Submitted:
26 April 2023
Posted:
27 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
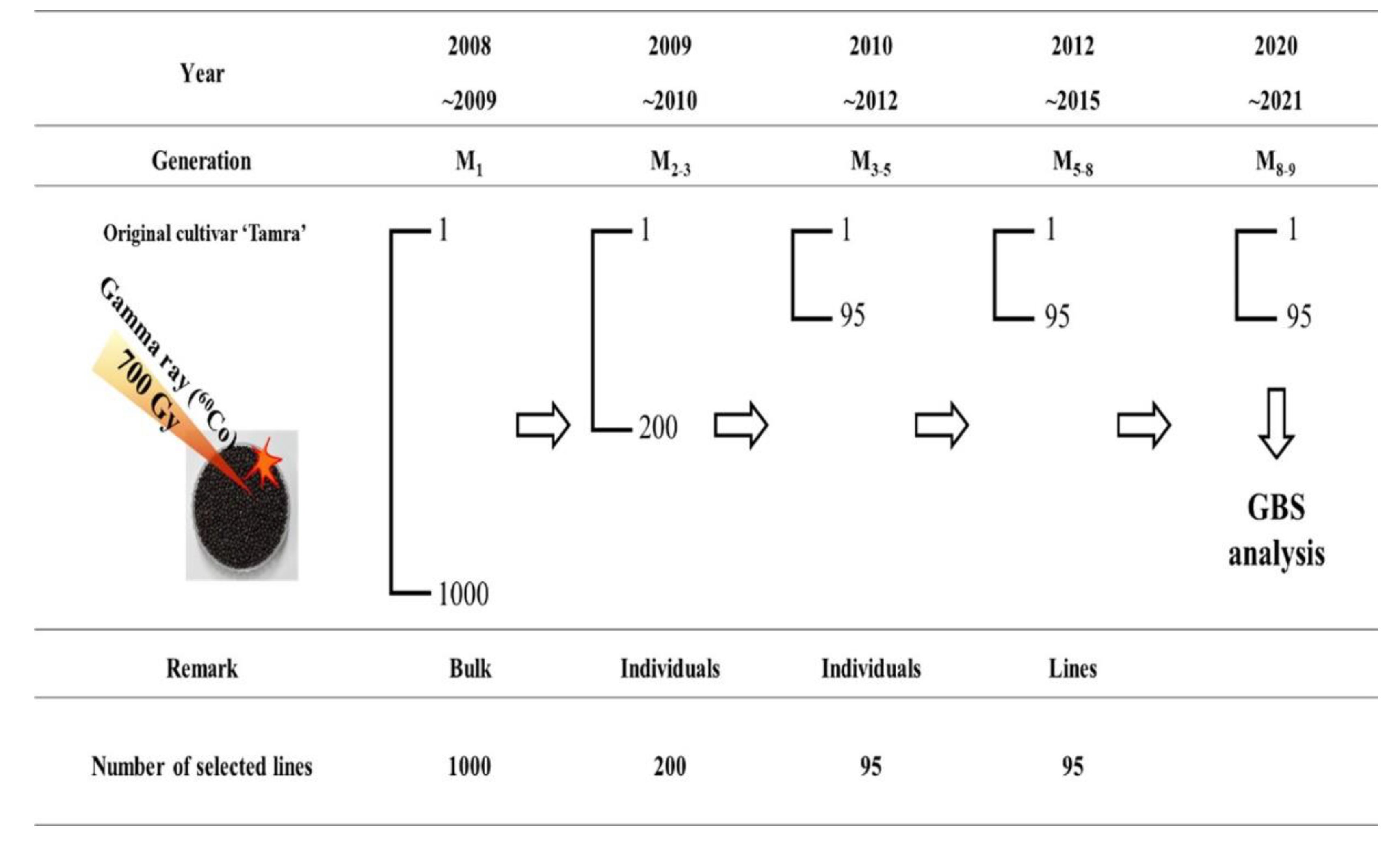
2.1. Plant Material
2.2. Determination of Fatty Acid Compositions and Crude Fat Contents
2.3. DNA Extraction
2.4. Library Construction and Genotyping-by-Sequencing (GBS)
2.5. Sequence Preprocessing and Alignment to Reference Genome Sequence
2.6. Raw SNP Detection and Consensus Sequence Extraction
2.7. Generate SNP Matrix
2.8. Gene Ontology (GO) Analysis of Genes with Polymorphic SNPs
2.9. GWAS with Agronomic Characteristics, Fatty Acid and Crude Fat
3. Results
3.1. Agronomic Characteristics
3.2. Fatty Acid Compositions and Crude Fat Contents
3.3. GBS Analysis of Rapeseed Mutant Lines
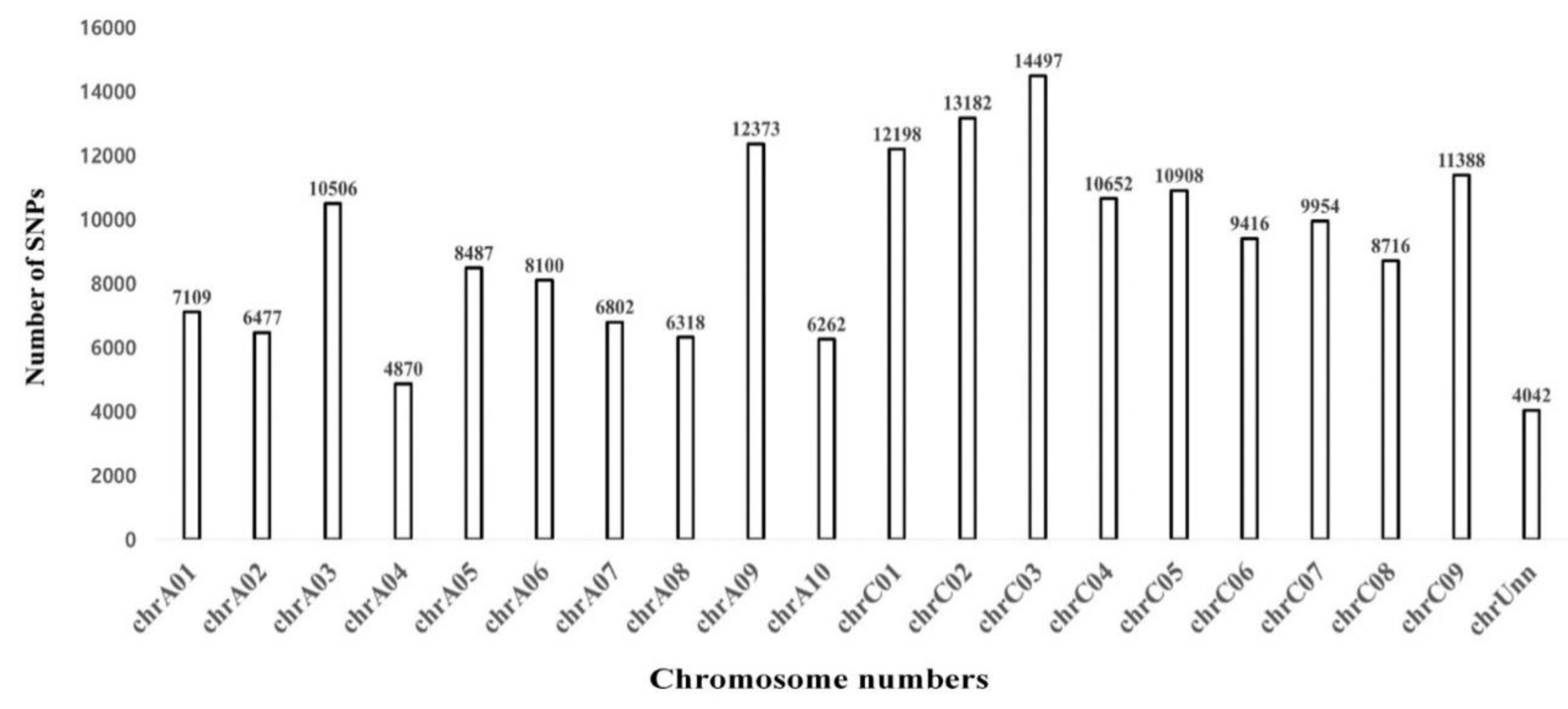
3.4. Identification of SNPs
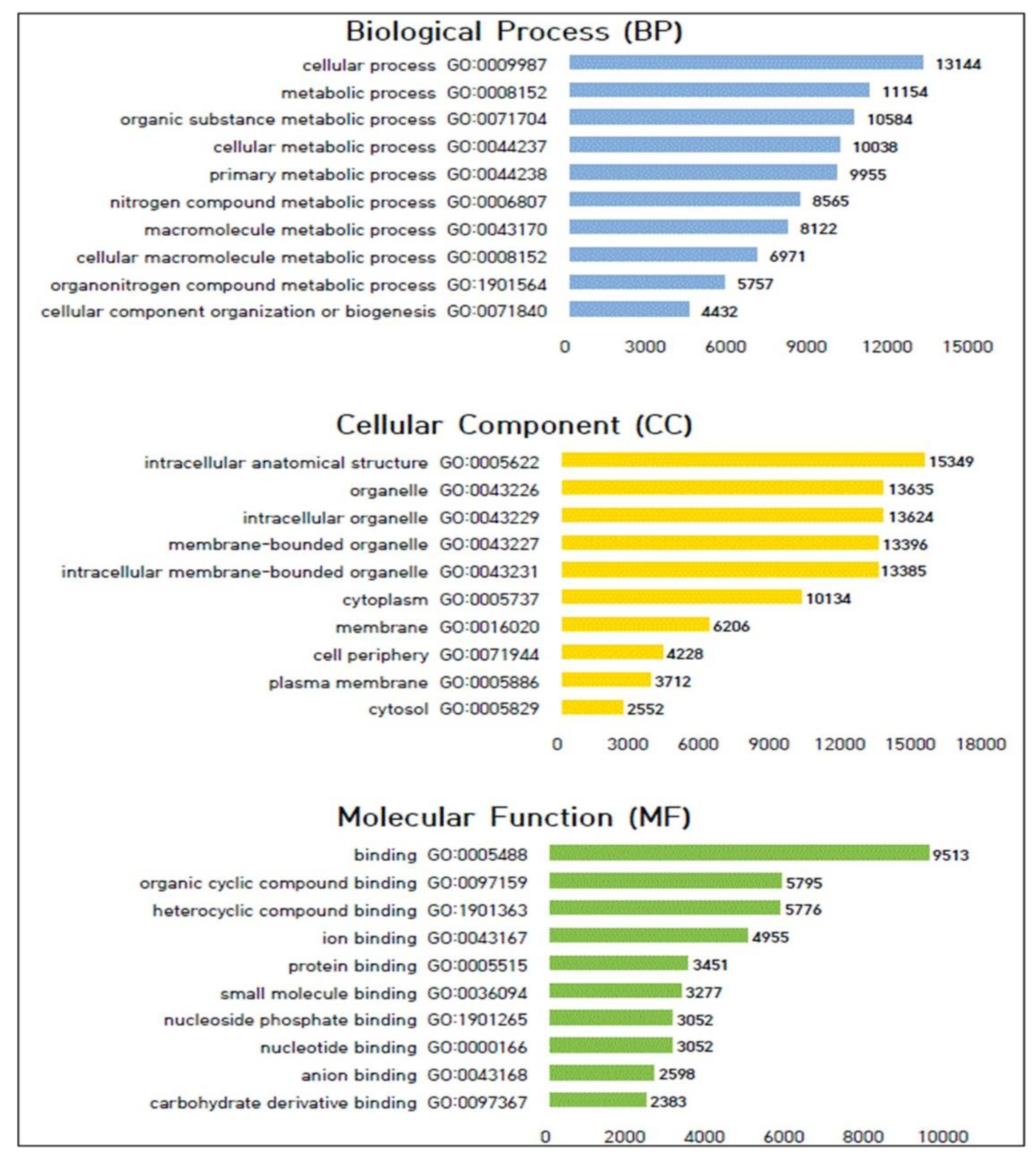
3.5. Gene Ontology (GO) Analysis of Genes with Polymorphic SNPs
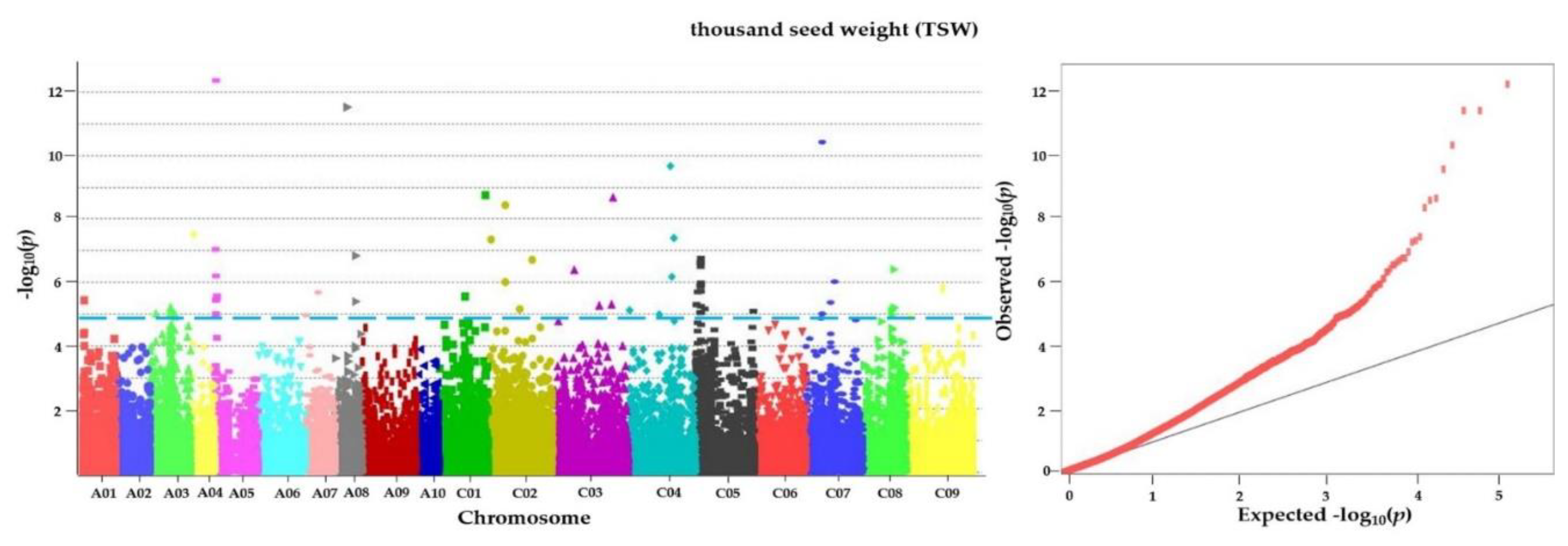
3.6. GWAS for Agronomic Characteristics
3.7. GWAS for Fatty Acid and Crude Fat
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gilchrist, E.J.; Sidebottom, C.H.D.; Koh, C.S.; MacInnes, T.; Sharpe, A.G.; Haughn, G.W. A Mutant Brassica napus (Canola) Population for the Identification of New Genetic Diversity via TILLING and Next Generation Sequencing. PLOS ONE 2013, 8, e84303, . [CrossRef]
- Malek, M.A.; Ismail, M.R.; Rafii, M.Y.; Rahman, M. SyntheticBrassica napusL.: Development and Studies on Morphological Characters, Yield Attributes, and Yield. 2012, 2012, 416901. [CrossRef]
- Zhu, Q.; King, G.J.; Liu, X.; Shan, N.; Borpatragohain, P.; Baten, A.; Wang, P.; Luo, S.; Zhou, Q. Identification of SNP loci and candidate genes related to four important fatty acid composition in Brassica napus using genome wide association study. PLOS ONE 2019, 14, e0221578, . [CrossRef]
- Ryu, J.; Lyu, J.I.; Kim, D.-G.; Koo, K.M.; Yang, B.; Jo, Y.D.; Kim, S.H.; Kwon, S.-J.; Ha, B.-K.; Kang, S.-Y.; et al. Single Nucleotide Polymorphism (SNP) Discovery and Association Study of Flowering Times, Crude Fat and Fatty Acid Composition in Rapeseed (Brassica napus L.) Mutant Lines Using Genotyping-by-Sequencing (GBS). Agronomy 2021, 11, 508, . [CrossRef]
- Geng, X.; Dong, N.; Wang, Y.; Li, G.; Wang, L.; Guo, X.; Li, J.; Wen, Z.; Wei, W. RNA-seq transcriptome analysis of the immature seeds of two Brassica napus lines with extremely different thousand-seed weight to identify the candidate genes related to seed weight. PLOS ONE 2018, 13, e0191297, . [CrossRef]
- Khan, S.U.; Yangmiao, J.; Liu, S.; Zhang, K.; Khan, M.H.U.; Zhai, Y.; Olalekan, A.; Fan, C.; Zhou, Y. Genome-wide association studies in the genetic dissection of ovule number, seed number, and seed weight in Brassica napus L.. Ind. Crop. Prod. 2019, 142, 111877, . [CrossRef]
- Li, N.; Peng, W.; Shi, J.; Wang, X.; Liu, G.; Wang, H. The Natural Variation of Seed Weight Is Mainly Controlled by Maternal Genotype in Rapeseed (Brassica napus L.). PLOS ONE 2015, 10, e0125360. [CrossRef]
- Long, W.; Hu, M.; Gao, J.; Chen, S.; Zhang, J.; Cheng, L.; Pu, H. Identification and Functional Analysis of Two New Mutant BnFAD2 Alleles That Confer Elevated Oleic Acid Content in Rapeseed. Front. Genet. 2018, 9, 399, . [CrossRef]
- Zhang, S.; Liao, X.; Zhang, C.; Xu, H. Influences of plant density on the seed yield and oil content of winter oilseed rape (Brassica napus L.). Ind. Crop. Prod. 2012, 40, 27–32, . [CrossRef]
- Wang, P.; Xiong, X.; Zhang, X.; Wu, G.; Liu, F. A Review of Erucic Acid Production in Brassicaceae Oilseeds: Progress and Prospects for the Genetic Engineering of High and Low-Erucic Acid Rapeseeds (Brassica napus). Front. Plant Sci. 2022, 13, 899076, . [CrossRef]
- Nath, U. K.; Kim, H.-T.; Khatun, K.; Park, J.-I.; Kang, K.-K.; Nou, I.-S. Modification of fatty acid profiles of rapeseed (Brassica napus L.) oil for using as food, industrial feed-stock and biodiesel. Plant Breed. Biotech. 2016, 4(2), 123-134.
- Sharafi, Y.; Majidi, M.M.; Goli, S.A.H.; Rashidi, F. Oil Content and Fatty Acids Composition inBrassicaSpecies. Int. J. Food Prop. 2015, 18, 2145–2154, . [CrossRef]
- Kim, S.H.; Ryu, J.; Kim, W.J.; Kang, R.; Seo, E.; Kim, G.; Kang, S.-Y.; Lee, J.-D.; Ha, B.-K. Identification of a new GmSACPD-C allele in high stearic acid mutant Hfa180 derived from gamma-ray irradiation. Mol. Breed. 2019, 39(2), 17.
- Anwar, M.M.; Ali, S.E.; Nasr, E.H. Improving the nutritional value of canola seed by gamma irradiation. J. Radiat. Res. Appl. Sci. 2015, 8, 328–333, . [CrossRef]
- Larik, A.S.; S. Memon.; Z.A. Soomro. Radiation induced polygenic mutations in Sorghum bicolor L. J. Agric. Res. 2009, 47, 11-19.
- He, J.; Zhao, X.; Laroche, A.; Lu, Z.-X.; Liu, H.K.; Li, Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 5, 484, . [CrossRef]
- Kim, D.-G.; Lyu, J.I.; Kim, J.M.; Seo, J.S.; Choi, H.-I.; Jo, Y.D.; Kim, S.H.; Eom, S.H.; Ahn, J.-W.; Bae, C.-H.; et al. Identification of Loci Governing Agronomic Traits and Mutation Hotspots via a GBS-Based Genome-Wide Association Study in a Soybean Mutant Diversity Pool. Int. J. Mol. Sci. 2022, 23, 10441, . [CrossRef]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A High Capacity Genotyping by Sequencing Analysis Pipeline. PLoS ONE 2014, 9, e90346, doi:10.1371/journal.pone.0090346.
- Kim, W.J.; Kang, B.H.; Moon, C.Y.; Kang, S.; Shin, S.; Chowdhury, S.; Jeong, S.-C.; Choi, M.-S.; Park, S.-K.; Moon, J.-K.; et al. Genome-Wide Association Study for Agronomic Traits in Wild Soybean (Glycine soja). Agronomy 2023, 13, 739, . [CrossRef]
- Hwang, S.-G.; Lee, S.C.; Lee, J.; Lee, J.W.; Choi, S.Y.; Kim, J.-B.; Choi, H.-I.; Jang, C.S. Resequencing of a Core Rice Mutant Population Induced by Gamma-Ray Irradiation and Its Application in a Genome-Wide Association Study. J. Plant Biol. 2020, 63, 463–472, . [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379, doi:10.1371/journal.pone.0019379.
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 2011, 17, pp. 10-12, . [CrossRef]
- Cox, M.P.; A Peterson, D.; Biggs, P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 485. [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2013, 25, 1754–1760, . [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079, . [CrossRef]
- Kim, J.-E.; Oh, S.-K.; Lee, J.-H.; Lee, B.-M.; Jo, S.-H. Genome-Wide SNP Calling Using Next Generation Sequencing Data in Tomato. Mol. Cells 2014, 37, 36–42, . [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29, doi:10.1038/75556.
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635, doi:10.1093/bioinformatics/btm308.
- Guo, Y.; Huang, Y.; Hou, L.; Ma, J.; Chen, C.; Ai, H.; Huang, L.; Ren, J. Genome-wide detection of genetic markers associated with growth and fatness in four pig populations using four approaches. Genet. Sel. Evol. 2017, 49, 21. [CrossRef]
- Zhou, X.; Zhang, H.; Wang, P.; Liu, Y.; Zhang, X.; Song, Y.; Wang, Z.; Ali, A.; Wan, L.; Yang, G.; et al. BnaC7.ROT3, the causal gene of cqSL-C7, mediates silique length by affecting cell elongation in Brassica napus. J. Exp. Bot. 2022, 73(1), 154-167.
- Kwon, D.-E.; Kim, K.-S.; Hwang, E.-J.; Park, J.-C.; Lee, J.-E.; Lee, Y.-H. Changes in growth and productivity characteristics by sowing date on spring sowing rapeseed (Brassica napus L.) in paddy field of southern region of South Korea. Korean J. Crop Sci. 2021, 66(1), 80-86.
- Hase, Y.; Satoh, K.; Seito, H.; Oono, Y. Genetic Consequences of Acute/Chronic Gamma and Carbon Ion Irradiation of Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 336, . [CrossRef]
- Chung, Y.S.; Silva, R.R.; Park, M.; Lee, S. Radiation-Induced Breeding in Camelina Sativa. J. Crop. Sci. Biotechnol. 2019, 22, 17–20, . [CrossRef]
- Channaoui, S.; Labhilili, M.; Mouhib, M.; Mazouz, H.; El Fechtali, M.; Nabloussi, A. Development and evaluation of diverse promising rapeseed (Brassica napus L.) mutants using physical and chemical mutagens. OCL. 2019, 26, 35.
- Guan, M.; Shi, X.; Chen, S.; Wan, Y.; Tang, Y.; Zhao, T.; Gao, L.; Sun, F.; Yin, N.; Zhao, H.; et al. Comparative transcriptome analysis identifies candidate genes related to seed coat color in rapeseed. Front. Plant Sci. 2023, 14, 1154208, . [CrossRef]
- Lee, C.W.; Yoo, W.; Park, S.-H.; Le, L.T.H.L.; Jeong, C.-S.; Ryu, B.H.; Shin, S.C.; Kim, H.-W.; Park, H.; Kim, K.K.; et al. Structural and functional characterization of a novel cold-active S-formylglutathione hydrolase (SfSFGH) homolog from Shewanella frigidimarina, a psychrophilic bacterium. Microb. Cell Factories 2019, 18, 140. [CrossRef]
- Soriano, J.M.; Colasuonno, P.; Marcotuli, I.; Gadaleta, A. Meta-QTL analysis and identification of candidate genes for quality, abiotic and biotic stress in durum wheat. Sci. Rep. 2021, 11, 11877. [CrossRef]
- Huang, Y.; Du, L.; Wang, M.; Ren, M.; Yu, S.; Yang, Q. Multifaceted roles of zinc finger proteins in regulating various agronomic traits in rice. Front. Plant Sci. 2022, 13, 974396, . [CrossRef]
- Basim, H.; Basim, E.; Tombuloglu, H.; Unver, T. Comparative transcriptome analysis of resistant and cultivated tomato lines in response to Clavibacter michiganensis subsp. michiganensis. Genomics 2021, 113, 2455–2467, . [CrossRef]
- Li, P.-C.; Ma, J.-J.; Zhou, X.-M.; Li, G.-H.; Zhao, C.-Z.; Xia, H.; Fan, S.-J.; Wang, X.-J. Arabidopsis MDN1 Is Involved in the Establishment of a Normal Seed Proteome and Seed Germination. Front. Plant Sci. 2019, 10, 1118, . [CrossRef]
- Guo, C.; Yao, L.; You, C.; Wang, S.; Cui, J.; Ge, X.; Ma, H. MID1plays an important role in response to drought stress during reproductive development. Plant J. 2016, 88, 280–293, . [CrossRef]
- Huang, S.; Qu, X.; Zhang, R. Plant villins: versatile actin regulatory proteins: Plant villins. J. Integr. Plant Biol. 2015, 57(1), 40–49.
- Zhang, H.; Qu, X.; Bao, C.; Khurana, P.; Wang, Q.; Xie, Y.; Zheng, Y.; Chen, N.; Blanchoin, L.; Staiger, C.J.; et al. ArabidopsisVILLIN5, an Actin Filament Bundling and Severing Protein, Is Necessary for Normal Pollen Tube Growth. Plant Cell 2010, 22, 2749–2767, . [CrossRef]
- Piombo, E.; Vetukuri, R.R.; Broberg, A.; Kalyandurg, P.B.; Kushwaha, S.; Jensen, D.F.; Karlsson, M.; Dubey, M. Role of Dicer-Dependent RNA Interference in Regulating Mycoparasitic Interactions. Microbiol. Spectr. 2021, 9, e0109921, . [CrossRef]
- Zhang, X.; Sun, J.; Cao, X.; Song, X. Epigenetic Mutation of RAV6 Affects Leaf Angle and Seed Size in Rice. Plant Physiol. 2015, 169, 2118–2128, doi:10.1104/pp.15.00836.
- Shimada, T.; Kunieda, T.; Sumi, S.; Koumoto, Y.; Tamura, K.; Hatano, K.; Ueda, H.; Hara-Nishimura, I. The AP-1 Complex is Required for Proper Mucilage Formation in Arabidopsis Seeds. Plant Cell Physiol. 2018, 59, 2331–2338, . [CrossRef]
- Shahnejat-Bushehri, S.; Tarkowska, D.; Sakuraba, Y.; Balazadeh, S. Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat. Plants 2016, 2, 16013, . [CrossRef]
- Licatalosi, D. D. Roles of RNA-binding proteins and post-transcriptional regulation in driving male germ cell development in the mouse. Adv. Exp. Med. Biol. 2016, 907, 123–151.
- Lou, L.; Ding, L.; Wang, T.; Xiang, Y. Emerging Roles of RNA-Binding Proteins in Seed Development and Performance. Int. J. Mol. Sci. 2020, 21, 6822, . [CrossRef]
- Li, X.; Huang, W.; Yang, H.; Jiang, R.; Sun, F.; Wang, H.; Zhao, J.; Xu, C.; Tan, B. EMP18 functions in mitochondrial atp6 and cox2 transcript editing and is essential to seed development in maize. New Phytol. 2018, 221, 896–907, . [CrossRef]
- Scholz, P.; Anstatt, J.; Krawczyk, H.E.; Ischebeck, T. Signalling Pinpointed to the Tip: The Complex Regulatory Network That Allows Pollen Tube Growth. Plants 2020, 9, 1098, . [CrossRef]
- Xiang, X.; Wu, Y.; Planta, J.; Messing, J.; Leustek, T. Overexpression of serine acetyltransferase in maize leaves increases seed-specific methionine-rich zeins. Plant Biotechnol. J. 2018, 16, 1057–1067, . [CrossRef]
- Pal, L.; Sandhu, S.K.; Bhatia, D.; Sethi, S. Genome-wide association study for candidate genes controlling seed yield and its components in rapeseed (Brassica napus subsp. napus). Physiol. Mol. Biol. Plants 2021, 27, 1933–1951, . [CrossRef]
- Fujiki, Y.; Teshima, H.; Kashiwao, S.; Kawano-Kawada, M.; Ohsumi, Y.; Kakinuma, Y.; Sekito, T. Functional identification of AtAVT3, a family of vacuolar amino acid transporters, in Arabidopsis. FEBS Lett. 2017, 591, 5–15, . [CrossRef]
- Crisci, A.; Raleff, F.; Bagdiul, I.; Raabe, M.; Urlaub, H.; Rain, J.-C.; Kramer, A. Mammalian splicing factor SF1 interacts with SURP domains of U2 snRNP-associated proteins. Nucleic Acids Res. 2015, 43(21), 10456–10473.
- Yang, Q.; Zhao, J.; Chen, D.; Wang, Y. E3 ubiquitin ligases: styles, structures and functions. Mol. Biomed. 2021, 2, 23. [CrossRef]
- Ando, H.; Watabe, H.; Valencia, J. C.; Yasumoto, K.-I.; Furumura, M.; Funasaka, Y.; Oka, M.; Ichihashi, M.; Hearing, V. J. Fatty acids regulate pigmentation via proteasomal degradation of tyrosinase: A new aspect of ubiquitin-proteasome function. J. Biol. Chem. 2004, 279(15), 15427–15433.
- Wang, H.; Ma, Z.-H.; Mao, J.; Chen, B.-H. Genome-wide identification and expression analysis of the EXO70 gene family in grape (Vitis vinifera L). PeerJ 2021, 9, e11176, . [CrossRef]
- Bao, Y.; Lopez, J.A.; James, D.E.; Hunziker, W. Snapin Interacts with the Exo70 Subunit of the Exocyst and Modulates GLUT4 Trafficking. J. Biol. Chem. 2008, 283, 324–331, . [CrossRef]
- Wang, S.; Crisman, L.; Miller, J.; Datta, I.; Gulbranson, D. R.; Tian, Y.; Yin, Q.; Yu, H.; Shen, J. Inducible Exoc7/Exo70 knockout reveals a critical role of the exocyst in insulin-regulated GLUT4 exocytosis. J. Biol. Chem. 2019, 294(52), 19988–19996.
- Vorwerk, S.; Schiff, C.; Santamaria, M.; Koh, S.; Nishimura, M.; Vogel, J.; Somerville, C.; Somerville, S. EDR2 negatively regulates salicylic acid-based defenses and cell death during powdery mildew infections of Arabidopsis thaliana. BMC Plant Biol. 2007, 7, 35. [CrossRef]
- Tang, D.; Ade, J.; Frye, C. A.; Innes, R. W. Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein: Regulation of plant defenses by EDR2. Plant J. 2005, 44(2), 245–257.
- Omnus, D.J.; Cadou, A.; Thomas, F.B.; Bader, J.M.; Soh, N.; Chung, G.H.C.; Vaughan, A.N.; Stefan, C.J. A heat-sensitive Osh protein controls PI4P polarity. BMC Biol. 2020, 18, 1–21, . [CrossRef]
- Delage, E.; Puyaubert, J.; Zachowski, A.; Ruelland, E. Signal transduction pathways involving phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: Convergences and divergences among eukaryotic kingdoms. Prog. Lipid Res. 2013, 52, 1–14, . [CrossRef]
- Massimo, A.; Maura, S.; Nicoletta, L.; Giulia, M.; Valentina, M.; Elena, C.; Alessandro, P.; Rosaria, B.; Sandro, S. Current and Novel Aspects on the Non-lysosomal β-Glucosylceramidase GBA2. Neurochem. Res. 2016, 41, 210–220, . [CrossRef]
- Gao, Y.; Li, H.; Deng, D.; Chen, S.; Jiang, W.; Chen, J. Characterization and expression analysis of the maize RING-H2 finger protein gene ZmXERICO responsive to plant hormones and abiotic stresses. Acta Physiol. Plant. 2012, 34, 1529–1535, . [CrossRef]
- Zeng, D.-E.; Hou, P.; Xiao, F.; Liu, Y. Overexpressing a novel RING-H2 finger protein gene, OsRHP1, enhances drought and salt tolerance in rice (Oryza sativa L.). J. Plant Biol. 2014, 57, 357–365, . [CrossRef]
- Qiu, Z.; Zhuang, K.; Liu, Y.; Ge, X.; Chen, C.; Hu, S.; Han, H. Functional characterization of C-TERMINALLY ENCODED PEPTIDE (CEP) family in Brassica rapa L. Plant Signal. Behav. 2022, 17, 2021365, . [CrossRef]
| Lines | Plant height (cm) | Ear length (cm) | TSWa (g) | Seed yield (kg/10a) |
|---|---|---|---|---|
| Tamra | 163.5 ± 3.50 | 55.5 ± 9.50 | 3.8 ± 0.17 | 309.4 ± 17.46 |
| Tr1 | 160.5 ± 3.00 | 54.5 ± 7.50 | 3.7 ± 0.12 | 346.1 ± 24.43 |
| Tr1-1 | 151.0 ± 2.00 | 50.5 ± 9.50 | 4.0 ± 0.12 | 303.0 ± 13.60 |
| Tr1-2 | 153.5 ± 1.50 | 49.0 ± 9.00 | 4.0 ± 0.06 | 254.0 ± 7.17 |
| Tr2-late | 159.5 ± 3.50 | 50.0 ± 8.00 | 5.0 ± 0.15 | 342.0 ± 4.27 |
| Tr2-1 | 158.0 ± 5.00 | 57.5 ± 4.50 | 3.7 ± 0.06 | 374.0 ± 23.51 |
| Tr2-2 | 134.0 ± 9.54 | 56.0 ± 7.00 | 4.0 ± 0.06 | 290.0 ± 8.62 |
| Tr2-3 | 171.0 ± 15.39 | 62.5 ± 7.50 | 4.0 ± 0.15 | 377.0 ± 11.60 |
| Tr3 | 162.0 ± 5.57 | 57.0 ± 8.00 | 3.5 ± 0.06 | 384.0 ± 6.99 |
| Tr4 | 151.0 ± 2.00 | 56.0 ± 9.00 | 3.7 ± 0.15 | 238.0 ± 11.92 |
| Tr4-2 | 160.0 ± 2.00 | 58.5 ± 7.50 | 3.9 ± 0.10 | 393.0 ± 9.13 |
| Tr5 | 147.7 ± 9.07 | 62.0 ± 10.00 | 3.9 ± 0.12 | 151.0 ± 25.88 |
| Tr5-1 | 154.0 ± 5.00 | 59.0 ± 9.00 | 4.0 ± 0.00 | 369.0 ± 5.83 |
| Tr5-3 | 155.3 ± 13.05 | 58.5 ± 10.50 | 4.0 ± 0.12 | 353.0 ± 8.94 |
| Tr6 | 158.0 ± 3.61 | 53.0 ± 10.00 | 4.3 ± 0.12 | 393.0 ± 8.12 |
| Tr7 | 157.0 ± 1.00 | 52.5 ± 6.50 | 4.1 ± 0.06 | 395.0 ± 39.89 |
| Tr14-6 | 161.0 ± 4.00 | 57.0 ± 3.00 | 4.1 ± 0.06 | 368.7 ± 30.21 |
| Tr14-8 | 167.3 ± 5.51 | 59.0 ± 8.00 | 3.9 ± 0.10 | 378.0 ± 5.29 |
| Tr14-9 | 161.0 ± 2.00 | 58.5 ± 10.50 | 4.6 ± 0.15 | 350.0 ± 8.26 |
| Tr14-10 | 163.7 ± 7.37 | 61.5 ± 8.50 | 4.3 ± 0.06 | 329.0 ± 22.38 |
| Tr14-11 | 150.0 ± 5.29 | 52.5 ± 3.50 | 4.3 ± 0.10 | 341.0 ± 4.77 |
| Tr14-13 | 158.0 ± 2.00 | 59.0 ± 7.00 | 3.9 ± 0.06 | 382.0 ± 2.35 |
| Tr14-16 | 162.7 ± 10.07 | 63.5 ± 5.50 | 5.2 ± 0.06 | 384.0 ± 8.49 |
| Tr14-20 | 151.0 ± 4.00 | 58.0 ± 11.00 | 4.3 ± 0.15 | 316.0 ± 13.07 |
| Tr18-4 | 159.0 ± 6.00 | 60.0 ± 8.00 | 4.2 ± 0.06 | 381.0 ± 3.33 |
| Tr18-5 | 170.5 ± 4.50 | 72.5 ± 11.50 | 4.3 ± 0.06 | 397.0 ± 4.96 |
| Tr18-7 | 163.3 ± 13.32 | 59.0 ± 8.00 | 4.1 ± 0.06 | 379.3 ± 3.25 |
| Tr25-5 | 142.3 ± 3.21 | 51.5 ± 3.50 | 5.1 ± 0.10 | 243.0 ± 5.02 |
| Tr25-6 | 159.0 ± 1.00 | 60.0 ± 0.00 | 4.4 ± 0.06 | 226.0 ± 9.20 |
| Tr25-7 | 143.3 ± 1.53 | 48.0 ± 4.00 | 3.9 ± 0.06 | 231.0 ± 6.76 |
| Tr25-9 | 153.5 ± 2.50 | 71.0 ± 11.00 | 4.2 ± 0.06 | 226.0 ± 13.16 |
| Tr25-10 | 160.0 ± 1.50 | 59.0 ± 12.00 | 4.5 ± 0.10 | 274.0 ± 7.16 |
| Tr25-11 | 160.0 ± 1.00 | 63.0 ± 0.00 | 3.8 ± 0.15 | 371.0 ± 9.38 |
| Tr25-12 | 146.5 ± 1.50 | 55.5 ± 10.50 | 3.7 ± 0.10 | 215.0 ± 10.86 |
| Tr25-14 | 175.0 ± 4.00 | 57.5 ± 7.50 | 4.4 ± 0.12 | 378.0 ± 17.22 |
| Tr25-16 | 143.5 ± 2.50 | 55.5 ± 7.50 | 3.6 ± 0.12 | 226.0 ± 12.90 |
| Tr38-4 | 157.5 ± 2.50 | 77.0 ± 15.00 | 4.1 ± 0.06 | 398.0 ± 8.32 |
| Tr38-7 | 160.5 ± 5.50 | 77.5 ± 18.00 | 4.0 ± 0.06 | 377.0 ± 3.58 |
| Tr38-9 | 164.0 ± 4.00 | 60.5 ± 10.50 | 3.6 ± 0.06 | 392.0 ± 7.20 |
| Tr38-16 | 147.7 ± 0.58 | 49.0 ± 7.00 | 4.1 ± 0.06 | 387.0 ± 50.84 |
| Tr38-17 | 148.0 ± 1.00 | 65.0 ± 4.00 | 3.7 ± 0.12 | 391.0 ± 28.98 |
| Tr65-7 | 151.0 ± 4.00 | 63.5 ± 6.50 | 4.1 ± 0.06 | 233.0 ± 59.31 |
| Tr65-9 | 164.0 ± 5.57 | 61.0 ± 10.00 | 3.7 ± 0.06 | 284.0 ± 10.60 |
| Tr65-10 | 170.0 ± 2.00 | 75.5 ± 15.50 | 4.5 ± 0.06 | 397.0 ± 21.53 |
| Tr65-11 | 148.0 ± 1.00 | 62.0 ± 8.00 | 4.3 ± 0.06 | 333.0 ± 7.67 |
| Tr138-L | 170.0 ± 2.00 | 45.0 ± 10.00 | 2.8 ± 0.10 | 234.0 ± 15.34 |
| Tr138-8 | 163.0 ± 2.00 | 61.0 ± 11.00 | 4.0 ± 0.06 | 388.0 ± 16.51 |
| Tr138-9 | 148.0 ± 1.00 | 61.0 ± 9.00 | 4.3 ± 0.15 | 198.0 ± 7.62 |
| Tr138-10 | 160.3 ± 0.58 | 61.5 ± 7.50 | 4.4 ± 0.06 | 269.0 ± 11.08 |
| Tr138-11 | 148.3 ± 4.73 | 56.0 ± 8.00 | 4.1 ± 0.15 | 197.0 ± 10.79 |
| Tr138-12 | 151.0 ± 2.00 | 66.0 ± 8.00 | 3.8 ± 0.06 | 222.0 ± 4.28 |
| Tr138-13 | 158.0 ± 4.00 | 63.5 ± 5.50 | 4.0 ± 0.15 | 382.0 ± 10.15 |
| Tr138-14 | 164.0 ± 2.00 | 67.5 ± 7.50 | 4.0 ± 0.06 | 215.0 ± 9.98 |
| Tr138-15 | 160.0 ± 1.00 | 66.0 ± 11.00 | 4.0 ± 0.10 | 380.0 ± 11.52 |
| Tr138-18 | 148.0 ± 3.00 | 58.0 ± 7.00 | 4.2 ± 0.06 | 384.0 ± 7.26 |
| Tr138-19 | 156.0 ± 3.00 | 64.0 ± 9.00 | 4.4 ± 0.12 | 376.0 ± 10.34 |
| Tr138-20 | 162.0 ± 1.00 | 66.5 ± 9.50 | 4.5 ± 0.06 | 281.0 ± 11.12 |
| Tr138-21 | 155.0 ± 11.36 | 51.0 ± 11.00 | 4.3 ± 0.10 | 204.0 ± 5.86 |
| Tr138-22 | 157.0 ± 1.53 | 67.0 ± 7.00 | 2.9 ± 0.06 | 179.0 ± 10.36 |
| Tr138-23 | 160.0 ± 1.50 | 61.0 ± 8.00 | 3.9 ± 0.06 | 236.0 ± 7.02 |
| Tr6-1-1 | 160.0 ± 2.00 | 69.0 ± 9.00 | 4.1 ± 0.06 | 313.0 ± 5.15 |
| Tr6-1-2 | 173.7 ± 9.07 | 63.0 ± 7.00 | 4.1 ± 0.06 | 376.0 ± 8.93 |
| Tr6-1-3 | 154.0 ± 12.00 | 62.0 ± 6.00 | 4.7 ± 0.10 | 184.0 ± 16.96 |
| Tr6-1-4 | 166.0 ± 2.00 | 61.0 ± 8.00 | 3.8 ± 0.06 | 380.0 ± 11.52 |
| Tr6-11-1 | 160.0 ± 3.00 | 64.5 ± 5.50 | 5.4 ± 0.06 | 221.0 ± 3.37 |
| Tr8-3-1 | 152.0 ± 2.50 | 35.0 ± 7.00 | 4.3 ± 0.06 | 230.0 ± 28.17 |
| Tr8-3-2 | 150.5 ± 7.50 | 60.5 ± 2.50 | 4.3 ± 0.06 | 383.0 ± 16.97 |
| Tr8-3-3 | 153.0 ± 7.00 | 61.5 ± 8.50 | 4.0 ± 0.10 | 278.0 ± 11.08 |
| Tr8-5-1 | 160.0 ± 1.00 | 60.0 ± 6.00 | 4.0 ± 0.10 | 337.0 ± 11.35 |
| Tr8-7-1 | 150.5 ± 0.50 | 61.0 ± 7.00 | 4.9 ± 0.06 | 156.0 ± 25.26 |
| Tr14-1 | 161.0 ± 3.00 | 65.0 ± 6.00 | 5.1 ± 0.06 | 392.0 ± 10.96 |
| Tr14-1-1 | 157.0 ± 2.00 | 66.0 ± 6.00 | 4.8 ± 0.10 | 393.0 ± 14.28 |
| Tr14-1-3 | 155.0 ± 1.00 | 58.5 ± 6.50 | 4.8 ± 0.06 | 307.0 ± 19.34 |
| Tr14-2 | 150.0 ± 3.00 | 64.5 ± 6.50 | 4.4 ± 0.06 | 198.0 ± 16.66 |
| Tr14-3 | 174.0 ± 9.00 | 61.0 ± 12.00 | 4.3 ± 0.06 | 144.0 ± 21.58 |
| Tr14-4 | 159.0 ± 1.00 | 57.0 ± 7.00 | 4.3 ± 0.06 | 392.0 ± 15.36 |
| Tr14-5 | 164.0 ± 2.00 | 58.0 ± 8.00 | 3.7 ± 0.12 | 327.0 ± 8.19 |
| Tr18-1 | 151.0 ± 0.00 | 61.5 ± 7.50 | 3.8 ± 0.06 | 228.0 ± 4.29 |
| Tr18-2 | 148.5 ± 1.50 | 64.5 ± 7.50 | 5.2 ± 0.06 | 245.0 ± 10.15 |
| Tr25-1 | 155.0 ± 3.00 | 57.0 ± 6.00 | 4.8 ± 0.06 | 188.0 ± 12.88 |
| Tr25-2 | 160.0 ± 0.50 | 57.5 ± 8.50 | 5.2 ± 0.06 | 224.0 ± 5.89 |
| Tr25-3 | 170.0 ± 2.00 | 57.5 ± 9.50 | 4.2 ± 0.06 | 395.0 ± 6.33 |
| Tr38-1 | 141.0 ± 8.19 | 58.0 ± 8.00 | 5.0 ± 0.06 | 181.0 ± 14.14 |
| Tr38-2 | 158.0 ± 3.00 | 55.0 ± 8.00 | 3.7 ± 0.06 | 168.0 ± 4.46 |
| Tr65-1 | 166.5 ± 1.50 | 62.0 ± 6.00 | 4.0 ± 0.06 | 397.0 ± 13.16 |
| Tr65-2 | 159.0 ± 1.00 | 54.0 ± 5.00 | 4.1 ± 0.06 | 259.0 ± 23.92 |
| Tr138-1 | 163.0 ± 4.00 | 57.5 ± 7.50 | 4.3 ± 0.10 | 145.0 ± 13.84 |
| Tr138-2 | 160.5 ± 0.50 | 62.0 ± 9.00 | 3.7 ± 0.12 | 346.0 ± 16.60 |
| Tr138-3 | 157.5 ± 1.50 | 59.0 ± 7.00 | 4.6 ± 0.10 | 393.0 ± 19.62 |
| Tr138-4 | 164.5 ± 4.50 | 59.5 ± 8.50 | 4.2 ± 0.06 | 373.7 ± 3.57 |
| Tr6-1 | 160.0 ± 2.00 | 66.0 ± 7.00 | 4.3 ± 0.15 | 377.0 ± 7.40 |
| Tr6-3 | 162.0 ± 2.00 | 63.0 ± 7.00 | 4.2 ± 0.06 | 344.0 ± 13.80 |
| Tr6-4 | 169.5 ± 3.50 | 58.0 ± 8.00 | 5.3 ± 0.10 | 377.0 ± 10.38 |
| Tr6-5 | 159.5 ± 3.50 | 56.5 ± 7.50 | 4.2 ± 0.12 | 393.0 ± 33.24 |
| Tr8-1 | 158.5 ± 8.50 | 60.5 ± 8.50 | 4.2 ± 0.00 | 228.0 ± 42.92 |
| Tr8-3 | 157.0 ± 3.00 | 60.5 ± 10.50 | 3.5 ± 0.06 | 188.0 ± 21.51 |
| Lines | Crude fat content (mg/100 g) |
Fatty acid composition (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| C16:0 | C16:1 | C18:0 | C18:1 | C18:2 | C18:3 | C20:1 | C22:1 | ||
| Tamra | 40.16 ± 1.25* | 5.5 ± 0.54 | 0.0 ± 0.00 | 1.9 ± 0.06 | 65.5 ± 1.80 | 20.5 ± 1.10 | 5.6 ± 0.38 | 1.0 ± 0.15 | 0.0 ± 0.00 |
| Tr1 | 37.55 ± 2.13 | 2.3 ± 0.72 | 0.0 ± 0.00 | 0.5 ± 0.07 | 53.8 ± 0.80 | 9.5 ± 1.85 | 5.8 ± 0.65 | 11.5 ± 0.40 | 16.6 ± 4.49 |
| Tr1-1 | 34.76 ± 0.69 | 2.1 ± 0.41 | 0.0 ± 0.00 | 0.6 ± 0.05 | 55.9 ± 4.26 | 7.9 ± 0.59 | 5.3 ± 0.00 | 10.8 ± 0.25 | 17.4 ± 5.04 |
| Tr1-2 | 36.72 ± 0.13 | 2.4 ± 0.40 | 0.0 ± 0.00 | 0.6 ± 0.03 | 54.9 ± 2.64 | 16.6 ± 0.92 | 5.7 ± 0.11 | 8.4 ± 0.30 | 11.5 ± 3.59 |
| Tr2-late | 38.66 ± 0.49 | 3.0 ± 0.57 | 0.2 ± 0.02 | 1.1 ± 0.04 | 72.9 ± 0.81 | 11.0 ± 0.75 | 3.0 ± 0.05 | 5.6 ± 0.09 | 3.1 ± 0.43 |
| Tr2-1 | 38.29 ± 1.93 | 3.5 ± 0.38 | 0.0 ± 0.00 | 1.4 ± 0.02 | 69.5 ± 1.05 | 11.4 ± 0.10 | 3.6 ± 0.31 | 6.6 ± 0.42 | 4.0 ± 0.62 |
| Tr2-2 | 39.50 ± 2.99 | 9.00 ± 0.88 | 0.0 ± 0.00 | 1.1 ± 0.01 | 55.0 ± 0.76 | 21.8 ± 1.16 | 5.4 ± 0.41 | 4.5 ± 0.71 | 3.2 ± 0.99 |
| Tr2-3 | 35.18 ± 1.39 | 9.0 ± 0.81 | 0.0 ± 0.00 | 1.0 ± 0.11 | 55.1 ± 1.08 | 22.0 ± 1.41 | 5.3 ± 0.38 | 4.2 ± 0.63 | 3.4 ± 0.99 |
| Tr3 | 38.01 ± 0.63 | 2.5 ± 0.37 | 0.0 ± 0.00 | 0.5 ± 0.02 | 78.7 ± 0.15 | 12.0 ± 0.17 | 5.0 ± 0.35 | 1.0 ± 0.02 | 0.4 ± 0.04 |
| Tr4 | 35.6 ± 1.16 | 2.2 ± 0.12 | 0.0 ± 0.00 | 0.5 ± 0.06 | 81.7 ± 3.13 | 10.5 ± 1.78 | 4.3 ± 1.06 | 0.7 ± 0.11 | 0.0 ± 0.00 |
| Tr4-2 | 39.20 ± 2.72 | 3.8 ± 0.65 | 0.0 ± 0.00 | 1.2 ± 0.01 | 69.7 ± 1.32 | 17.2 ± 0.45 | 7.2 ± 0.15 | 1.0 ± 0.07 | 0.0 ± 0.14 |
| Tr5 | 38.61 ± 1.12 | 2.4 ± 0.30 | 0.0 ± 0.00 | 0.4 ± 0.00 | 79.4 ± 0.43 | 13.8 ± 0.40 | 3.3 ± 0.32 | 0.7 ± 0.00 | 0.2 ± 0.02 |
| Tr5-1 | 40.41 ± 2.37 | 0.9 ± 0.13 | 0.0 ± 0.00 | 0.2 ± 0.03 | 74.4 ± 0.22 | 19.7 ± 0.17 | 4.3 ± 0.12 | 0.6 ± 0.02 | 0.0 ± 0.00 |
| Tr5-3 | 39.03 ± 1.42 | 10.2 ± 2.72 | 0.8 ± 0.25 | 2.9 ± 0.36 | 42.3 ± 7.53 | 29.1 ± 2.98 | 13.7 ± 0.08 | 0.8 ± 1.17 | 0.3 ± 0.01 |
| Tr6 | 38.79 ± 2.01 | 2.1 ± 0.37 | 0.0 ± 0.00 | 0.4 ± 0.05 | 40.6 ± 1.75 | 23.5 ± 1.65 | 7.6 ± 0.11 | 8.5 ± 0.22 | 17.3 ± 3.70 |
| Tr7 | 37.80 ± 3.33 | 2.7 ± 0.42 | 0.0 ± 0.00 | 0.4 ± 0.01 | 78.7 ± 0.38 | 13.2 ± 0.22 | 3.8 ± 0.25 | 0.9 ± 0.01 | 0.3 ± 0.04 |
| Tr14-6 | 35.35 ± 1.35 | 4.4f ± 0.43 | 0.2 ± 0.04 | 2.7 ± 0.08 | 67.5 ± 1.49 | 17.3 ± 0.81 | 7.0 ± 0.44 | 1.0 ± 0.15 | 0.0 ± 0.00 |
| Tr14-8 | 40.29 ± 1.16 | 4.5 ± 0.52 | 0.2 ± 0.05 | 2.7 ± 0.07 | 66.3 ± 1.65 | 17.4 ± 0.85 | 7.2 ± 0.12 | 0.6 ± 0.54 | 1.1 ± 0.13 |
| Tr14-9 | 40.53 ± 2.25 | 5.0 ± 0.53 | 0.3 ± 0.06 | 1.6 ± 0.04 | 64.6 ± 1.82 | 19.8 ± 0.94 | 7.5 ± 0.51 | 1.2 ± 0.17 | 0.0 ± 0.00 |
| Tr14-10 | 38.95 ± 2.76 | 4.2 ± 0.46 | 0.2 ± 0.05 | 2.7 ± 0.01 | 67.4 ± 1.51 | 16.8 ± 0.73 | 7.2 ± 0.09 | 0.6 ± 0.50 | 1.0 ± 0.15 |
| Tr14-11 | 39.40 ± 1.17 | 4.5 ± 0.42 | 0.0 ± 0.00 | 1.9 ± 0.12 | 68.4 ± 1.08 | 18.1 ± 0.54 | 6.3 ± 0.36 | 0.9 ± 0.11 | 0.0 ± 0.00 |
| Tr14-13 | 38.25 ± 2.19 | 5.7 ± 0.59 | 0.0 ± 0.00 | 1.9 ± 0.00 | 65.3 ± 1.62 | 20.6 ± 0.79 | 5.6 ± 0.37 | 0.9 ± 0.12 | 0.0 ± 0.00 |
| Tr14-16 | 36.99 ± 1.11 | 4.5 ± 0.47 | 0.0 ± 0.00 | 1.8 ± 0.04 | 67.9 ± 1.17 | 18.0 ± 0.62 | 0.4 ± 0.08 | 6.4 ± 0.35 | 1.0 ± 0.14 |
| Tr14-20 | 40.93 ± 1.78 | 5.8 ± 0.57 | 0.2 ± 0.06 | 2.1 ± 0.07 | 64.6 ± 1.72 | 20.7 ± 0.92 | 5.7 ± 0.39 | 1.0 ± 0.15 | 0.0 ± 0.00 |
| Tr18-4 | 38.43 ± 0.47 | 4.2 ± 0.42 | 0.3 ± 0.06 | 1.7 ± 0.07 | 66.2 ± 1.60 | 18.7 ± 0.86 | 7.7 ± 0.53 | 1.2 ± 0.19 | 0.0 ± 0.00 |
| Tr18-5 | 39.42 ± 1.93 | 4.2f ± 0.43 | 0.3 ± 0.07 | 1.6 ± 0.00 | 66.9 ± 1.57 | 18.3 ± 0.77 | 7.7 ± 0.45 | 1.1 ± 0.14 | 0.0 ± 0.00 |
| Tr18-7 | 39.55 ± 1.45 | 9.2 ± 1.82 | 0.3 ± 0.11 | 2.3 ± 0.21 | 60.2 ± 3.50 | 20.9 ± 1.23 | 6.6 ± 0.12 | 0.5 ± 0.01 | 0.0 ± 0.00 |
| Tr25-5 | 40.14 ± 1.98 | 5.9 ± 1.13 | 0.2 ± 0.07 | 2.3 ± 0.18 | 64.3 ± 3.04 | 16.5 ± 0.95 | 8.4 ± 0.84 | 2.0 ± 0.04 | 0.4 ± 0.08 |
| Tr25-6 | 40.31 ± 0.52 | 6.4 ± 0.96 | 0.2 ± 0.07 | 2.2 ± 0.06 | 53.9 ± 2.35 | 24.9 ± 0.44 | 10.7 ± 0.81 | 1.6 ± 0.02 | 0.0 ± 0.00 |
| Tr25-7 | 40.10 ± 1.28 | 9.0b ± 2.27 | 0.3 ± 0.12 | 3.7 ± 0.46 | 40.6 ± 5.32 | 31.2 ± 0.70 | 10.7 ± 1.68 | 3.2 ± 0.26 | 1.2 ± 0.18 |
| Tr25-9 | 39.96 ± 0.47 | 10.3 ± 2.47 | 0.4 ± 0.14 | 3.8 ± 0.48 | 39.9 ± 7.03 | 26.9 ± 1.60 | 14.7 ± 2.06 | 3.2 ± 0.40 | 0.8 ± 0.11 |
| Tr25-10 | 40.28 ± 1.24 | 3.6 ± 0.36 | 0.1 ± 0.03 | 1.5 ± 0.01 | 70.7 ± 1.32 | 15.3 ± 0.63 | 7.6 ± 0.48 | 1.2 ± 0.16 | 0.0 ± 0.00 |
| Tr25-11 | 37.92 ± 0.65 | 8.1 ± 1.25 | 0.2 ± 0.08 | 2.5 ± 0.11 | 52.8 ± 2.70 | 23.1 ± 0.41 | 11.1 ± 0.84 | 2.2 ± 0.01 | 0.0 ± 0.00 |
| Tr25-12 | 37.81 ± 1.24 | 8.5 ± 1.64 | 0.5 ± 0.07 | 3.3 ± 0.17 | 48.0 ± 0.48 | 28.2 ± 1.34 | 11.5 ± 0.06 | 0.0 ± 0.00 | 0.0 ± 0.00 |
| Tr25-14 | 39.34 ± 1.04 | 5.6 ± 0.97 | 0.2 ± 0.06 | 1.8 ± 0.13 | 67.1 ± 2.60 | 16.2 ± 0.90 | 7.4 ± 0.67 | 1.5 ± 0.06 | 0.3 ± 0.07 |
| Tr25-16 | 37.86 ± 1.19 | 9.0 ± 1.21 | 0.3 ± 0.09 | 4.4 ± 0.11 | 48.6 ± 0.11 | 24.7 ± 1.36 | 10.8 ± 0.19 | 2.2 ± 0.09 | 0.0 ± 0.00 |
| Tr38-4 | 38.36 ± 1.93 | 8.5 ± 1.20 | 0.0 ± 0.00 | 3.7 ± 0.04 | 49.6 ± 0.87 | 26.2 ± 0.72 | 10.13 ± 0.45 | 1.9 ± 0.01 | 0.0 ± 0.00 |
| Tr38-7 | 35.47 ± 1.31 | 7.3 ± 0.57 | 0.0 ± 0.00 | 2.0 ± 0.08 | 66.4 ± 1.54 | 19.0 ± 0.68 | 5.3 ± 0.38 | 0.0 ± 0.00 | 0.0 ± 0.00 |
| Tr38-9 | 36.86 ± 1.92 | 6.3 ± 0.66 | 0.0 ± 0.00 | 2.0 ± 0.08 | 66.1 ± 1.72 | 18.7 ± 0.94 | 5.8 ± 0.41 | 1.2 ± 0.22 | 0.0 ± 0.00 |
| Tr38-16 | 39.56 ± 0.07 | 7.6 ± 0.25 | 0.7 ± 0.10 | 1.5 ± 0.08 | 60.4 ± 1.62 | 25.1 ± 0.97 | 4.7 ± 0.38 | 0.0 ± 0.00 | 0.0 ± 0.00 |
| Tr38-17 | 40.19 ± 2.04 | 7.4 ± 0.58 | 0.0 ± 0.00 | 1.4 ± 0.04 | 64.3 ± 2.06 | 22.0 ± 1.03 | 5.0 ± 0.40 | 0.0 ± 0.00 | 0.0 ± 0.00 |
| Tr65-7 | 38.90 ± 0.54 | 8.3 ± 0.66 | 0.0 ± 0.00 | 1.4 ± 0.08 | 66.2 ± 1.97 | 20.1 ± 0.86 | 4.0 ± 0.37 | 0.0 ± 0.00 | 0.0 ± 0.00 |
| Tr65-9 | 38.15 ± 0.50 | 7.4 ± 0.59 | 0.0 ± 0.00 | 2.1 ± 0.17 | 66.1 ± 1.64 | 19.1 ± 0.86 | 5.3 ± 0.35 | 0.0 ± 0.00 | 0.0 ± 0.00 |
| Tr65-10 | 40.21 ± 1.67 | 7.3 ± 0.55 | 0.6 ± 0.05 | 1.6 ± 0.05 | 63.8 ± 1.58 | 21.7 ± 0.82 | 5.0 ± 0.32 | 0.0 ± 0.00 | 0.0 ± 0.00 |
| Tr65-11 | 38.07 ± 0.70 | 7.2 ± 0.59 | 0.2 ± 0.04 | 1.3 ± 0.02 | 60.4 ± 1.56 | 25.3 ± 0.81 | 5.0 ± 0.23 | 0.7 ± 0.12 | 0.0 ± 0.00 |
| Tr138-L | 39.13 ± 0.14 | 5.9 ± 0.47 | 0.3 ± 0.14 | 1.9 ± 0.06 | 68.3 ± 1.41 | 17.6 ± 0.67 | 5.3 ± 0.32 | 0.8 ± 0.12 | 0.0 ± 0.00 |
| Tr138-8 | 39.24 ± 1.32 | 4.2 ± 0.42 | 0.0 ± 0.00 | 1.7 ± 0.02 | 68.1 ± 1.39 | 17.4 ± 0.54 | 7.8 ± 0.55 | 0.9 ± 0.10 | 0.0 ± 0.00 |
| Tr138-9 | 39.08 ± 1.29 | 4.1 ± 0.41 | 0.0 ± 0.00 | 1.6 ± 0.02 | 67.9 ± 1.40 | 17.6 ± 0.68 | 7.8 ± 0.49 | 1.0 ± 0.17 | 0.0 ± 0.00 |
| Tr138-10 | 39.95 ± 2.43 | 5.2 ± 0.64 | 0.2 ± 0.05 | 1.9 ± 0.02 | 62.3 ± 2.25 | 22.1 ± 1.15 | 7.3 ± 0.53 | 1.0 ± 0.09 | 0.0 ± 0.00 |
| Tr138-11 | 39.42 ± 1.50 | 8.0 ± 0.68 | 0.2 ± 0.08 | 1.8 ± 0.02 | 60.6 ± 2.01 | 23.7 ± 0.99 | 5.1 ± 0.32 | 0.6 ± 0.03 | 0.0 ± 0.00 |
| Tr138-12 | 39.82 ± 0.66 | 8.1 ± 0.64 | 0.0 ± 0.00 | 1.9 ± 0.11 | 61.0 ± 1.63 | 23.4 ± 0.87 | 5.0 ± 0.29 | 0.7 ± 0.06 | 0.0 ± 0.00 |
| Tr138-13 | 40.26 ± 1.16 | 7.0 ± 0.65 | 0.0 ± 0.00 | 1.8 ± 0.06 | 51.3 ± 0.27 | 21.0 ± 1.71 | 5.5 ± 0.56 | 8.3 ± 1.26 | 5.3 ± 1.88 |
| Tr138-14 | 39.51 ± 0.55 | 5.1 ± 0.79 | 0.2 ± 0.05 | 1.7 ± 0.09 | 62.7 ± 2.52 | 20.6 ± 0.96 | 8.4 ± 0.71 | 1.3 ± 0.08 | 0.0 ± 0.00 |
| Tr138-15 | 39.15 ± 0.97 | 8.9 ± 2.25 | 0.3 ± 0.12 | 3.7 ± 0.46 | 38.6 ± 6.95 | 33.5 ± 2.37 | 10.6 ± 1.67 | 3.2 ± 0.27 | 1.2 ± 0.18 |
| Tr138-18 | 40.19 ± 0.28 | 5.9 ± 1.01 | 0.3 ± 0.07 | 2.2 ± 0.12 | 60.1 ± 2.92 | 22.6 ± 1.00 | 7.8 ± 0.73 | 1.1 ± 0.01 | 0.0 ± 0.00 |
| Tr138-19 | 38.07 ± 1.38 | 6.2 ± 2.02 | 0.0 ± 0.00 | 1.4 ± 0.81 | 58.8 ± 6.43 | 21.7 ± 1.67 | 10.5 ± 1.67 | 1.4 ± 0.27 | 0.0 ± 0.00 |
| Tr138-20 | 39.35 ± 1.90 | 4.3 ± 0.40 | 0.1 ± 0.04 | 2.0 ± 0.06 | 69.2 ± 1.30 | 16.4 ± 0.65 | 7.0 ± 0.46 | 1.1 ± 0.18 | 0.0 ± 0.00 |
| Tr138-21 | 38.87 ± 1.31 | 6.9 ± 0.84 | 0.0 ± 0.00 | 1.3 ± 0.03 | 44.5 ± 0.44 | 24.7 ± 1.74 | 6.8 ± 0.61 | 7.3 ± 1.10 | 8.6 ± 2.55 |
| Tr138-22 | 36.65 ± 1.63 | 5.6 ± 1.11 | 0.2 ± 0.07 | 2.5 ± 0.19 | 60.9 ± 3.08 | 21.6 ± 1.23 | 6.5 ± 0.68 | 2.0 ± 0.06 | 0.8 ± 0.15 |
| Tr138-23 | 36.50 ± 0.98 | 6.8 ± 0.73 | 0.0 ± 0.00 | 1.5 ± 0.02 | 50.4 ± 0.41 | 21.5 ± 1.67 | 5.5 ± 0.51 | 8.8 ± 1.37 | 5.7 ± 1.93 |
| Tr6-1-1 | 38.55 ± 0.55 | 3.5 ± 1.34 | 0.1 ± 0.04 | 0.6 ± 0.10 | 68.8 ± 6.87 | 21.2 ± 4.35 | 5.0 ± 0.85 | 0.6 ± 0.19 | 0.2 ± 0.00 |
| Tr6-1-2 | 38.56 ± 0.67 | 4.0 ± 0.38 | 0.0 ± 0.00 | 0.9 ± 0.06 | 44.5 ± 2.18 | 20.3 ± 0.12 | 8.1 ± 0.58 | 10.0 ± 0.39 | 12.3 ± 1.77 |
| Tr6-1-3 | 39.24 ± 1.13 | 4.0 ± 1.52 | 0.1 ± 0.04 | 0.5 ± 0.10 | 54.8 ± 6.51 | 17.7 ± 3.08 | 8.9 ± 1.30 | 8.8 ± 1.12 | 5.2 ± 0.65 |
| Tr6-1-4 | 36.20 ± 1.05 | 3.1 ± 0.34 | 0.0 ± 0.00 | 0.9 ± 0.05 | 53.5 ± 2.53 | 15.8 ± 0.00 | 7.2 ± 0.46 | 8.6 ± 0.44 | 11.0 ± 2.03 |
| Tr6-11-1 | 38.63 ± 0.15 | 3.2 ± 0.93 | 0.1 ± 0.04 | 0.8 ± 0.09 | 64.2 ± 4.71 | 24.9 ± 2.95 | 5.2 ± 0.51 | 1.1 ± 0.18 | 0.5 ± 0.01 |
| Tr8-3-1 | 38.60 ± 1.36 | 3.5 ± 1.26 | 0.1 ± 0.04 | 0.8 ± 0.13 | 70.4 ± 6.50 | 19.0 ± 4.00 | 5.0 ± 0.85 | 1.0 ± 0.25 | 0.3 ± 0.02 |
| Tr8-3-2 | 38.29 ± 1.42 | 3.0 ± 1.06 | 0.1 ± 0.04 | 0.8 ± 0.13 | 69.4 ± 6.14 | 19.2 ± 3.70 | 6.4 ± 0.99 | 0.8 ± 0.24 | 0.3 ± 0.01 |
| Tr8-3-3 | 39.70 ± 1.92 | 3.1 ± 0.87 | 0.0 ± 0.07 | 0.6 ± 0.06 | 47.3 ± ±2.09 | 20.9 ± 0.56 | 7.7 ± 0.41 | 10.0 ± 0.02 | 10.4 ± 1.84 |
| Tr8-5-1 | 40.98 ± 1.98 | 3.5 ± 1.30 | 0.1 ± 0.04 | 0.7 ± 0.09 | 59.2 ± 7.55 | 18.5 ± 3.51 | 8.6 ± 1.41 | 6.5 ± 1.30 | 2.8 ± 0.10 |
| Tr8-7-1 | 38.19 ± 0.91 | 3.0 ± 0.51 | 0.0 ± 0.00 | 0.7 ± 0.06 | 50.4 ± 0.06 | 20.1 ± 1.12 | 7.3 ± 0.07 | 8.7 ± 0.09 | 9.9 ± 1.77 |
| Tr14-1 | 34.99 ± 1.51 | 5.0 ± 0.94 | 0.0 ± 0.00 | 1.7 ± 0.11 | 32.4 ± 2.24 | 15.3 ± 2.16 | 8.9 ± 1.52 | 13.1 ± 1.23 | 23.6 ± 5.74 |
| Tr14-1-1 | 36.93 ± 1.07 | 8.9 ± 1.80 | 0.7 ± 0.17 | 2.5 ± 0.20 | 50.1 ± 3.16 | 25.3 ± 0.77 | 12.0 ± 0.02 | 0.7 ± 0.20 | 0.0 ± 0.00 |
| Tr14-1-3 | 38.23 ± 0.16 | 10.5 ± 1.06 | 0.5 ± 0.07 | 2.7 ± 0.10 | 53.6 ± 0.50 | 24.4 ± 0.85 | 7.7 ± 0.63 | 0.6 ± 0.06 | 0.0 ± 0.00 |
| Tr14-2 | 29.68 ± 3.07 | 3.9 ± 0.60 | 0.0 ± 0.00 | 1.8 ± 0.05 | 42.9 ± 1.86 | 17.5 ± 1.82 | 7.9 ± 1.01 | 12.0 ± 1.30 | 14.1 ± 4.03 |
| Tr14-3 | 28.83 ± 1.85 | 4.5 ± 0.73 | 0.0 ± 0.00 | 1.5 ± 0.07 | 41.2 ± 2.39 | 16.8 ± 2.04 | 6.3 ± 0.84 | 13.0 ± 1.39 | 16.7 ± 4.67 |
| Tr14-4 | 33.35 ± 0.75 | 6.4 ± 1.61 | 0.0 ± 0.00 | 0.9 ± 0.29 | 60.7 ± 5.17 | 18.7 ± 2.09 | 10.2 ± 1.39 | 1.4 ± 0.20 | 1.7 ± 0.01 |
| Tr14-5 | 24.07 ± 0.49 | 1.4 ± 0.37 | 0.0 ± 0.00 | 0.3 ± 0.04 | 32.8 ± 2.45 | 17.6 ± 2.34 | 7.2 ± 3.97 | 11.7 ± 2.68 | 29.0 ± 10.02 |
| Tr18-1 | 39.29 ± 0.95 | 4.8 ± 1.05 | 0.2 ± 0.02 | 1.0 ± 0.07 | 66.5 ± 3.01 | 21.9 ± 1.57 | 5.1 ± 0.32 | 0.7 ± 0.01 | 0.0 ± 0.00 |
| Tr18-2 | 21.31 ± 0.75 | 6.0 ± 0.30 | 0.4 ± 0.07 | 1.8 ± 0.20 | 66.3 ± 1.19 | 18.2 ± 0.79 | 6.2 ± 0.33 | 1.0 ± 0.11 | 0.0 ± 0.00 |
| Tr25-1 | 24.06 ± 0.95 | 7.1 ± 0.78 | 0.0 ± 0.00 | 1.2 ± 0.06 | 44.7 ± 0.32 | 24.9 ± 1.83 | 6.9 ± 0.63 | 7.1 ± 1.08 | 8.2 ± 2.54 |
| Tr25-2 | 26.76 ± 0.83 | 6.6 ± 1.65 | 0.0 ± 0.00 | 2.3 ± 0.97 | 62.7 ± 6.00 | 18.6 ± 2.16 | 8.5 ± 1.30 | 1.4 ± 0.07 | 0.0 ± 0.00 |
| Tr25-3 | 28.60 ± 1.00 | 5.7 ± 0.58 | 0.3 ± 0.07 | 2.0 ± 0.05 | 68.2 ± 0.05 | 16.6 ± 1.75 | 6.5 ± 0.78 | 0.8 ± 0.45 | 0.0 ± 0.08 |
| Tr38-1 | 29.01 ± 1.36 | 9.2 ± 4.49 | 0.0 ± 0.00 | 0.8 ± 0.32 | 48.6 ± 8.30 | 29.5 ± 1.04 | 10.9 ± 2.11 | 1.0 ± 0.36 | 0.0 ± 0.00 |
| Tr38-2 | 28.88 ± 0.88 | 4.6 ± 0.61 | 0.0 ± 0.00 | 1.7 ± 0.02 | 51.2 ± 1.34 | 17.1 ± 1.41 | 6.2 ± 0.61 | 10.7 ± 1.38 | 8.6 ± 2.61 |
| Tr65-1 | 36.33 ± 0.37 | 4.6 ± 0.52 | 0.2 ± 0.05 | 2.7 ± 0.08 | 66.7 ± 1.73 | 17.5 ± 0.83 | 7.3 ± 0.54 | 1.1 ± 0.13 | 0.0 ± 0.00 |
| Tr65-2 | 37.88 ± 2.38 | 5.1 ± 0.55 | 0.2 ± 0.04 | 1.7 ± 0.04 | 69.6 ± 1.75 | 15.7 ± 0.86 | 6.5 ± 0.50 | 1.4 ± 0.16 | 0.0 ± 0.00 |
| Tr138-1 | 35.14 ± 1.82 | 7.1 ± 2.17 | 0.2 ± 0.12 | 2.0 ± 0.46 | 56.4 ± 7.43 | 19.9 ± 2.42 | 11.6 ± 1.79 | 2.8 ± 0.49 | 0.0 ± 0.00 |
| Tr138-2 | 38.16 ± 0.16 | 5.6 ± 0.50 | 0.2 ± 0.05 | 1.8 ± 0.03 | 60.7 ± 1.44 | 23.9 ± 0.73 | 7.0 ± 0.35 | 1.0 ± 0.16 | 0.0 ± 0.00 |
| Tr138-3 | 33.04 ± 0.08 | 7.4 ± 2.54 | 0.4 ± 0.18 | 1.0 ± 0.40 | 62.7 ± 5.21 | 19.4 ± 0.80 | 8.3 ± 1.07 | 0.9 ± 0.23 | 0.0 ± 0.00 |
| Tr138-4 | 34.38 ± 0.31 | 6.2 ± 1.81 | 0.2 ± 0.06 | 1.3 ± 0.11 | 58.6 ± 5.23 | 23.2 ± 2.08 | 8.7 ± 1.07 | 1.1 ± 0.12 | 0.7 ± 0.02 |
| Tr6-1 | 37.56 ± 1.08 | 7.1 ± 1.28 | 0.2 ± 0.06 | 2.2 ± 0.13 | 65.8 ± 2.73 | 17.3 ± 1.20 | 7.1 ± 0.07 | 0.4 ± 0.00 | 0.0 ± 0.00 |
| Tr6-3 | 37.04 ± 0.06 | 5.6 ± 0.55 | 0.2 ± 0.04 | 2.0 ± 0.08 | 59.9 ± 1.85 | 24.1 ± 1.04 | 7.2 ± 0.43 | 1.0 ± 0.13 | 0.0 ± 0.00 |
| Tr6-4 | 39.25 ± 0.17 | 4.5 ± 0.47 | 0.3 ± 0.06 | 1.6 ± 0.06 | 71.1 ± 1.41 | 16.1 ± 0.69 | 5.6 ± 0.38 | 0.9 ± 0.11 | 0.0 ± 0.00 |
| Tr6-5 | 40.17 ± 0.16 | 8.3 ± 2.99 | 0.0 ± 0.00 | 1.9 ± 0.30 | 54.0 ± 6.78 | 25.7 ± 1.96 | 8.6 ± 1.28 | 0.6 ± 0.11 | 0.9 ± 0.16 |
| Tr8-1 | 31.93 ± 1.27 | 3.6 ± 0.36 | 0.0 ± 0.00 | 1.5 ± 0.01 | 70.8 ± 1.30 | 15.3 ± 0.63 | 7.6 ± 0.48 | 1.2 ± 0.16 | 0.0 ± 0.00 |
| Tr8-3 | 40.00 ± 0.01 | 5.9 ± 1.48 | 0.0 ± 0.00 | 1.7 ± 0.58 | 65.3 ± 4.82 | 17.6 ± 1.34 | 8.5 ± 1.12 | 1.1 ± 0.30 | 0.0 ± 0.00 |
| Total | Average/Plant | |
|---|---|---|
| Raw data | ||
| Reads | 715,259,878 | 7,450,624 |
| Bases (bp) | 108,004,241,578 | 1,125,044,183 |
| After trimming | ||
| Reads | 655,243,166 | 6,825,450 |
| Bases (bp) | 74,621,891,073 | 777,311,365 |
| Mapped reads on reference genome* | ||
| Reads | 651,288,040 | 6,784,250 |
| Bases (bp) | 3,163,629,539 | 32,954,474 |
| Reference genome coverage (%) | 3.5673% | |
| Traits | Chr_position | -log10(p) | Gene ID | Description | TAIR ID | Allele |
|---|---|---|---|---|---|---|
| TSW | chrA05_1312542 | 12.37 | BnaA05p02350D | S-formylglutathione hydrolase-like | AT2G41530.1 | A/G |
| TSW | chrC01_45614214 | 8.75 | BnaC01p49650D | zinc finger BED domain-containing protein RICESLEEPER 1-like | AT3G14800.2 | G/T |
| TSW | chrC02_16544482 | 8.44 | BnaC02p22490D | midasin-like | AT1G67120.2 | G/A |
| TSW | chrC08_30501410 | 6.41 | BnaC08p32830D | villin-3-like isoform X2 | AT3G57410.9 | A/G |
| TSW | chrC03_21138185 | 6.40 | BnaC03p36140D | death-associated inhibitor of apoptosis 1 | AT4G03965.1 | C/T |
| TSW | chrC05_4473754 | 5.90 | BnaC05p08830D | ubiquitin-like-specific protease 1C | AT1G10570.3 | G/T |
| TSW | chrA05_2029410 | 5.58 | BnaA05p03850D | chitinase 1 | AT2G43570.1 | T/A |
| TSW | chrA05_1197170 | 5.46 | BnaA05p02100D | amino acid transporter AVT1A | AT2G41190.2 | A/C |
| TSW | chrC05_439211 | 5.30 | BnaC05p00900D | endoribonuclease Dicer homolog 1-like | AT1G01040.2 | T/C |
| TSW | chrC05_4629489 | 5.25 | BnaC05p09110D | AP-1 complex subunit mu-2 | AT1G10730.1 | C/T |
| TSW | chrC08_28339798 | 5.12 | BnaC08p29480D | RNA-binding protein 5 | AT3G54230.5 | G/A |
| TSW | chrA03_20827042 | 5.07 | BnaA03p44720D | ATPase 6, plasma membrane-type | AT2G07560.1 | T/A |
| TSW | chrA03_5465396 | 5.02 | BnaA03p13200D | serine acetyltransferase 5 | AT5G56760.1 | G/A |
| TSW | chrC09_4097062 | 4.91 | BnaC09p07010D | U2 snRNP-associated SURP motif-containing protein isoform X2 | AT5G25060.1 | T/G |
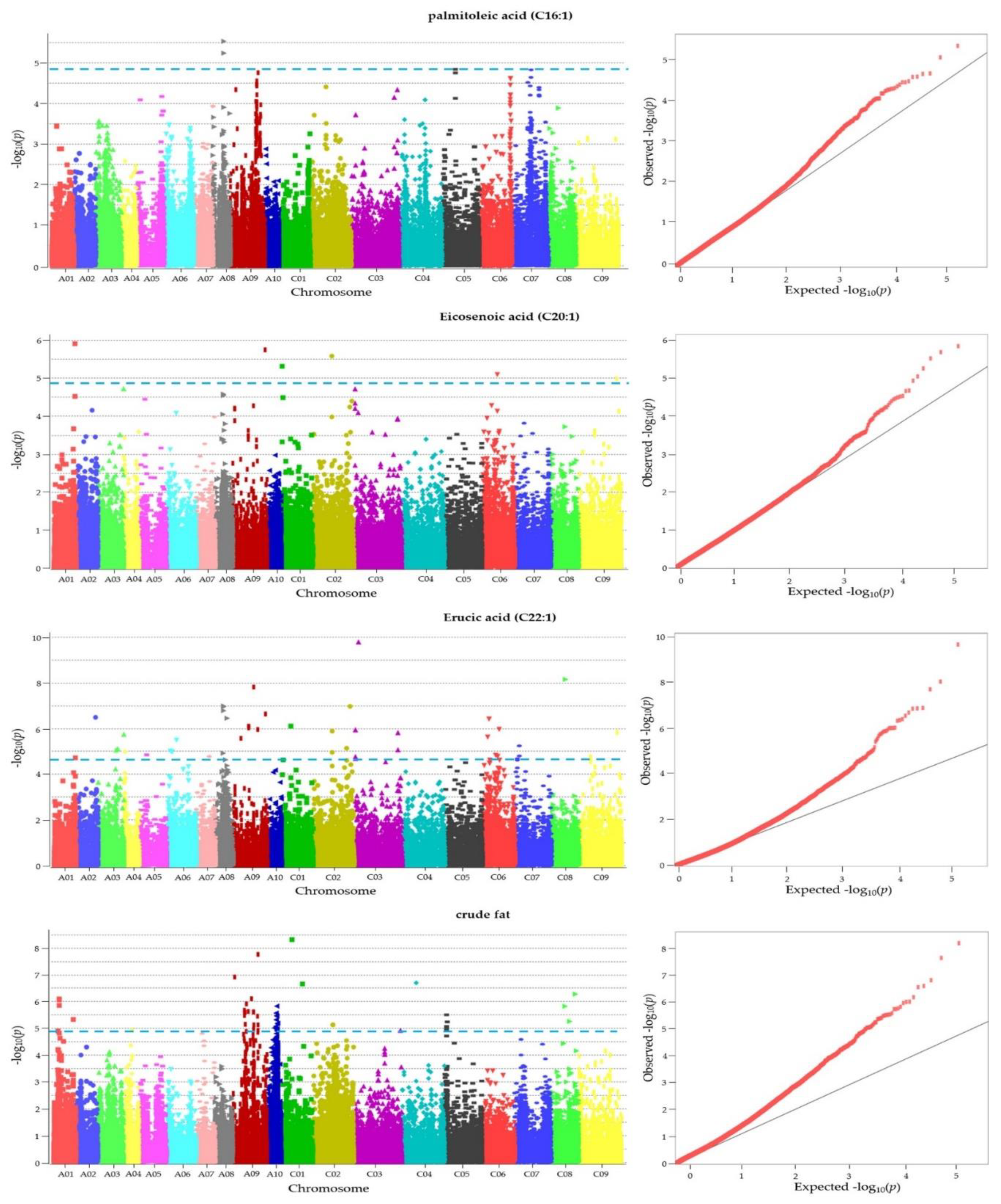
| Traits | Chr_position | -log10(p) | Gene ID | Description | TAIR ID | Allele |
|---|---|---|---|---|---|---|
| C16:1 | chrA08_16580415 | 5.244 | BnaA08p17150D | Encodes a putative E3 ubiquitin ligase | AT4G34100.1 | C/T |
| C20:1 | chrC01_1257401 | 5.317 | BnaC01p02330D | probable Xaa-Pro aminopeptidase P isoform X2 | AT4G36760.2 | C/A |
| C22:1 | chrC03_6878724 | 9.783 | BnaC03p14350D | EXO70 gene family | AT5G58430.1 | C/T |
| C22:1 | chrC08_21249315 | 8.150 | BnaC08p19880D | paramyosin-like | AT1G19980.1 | C/T |
| C22:1 | chrA08_12298481 | 6.984 | BnaA08p10580D | mediator of RNA polymerase II transcription subunit 34 | AT1G31360.3 | C/A |
| C22:1 | chrC02_56355748 | 6.971 | BnaC02p56750D | cyclin-C1-1 | AT5G48640.1 | A/G |
| C22:1 | chrC02_51025897 | 5.142 | BnaC02p51020D | ribonuclease T2 family | AT2G02990.1 | G/A |
| C22:1 | chrA08_11469852 | 4.922 | BnaA08p09630D | protein ENHANCED DISEASE RESISTANCE 2-like isoform X2 | AT5G35180.4 | G/A |
| Crude fat | chrA09_3387964 | 6.924 | BnaA09p05630D | Part of a nanodomain complex that tethers PI4Kα1 to the plasma membrane | AT5G26850.1 | C/A |
| Crude fat | chrA09_18251187 | 5.590 | BnaA09p27860D | 1-aminocyclopropane-1-carboxylate synthase 6 | AT4G11280.1 | A/G |
| Crude fat | chrC05_2572075 | 5.511 | BnaC05p05120D | mitoferrin | AT1G07030.1 | G/C |
| Crude fat | chrA09_38265711 | 5.460 | BnaA09p39540D | Transmembrane amino acid transporter family protein | AT1G25530.1 | C/T |
| Crude fat | chrA10_15664715 | 5.203 | BnaA10p21730D | chromo domain-containing protein LHP1-like | AT5G17690.3 | T/C |
| Crude fat | chrA09_32798569 | 5.137 | BnaA09p32690D | non-lysosomal glucosylceramidase | AT1G33700.1 | T/G |
| Crude fat | chrA09_39092505 | 5.136 | BnaA09p40920D | RING-H2 finger protein ATL47-like | AT1G23980.1 | C/T |
| Crude fat | chrA09_32277805 | 4.977 | BnaA09p32130D | embryogenesis-associated protein EMB8 | AT1G34340.1 | T/C |
| Crude fat | chrC05_2503939 | 4.970 | BnaC05p04980D | CEP DOWNSTREAM 1 | AT1G06830.1 | A/G |
| rude fat | chrC03_71451788 | 4.926 | BnaC03p90410D | protein CHUP1, chloroplastic-like | AT1G52080.1 | A/T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





