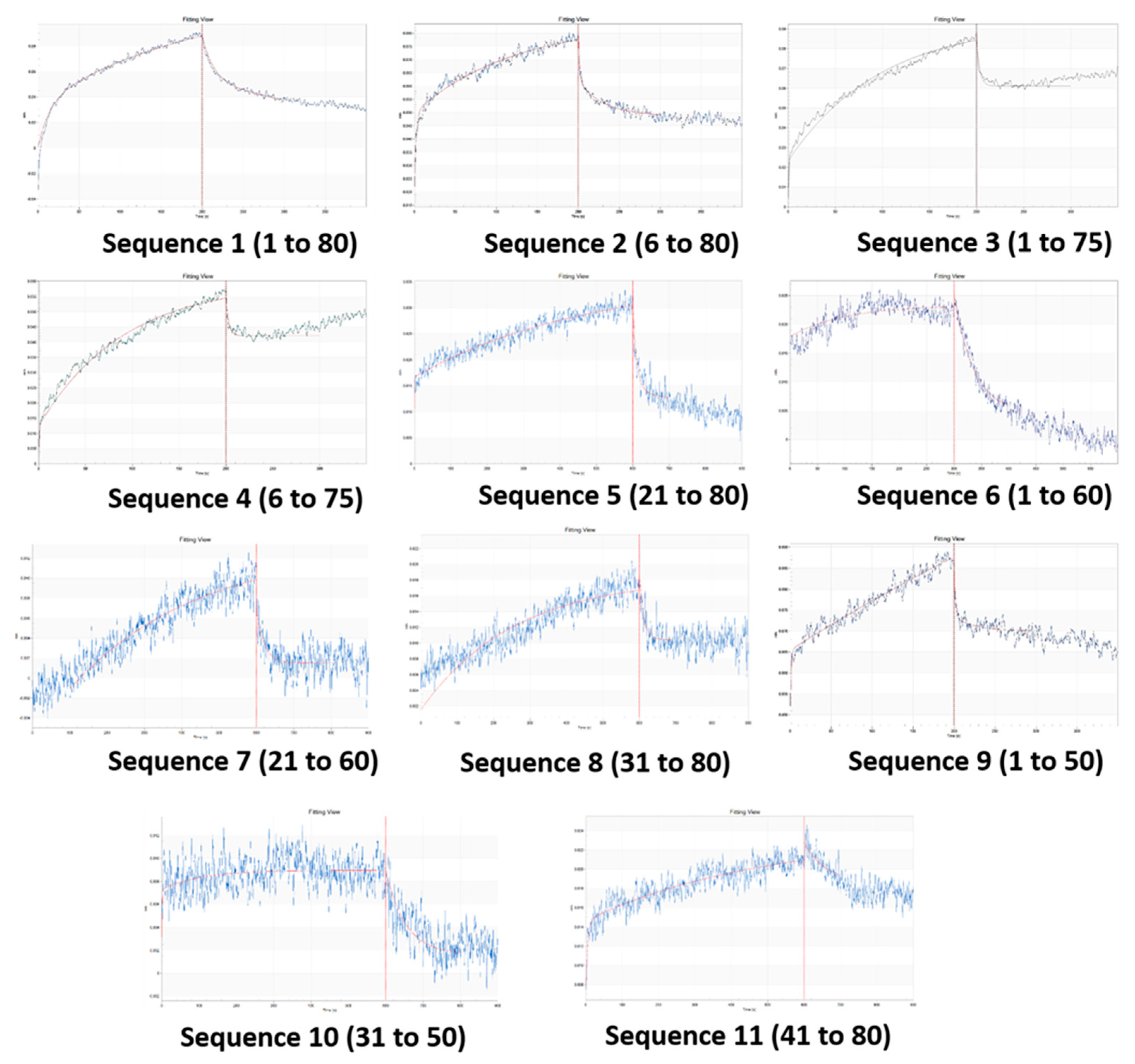

| Sl. no | Sequence (5’ → 3’) | No. of bases removed | Terminal/ Seq base numbers |
| 1 | AGCAGCACAGAGGTCAGATGACTTCAGTGAGTTGTCCCACGGTCGGCGAGTCGGTGGTAGCCTATGCGTGCTACCGTGAA (original) | 0 | - 1 to 80 |
| 2 | CACAGAGGTCAGATGACTTCAGTGAGTTGTCCCACGGTCGGCGAGTCGGTGGTAGCCTATGCGTGCTACCGTGAA | 5 | 5’ 6 to 80 |
| 3 | AGCAGCACAGAGGTCAGATGACTTCAGTGAGTTGTCCCACGGTCGGCGAGTCGGTGGTAGCCTATGCGTGCTACC | 5 | 3’ 1 to 75 |
| 4 | CACAGAGGTCAGATGACTTCAGTGAGTTGTCCCACGGTCGGCGAGTCGGTGGTAGCCTATGCGTGCTACC | 10 | 5’& 3’ 6 to 75 |
| 5 | ACTTCAGTGAGTTGTCCCACGGTCGGCGAGTCGGTGGTAGCCTATGCGTGCTACCGTGAA | 20 | 5’ 21 to 80 |
| 6 | AGCAGCACAGAGGTCAGATGACTTCAGTGAGTTGTCCCACGGTCGGCGAGTCGGTGGTAG | 20 | 3’ 1 to 60 |
| 7 | ACTTCAGTGAGTTGTCCCACGGTCGGCGAGTCGGTGGTAG | 40 | 5’ & 3’ 21 to 60 |
| 8 | GTTGTCCCACGGTCGGCGAGTCGGTGGTAGCCTATGCGTGCTACCGTGAA | 30 | 5’ 31 to 80 |
| 9 | AGCAGCACAGAGGTCAGATGACTTCAGTGAGTTGTCCCACGGTCGGCGAG | 30 | 3’ 1 to 50 |
| 10 | GTTGTCCCACGGTCGGCGAG | 60 | 5’ & 3’ 31 to 50 |
| 11 | GGTCGGCGAGTCGGTGGTAGCCTATGCGTGCTACCGTGAA | 40 | 5’ 41 to 80 |
| Sl.no. |
Sequence number |
Kd (µM) | R2 |
|---|---|---|---|
| 1 | 1 | 0.704 (previously reported value in literature is 0.766) |
0.993 |
| 2 | 2 | 0.877 | 0.973 |
| 3 | 3 | 0.645 | 0.946 |
| 4 | 4 | 0.661 | 0.945 |
| 5 | 5 | 1.712 | 0.924 |
| 6 | 6 | 4.014 | 0.905 |
| 7 | 7 | 0.370 | 0.882 |
| 8 | 8 | 0.153 | 0.853 |
| 9 | 9 | 0.092 | 0.914 |
| 10 | 10 | 3.109 | 0.699 |
| 11 | 11 | 0.426 | 0.757 |
| Sl.no. | Spiked amount (pg mL-1) | Aptasensing (pg mL-1) | HPLC (pg mL-1) | Recovery (%) ± SD (with respect to spiked amount) |
|---|---|---|---|---|
| 1 | 0 | Not detected | Not detected | - |
| 2 | 10 | 9.66 | Not detected | 96.6 ± 2.7 |
| 3 | 100 | 94.77 | Not detected | 94.7 ± 3.8 |
| 4 | 1000 | 1004.28 | Not detected | 100.4 ± 6.9 |
| 5 | 10000 | 9812.50 | 9766.66 | 98.12 ± 1.1 |
| 6 | 50000 | 49605.71 | 48942.94 | 97.88 ± 8.6 |
| Sl. no. | Detection principle | Significant features | Limit of detection (pg mL -1) Real sample |
Reference |
|---|---|---|---|---|
| 1 | Competitive Signalling molecule: TMB chromogen (enzymatic) |
Capture probe attached to core-shell Fe@Au nanoparticles CAP competes with HRP labelled probe Requirement of functionalized core shell iron nanoparticles, gold nanoparticles, cDNA, HRP, substrates Multistep detection with magnetic separation |
20 pg mL-1 (buffer) Fish Pork |
9 |
| 2 | Competitive Signalling molecule: TMB chromogen (enzymatic) |
cDNA (complementary to aptamer) attached to Fe@Au nanoparticles. In presence of CAP, HRP labelled probe (with antibody) attaches to cDNA. Requirement of functionalized core-shell iron nanoparticles, cDNA, HRP, antibodies, substrates Multistep detection with magnetic separation |
3 pg mL-1 (buffer) Fish Duck |
10 |
| 3 | Competitive Signalling molecule: TMB chromogen (enzymatic) |
Aptamer attached to Fe@Au nanoparticles In presence of CAP, HRP labelled probe (with antibody) detaches from aptamer Requirement of functionalized core-shell iron nanoparticles, cDNA, HRP, antibodies, substrates Multistep detection with magnetic separation |
15 pg mL-1 Fish |
11 |
| 4 | Competitive Signalling molecule: TMB chromogen (enzymatic) |
Aptamer-cDNA-Pt-HRP conjugate attached to antibodies on Fe@Au nanoparticles In presence of CAP, cDNA-Pt-HRP HRP probe detaches from aptamer Exonuclease I used to cleave single strands releasing CAP (for recycling) and Pt-HRP (for signalling) Requirement of functionalized core-shell iron nanoparticles, Pt nanoparticles, cDNA, HRP and Exo I, antibodies, substrates Multistep detection with magnetic separation Recycling of target increases signal |
0.30 pg mL-1 Milk | 12 |
| 5 | Competitive Signalling molecule: Gold nanoparticles |
Free biotinylated aptamer binds to BSA on solid support, streptavidin-modified-DNA-nanoparticle conjugate attaches giving red colour. Colour fades when aptamer engaged by CAP Requirement of solid support, modification of aptamer (thiol, biotin), DNA to bind nanoparticles |
145.67 pg mL-1 (buffer) 72.352 pg mL-1 (milk) 194.123 pg mL-1 (rat serum) |
13 |
| 6 | Protection of nanoparticles by aptamers Signalling molecule: Gold nanoparticles |
Aptamers protected triangular nanoparticles are not etched to spherical particles Cu2+-assisted I−mediated method Simple one-step method with minimum components |
1.62 x 106 pg mL-1 (buffer) |
14 |
| 7 | Competitive Signalling molecule: TMB chromogen (enzymatic) |
Single stranded binding protein attached to Fe@Au nanoparticles Aptamer attached to SiO2@Au-HRP probe In presence of CAP, probe detached from magnetic nanoparticle complex Requirement of functionalized core-shell iron nanoparticles, functionalized core shell silica nanoparticles, SSB protein, HRP, substrates Multistep detection with magnetic separation |
20 pg mL-1 Milk |
15 |
| 8 | Protection of nanoparticles by ssDNA Signalling molecule: Gold nanoparticles |
Aptamer locked by short ssDNA in absence of CAP, leaving gold nanoparticles free for salt-induced aggregation Simple, one-step method, providing solution for long length of CAP aptamer |
9.69 pg mL-1 (buffer) Milk |
16 |
| 9 | Competitive between CAP-base and CAP Signalling molecule: Gold nanoparticles |
Negatively charged aptamer-functionalized gold nanoparticles aggregate in presence of positive CAP-base Presence of CAP leads to de-aggregation Simple, one-step detection |
7.11 x 103 pg mL-1 (buffer) Spiked environmental water |
17 |
| 10 | Competitive Signalling molecule: TMB chromogen (enzymatic) |
cDNA capture probe bound to microplate Aptamer tagged with HRP Requirement of binding to solid support by streptavidin, HRP, substrate |
3.10 pg mL-1 Honey Fish |
18 |
| 11 | Competitive Signalling molecule: TMB chromogen (enzymatic) |
Magnetic bead functionalized with aptamer cDNA-gold nanoparticle-hemin/G-quadruplex DNAzyme catalysed TMB conversion Requirement of complex functionalization of gold nanoparticles, cDNA, hemin, DNAzyme, substrates Multistep detection with magnetic separation |
0.13 pg mL-1 (buffer) Milk |
19 |
| 12 | Competitive Signalling molecule: TMB chromogen (enzymatic) |
Fe-based metal organic framework catalyses TMB conversion. Catalysis reduced if gold nanoparticle-aptamer-CAP complex binds to it Simple, easy transduction |
8.1 x 103 pg mL-1 (buffer) Spiked tap water |
20 |
| 13 | Protection of nanoparticles by ssDNA Signalling molecule: Gold nanoparticles |
Lanthanide attaches to aptamer functionalized gold nanoparticles and assists aggregation In presence of CAP, aptamer detaches from nanoparticles Simple, one-step detection Can be detected through instrument and smartphone imaging app |
2.471 x 103 pg mL-1 (spectro-photometer) 1.899 x 103 pg mL-1 (smartphone app) Solid milk Chicken |
21 |
| 14 | Protection of nanoparticles by aptamers Signalling molecule: Gold nanoparticles |
Truncated aptamers bind to gold nanospheres in absence of CAP Unbound nanospheres aggregate in presence of salt, changing colour of colloid |
1.67 pg mL-1 (buffer) Honey |
Present work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




